
中国水稻科学 ›› 2025, Vol. 39 ›› Issue (6): 813-824.DOI: 10.16819/j.1001-7216.2025.241003
陆帅1, 陶涛1, 刘冉1, 周文玉1, 曹蕾1, 杨青青1, 张明秋1, 任鑫哲1, 杨芝笛1, 徐福祥1, 环海东1, 龚远航1, 张皓程1, 金素奎3, 蔡秀玲1,2, 高继平1,2, 冷语佳1,2,*( )
)
收稿日期:2024-10-11
修回日期:2024-12-02
出版日期:2025-11-10
发布日期:2025-11-19
通讯作者:
* email:yujialeng@yzu.edu.cn
基金资助:
LU Shuai1, TAO Tao1, LIU Ran1, ZHOU Wenyu1, CAO Lei1, YANG Qingqing1, ZHANG Mingqiu1, REN Xinzhe1, YANG Zhidi1, XU Fuxiang1, HUAN Haidong1, GONG Yuanhang1, ZHANG Haocheng1, JIN Sukui3, CAI Xiuling1,2, GAO Jiping1,2, LENG Yujia1,2,*( )
)
Received:2024-10-11
Revised:2024-12-02
Online:2025-11-10
Published:2025-11-19
Contact:
* email:yujialeng@yzu.edu.cn
摘要:
【目的】通过对一个水稻长护颖、小粒突变体的表型鉴定与基因克隆,探明控制该性状的遗传基础和分子机制。【方法】利用甲基磺酸乙酯(EMS)诱变粳稻品种武育粳27(WYJ27),从中获得一个稳定遗传的长护颖、小粒突变体,将该突变体命名为lsg8 ( long sterile lemma and small grain on chromosome 8)。观察并统计野生型和突变体的农艺性状变化;利用透射电镜观察颖壳外表皮细胞的变化;将突变体lsg8与籼稻品种IR36进行杂交,构建F2群体并进行遗传分析,利用图位克隆对LSG8基因进行定位,通过对候选基因测序和表达分析进一步确定候选基因;利用RT-qPCR分析细胞扩展相关基因和花器官特征基因的表达量。【结果】与野生型相比,突变体lsg8护颖长度明显变长,粒宽和粒厚显著下降,从而导致千粒重下降。此外,突变体lsg8的株高、穗长、倒1节间长、倒2节间长、倒4节间长、一次枝梗数、二次枝梗数、每穗粒数和结实率均较野生型WYJ27显著降低。颖壳外表皮扫描电镜观察结果表明,突变体lsg8的细胞长度较野生型显著变短,细胞宽度较野生型显著变窄。遗传分析表明,突变体lsg8受一对隐性核基因控制。通过图位克隆将LSG8基因定位在8号染色体标记M5和标记M6之间,物理距离约为276 kb,该区间包含42个开放阅读框。通过对候选基因测序分析,发现ORF18(LOC_Os08g06480)在野生型和突变体之间出现了一个碱基的差异,认为该基因可能是控制长护颖、小粒表型的候选基因。RT-qPCR分析表明,LSG8在不同时期各个组织中均有表达,其中在成熟期的穗中表达量最高,而在成熟期的叶鞘中表达量最低。此外,突变体lsg8在细胞扩展相关基因、护颖发育调控基因和颖壳特征基因的表达量也发生了显著的变化。【结论】水稻长护颖、小粒突变体lsg8是已报道基因ASP1的新等位基因,该基因突变导致护颖变长、种子变小,对于维持水稻护颖的形态建成及籽粒形态起到重要的作用。
陆帅, 陶涛, 刘冉, 周文玉, 曹蕾, 杨青青, 张明秋, 任鑫哲, 杨芝笛, 徐福祥, 环海东, 龚远航, 张皓程, 金素奎, 蔡秀玲, 高继平, 冷语佳. 水稻长护颖小粒突变体lsg8的表型鉴定与基因克隆[J]. 中国水稻科学, 2025, 39(6): 813-824.
LU Shuai, TAO Tao, LIU Ran, ZHOU Wenyu, CAO Lei, YANG Qingqing, ZHANG Mingqiu, REN Xinzhe, YANG Zhidi, XU Fuxiang, HUAN Haidong, GONG Yuanhang, ZHANG Haocheng, JIN Sukui, CAI Xiuling, GAO Jiping, LENG Yujia. Identification and Gene Cloning of a Long Sterile Lemma and Small Grain Mutant lsg8 in Rice (Oryza sativa L.)[J]. Chinese Journal OF Rice Science, 2025, 39(6): 813-824.
| 引物名称 Primer name | 物理位置 Physical location(bp) | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|---|
| M1 | 201350 | GTTTGAACAGTAGGACTTGT | AGAACATCTCACACTTCTCT |
| M2 | 4588509 | ATCTCCCTCCCTCTCCTCAC | TCCACACCTTCACAGTTGAC |
| M3 | 1135865 | TGGGATAGGAGTAGCACTTTTGG | CCATACATTCCAAACCATCCTAG |
| M4 | 2372755 | GCAAACTGTGAGCAACAATGG | AAACAAGACCATGCTCGTCGG |
| M5 | 3502364 | AATTTTACACCGGATCTAAACAC | ATGGAAATGCAAATTAAGAACAC |
| M6 | 3778634 | GTTCTGTTTCTTGCCCGACCTTT | GAATATGACCCACATGCCGACTC |
| M7 | 4092309 | ATCTAAAGGAGATCGGATGGTAT | AAGCATCCAGAGGTCGCAGCAAA |
表1 基因定位所用引物及序列
Table 1. Primers and sequences used in gene mapping
| 引物名称 Primer name | 物理位置 Physical location(bp) | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|---|
| M1 | 201350 | GTTTGAACAGTAGGACTTGT | AGAACATCTCACACTTCTCT |
| M2 | 4588509 | ATCTCCCTCCCTCTCCTCAC | TCCACACCTTCACAGTTGAC |
| M3 | 1135865 | TGGGATAGGAGTAGCACTTTTGG | CCATACATTCCAAACCATCCTAG |
| M4 | 2372755 | GCAAACTGTGAGCAACAATGG | AAACAAGACCATGCTCGTCGG |
| M5 | 3502364 | AATTTTACACCGGATCTAAACAC | ATGGAAATGCAAATTAAGAACAC |
| M6 | 3778634 | GTTCTGTTTCTTGCCCGACCTTT | GAATATGACCCACATGCCGACTC |
| M7 | 4092309 | ATCTAAAGGAGATCGGATGGTAT | AAGCATCCAGAGGTCGCAGCAAA |
| 引物名称 Primer name | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| CL-1 | CCTCATCCTGCAGTTCCTCG | AACCTCAGCATATTTGGCGT |
| CL-2 | ATGGGCATGCTCCAACCAAT | GAGCCAACTTCCCATATCCCA |
| CL-3 | TCTTTGTCCATGTTCTGTCCACT | CGCATTGCAAATTAAAACGGCA |
| CL-4 | TGAAGGTCACGAAGCTCCAG | ATTTGTTGAGGTGGCCCTCG |
| CL-5 | TAATTTTGCAGGCGAGCCCA | CGTTGCCATTTCCACAGCTT |
| CL-6 | AGGCTGTTCAGTTCTGAGCAATA | TATAGTCCACTTGCCACCGC |
| CL-7 | TGCTCAGTGACCTTCAACTAACT | CCCCACTTTGGGTCTGAGTC |
表2 LSG8基因测序引物
Table 2. Primer sequences used for LSG8 gene sequencing
| 引物名称 Primer name | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| CL-1 | CCTCATCCTGCAGTTCCTCG | AACCTCAGCATATTTGGCGT |
| CL-2 | ATGGGCATGCTCCAACCAAT | GAGCCAACTTCCCATATCCCA |
| CL-3 | TCTTTGTCCATGTTCTGTCCACT | CGCATTGCAAATTAAAACGGCA |
| CL-4 | TGAAGGTCACGAAGCTCCAG | ATTTGTTGAGGTGGCCCTCG |
| CL-5 | TAATTTTGCAGGCGAGCCCA | CGTTGCCATTTCCACAGCTT |
| CL-6 | AGGCTGTTCAGTTCTGAGCAATA | TATAGTCCACTTGCCACCGC |
| CL-7 | TGCTCAGTGACCTTCAACTAACT | CCCCACTTTGGGTCTGAGTC |
| 基因 Gene | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| OsMADS34 | AGGAATATGTGAACTTGAA | TTCTGACTACTTGACTCT |
| SAD1 | CCAGGGGAGAAATCCAAGA | CTGTCGACCAAGCTTCAGG |
| LRG1 | ATGGAGTTTGGGATGGTGGAG | CAGCTTGTGCGCGTTCTGGT |
| EG1 | AACGTACACGACCCGATCAC | GACGTGGGTGTAGCAGGAGT |
| ASP1 | TCTTGTTCAACCTCCAAACACAGC | CAATGGCAGGAGCACTTGTTGG |
| OsVIL2 | GGAGTATGCTTTCCGGATCA | GTGGGAAACAACATGTGCAG |
| OsIG1 | TTCATCAACGTCGGGCACT | CTCCCCTTCGTAGCTCCTC |
| SNB | ACCACGAAGTAGGGAACGACTGGG | CAGCCAATAAGTCCTCAGTGGCCTG |
| OsIDS1 | CTGGCCTCCAGTTAACTTGT | GGCGCCGGCAGAGAATCCT |
| MFS1 | CGGCTCGTGATCTCGACACGTAC | CACAGCCGGACCAGTGCTCTC |
| LSG8 | TCTTGTTCAACCTCCAAACACAGC | CAATGGCAGGAGCACTTGTTGG |
| OsMADS1 | CCAAGCCACTCTTCTTGTTCG | TGATGGTGAGCATGAGGGTG |
| OsMADS6 | CCAACAATGCACTTTCTGAAAC | GGAGGCTTGCTGCATGGC |
| OsMADS14 | CCATTAACGAGCTTCAACGG | TGGTATGGATCTGAAGCCTCC |
| OsMADS15 | AGTACGCCACTGACTCCAGG | TGCTGGCCCCTCACATTC |
| DL | CCCATCTGCTTACAACCGCTT | GTTGGAGGTGGAAACCGTCG |
| OsEXPA1 | TGCAGAGCCTCCAATAGTAGTCCA | GGTACATCAAGCCTCTGTAGTGCAA |
| OsEXPB5 | TGTTTGTTAACGTCGCCGCGATAG | TCACTAGAAGCAGCTCTGCAAACG |
| OsEXPA10 | TCTTGTGCTCGTGACAAACGTTGC | CATTGGCATCCAGTCGGTTGAGTT |
| OsEXPB11 | GCAGTGCAGAGTTGCGGTAAATTG | ATCGACGACGACACAGTCACATCA |
| OsEXPA25 | TGGATCACGCTGAACCGGAACT | TGTAGATGTAGAGCGTCTGGCCG |
| UBQ | ACCACTTCGACCGCCACTACT | ACGCCTAAGCCTGCTGGTT |
表3 RT-qPCR中所用到的引物
Table 3. Primers for RT-qPCR
| 基因 Gene | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| OsMADS34 | AGGAATATGTGAACTTGAA | TTCTGACTACTTGACTCT |
| SAD1 | CCAGGGGAGAAATCCAAGA | CTGTCGACCAAGCTTCAGG |
| LRG1 | ATGGAGTTTGGGATGGTGGAG | CAGCTTGTGCGCGTTCTGGT |
| EG1 | AACGTACACGACCCGATCAC | GACGTGGGTGTAGCAGGAGT |
| ASP1 | TCTTGTTCAACCTCCAAACACAGC | CAATGGCAGGAGCACTTGTTGG |
| OsVIL2 | GGAGTATGCTTTCCGGATCA | GTGGGAAACAACATGTGCAG |
| OsIG1 | TTCATCAACGTCGGGCACT | CTCCCCTTCGTAGCTCCTC |
| SNB | ACCACGAAGTAGGGAACGACTGGG | CAGCCAATAAGTCCTCAGTGGCCTG |
| OsIDS1 | CTGGCCTCCAGTTAACTTGT | GGCGCCGGCAGAGAATCCT |
| MFS1 | CGGCTCGTGATCTCGACACGTAC | CACAGCCGGACCAGTGCTCTC |
| LSG8 | TCTTGTTCAACCTCCAAACACAGC | CAATGGCAGGAGCACTTGTTGG |
| OsMADS1 | CCAAGCCACTCTTCTTGTTCG | TGATGGTGAGCATGAGGGTG |
| OsMADS6 | CCAACAATGCACTTTCTGAAAC | GGAGGCTTGCTGCATGGC |
| OsMADS14 | CCATTAACGAGCTTCAACGG | TGGTATGGATCTGAAGCCTCC |
| OsMADS15 | AGTACGCCACTGACTCCAGG | TGCTGGCCCCTCACATTC |
| DL | CCCATCTGCTTACAACCGCTT | GTTGGAGGTGGAAACCGTCG |
| OsEXPA1 | TGCAGAGCCTCCAATAGTAGTCCA | GGTACATCAAGCCTCTGTAGTGCAA |
| OsEXPB5 | TGTTTGTTAACGTCGCCGCGATAG | TCACTAGAAGCAGCTCTGCAAACG |
| OsEXPA10 | TCTTGTGCTCGTGACAAACGTTGC | CATTGGCATCCAGTCGGTTGAGTT |
| OsEXPB11 | GCAGTGCAGAGTTGCGGTAAATTG | ATCGACGACGACACAGTCACATCA |
| OsEXPA25 | TGGATCACGCTGAACCGGAACT | TGTAGATGTAGAGCGTCTGGCCG |
| UBQ | ACCACTTCGACCGCCACTACT | ACGCCTAAGCCTGCTGGTT |
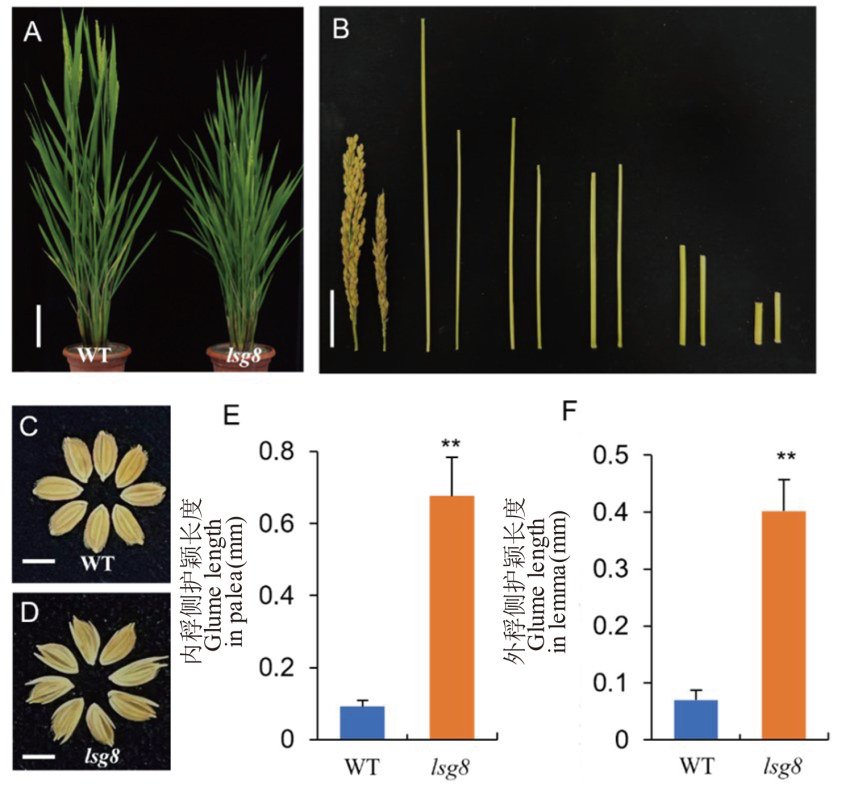
图1 野生型WYJ27与突变体lsg8的表型对比 A: 野生型WYJ27和突变体lsg8抽穗期株型比较,标尺=10 cm。B: 野生型WYJ27和突变体lsg8穗及各节间长对比。左边为野生型WYJ27, 右边为突变体lsg8。标尺=5 cm。C: 野生型WYJ27成熟种子,标尺=5 cm。D: 突变体lsg8成熟种子,标尺=5 cm。E: 野生型WYJ27和突变体lsg8内稃侧护颖长度比较。F: 野生型WYJ27和突变体lsg8外稃侧护颖长度比较。用t检验进行显著性分析, **表示P<0.01水平上差异显著。
Fig. 1. Comparison of phenotypic traits between the wild type WYJ27 and lsg8 A, Phenotypic comparison of the wild type WYJ27 and lsg8 at heading stage, scale bar = 10 cm. B, Comparison of spikelet and internode length between the wild type WYJ27 and lsg8. The left is the wild type WYJ27, and the right is lsg8, scale bar=5 cm. C, Mature seeds of wild type WYJ27, scale bar=5 cm. D, Mature seeds of lsg8, scale bar=5 cm. E, Comparison of glume length on palea between wild type WYJ27 and lsg8. F, Comparison of glume length on lemma between wild type WYJ27 and lsg8. The t-test was used for significance analysis, and ** means significant difference at P < 0.01.
| 性状 Trait | WYJ27 | lsg8 |
|---|---|---|
| 株高 Plant height (cm) | 89.1 ± 2.8 | 76.8 ± 1.3** |
| 穗长 Panicle length (cm) | 17.0 ± 1.2 | 15.2 ± 0.9** |
| 倒1节长The 1st internode from the top (cm) | 26.9 ± 1.6 | 19.5 ± 1.4** |
| 倒2节长The 2nd internode from the top (cm) | 19.7 ± 0.8 | 17.6 ± 0.2** |
| 倒3节长The 3rd internode from the top (cm) | 12.9 ± 0.9 | 13.0 ± 1.2 |
| 倒4节长The 4th internode from the top (cm) | 9.4 ± 1.0 | 8.4 ± 1.0* |
| 倒5节长The 5th internode from the top (cm) | 4.3 ± 1.6 | 3.5 ± 1.1 |
| 分蘖数 No. of tillers per plant | 7.4 ± 1.4 | 16.4 ± 2.9** |
| 一次枝梗数 No. of primary rachis branches per panicle | 13.9 ± 1.0 | 10.1 ± 2.3** |
| 二次枝梗数No. of secondary rachis branches per panicle | 35.8 ± 5.4 | 24.9 ± 6.8** |
| 每穗粒数No. of spikelets per panicle | 208.1 ± 14.1 | 115.9 ± 8.9** |
| 结实率 Seed-setting rate (%) | 93.8 ± 2.3 | 61.5 ± 9.9** |
表4 野生型WYJ27与突变体lsg8的农艺性状对比
Table 4. Comparison of agronomic traits between WYJ27 and lsg8
| 性状 Trait | WYJ27 | lsg8 |
|---|---|---|
| 株高 Plant height (cm) | 89.1 ± 2.8 | 76.8 ± 1.3** |
| 穗长 Panicle length (cm) | 17.0 ± 1.2 | 15.2 ± 0.9** |
| 倒1节长The 1st internode from the top (cm) | 26.9 ± 1.6 | 19.5 ± 1.4** |
| 倒2节长The 2nd internode from the top (cm) | 19.7 ± 0.8 | 17.6 ± 0.2** |
| 倒3节长The 3rd internode from the top (cm) | 12.9 ± 0.9 | 13.0 ± 1.2 |
| 倒4节长The 4th internode from the top (cm) | 9.4 ± 1.0 | 8.4 ± 1.0* |
| 倒5节长The 5th internode from the top (cm) | 4.3 ± 1.6 | 3.5 ± 1.1 |
| 分蘖数 No. of tillers per plant | 7.4 ± 1.4 | 16.4 ± 2.9** |
| 一次枝梗数 No. of primary rachis branches per panicle | 13.9 ± 1.0 | 10.1 ± 2.3** |
| 二次枝梗数No. of secondary rachis branches per panicle | 35.8 ± 5.4 | 24.9 ± 6.8** |
| 每穗粒数No. of spikelets per panicle | 208.1 ± 14.1 | 115.9 ± 8.9** |
| 结实率 Seed-setting rate (%) | 93.8 ± 2.3 | 61.5 ± 9.9** |
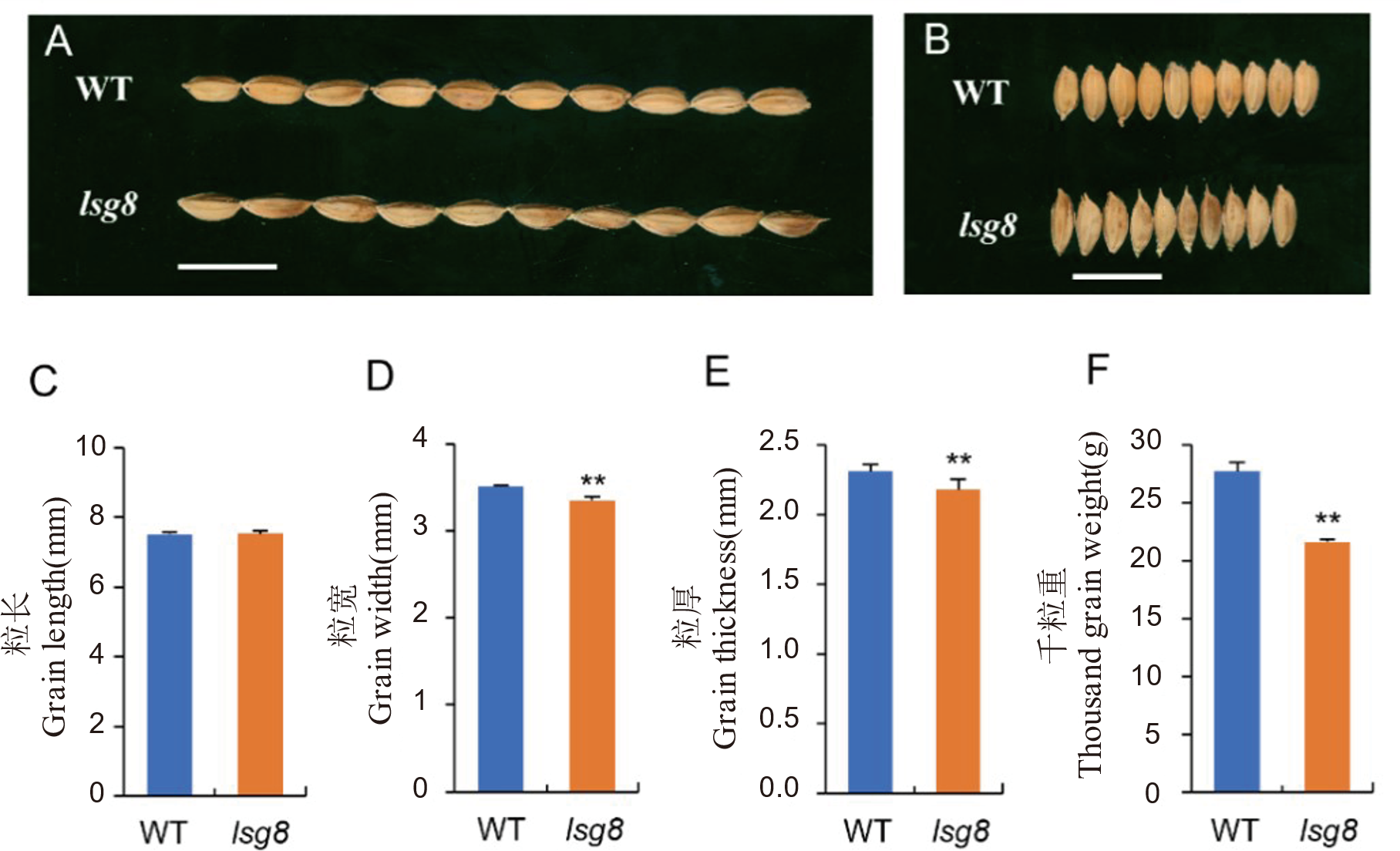
图2 野生型WYJ27与突变体lsg8粒型对比 A: 野生型WYJ27与突变体lsg8成熟种子粒长的比较。标尺= 1 cm。B: 野生型WYJ27与突变体lsg8成熟种子粒宽的比较。标尺= 0.5 cm。C-F: 野生型WYJ27与突变体lsg8的粒长(C)、粒宽(D)、粒厚(E)和千粒重(F)统计对比。用t检验进行显著性分析, **表示P<0.01上差异显著。
Fig. 2. Comparison of grain size between the wild type WYJ27 and lsg8 A, Comparison of grain length between the wild type WYJ27 and the lsg8 mutant, scale bar=1 cm. B, Comparison of grain width between the wild type WYJ27 and lsg8, scale bar=0.5 cm. C-F, Comparison of grain length (C), grain size (D), grain thickness (E) and 1000-grain weight (F) between the wild type WYJ27 and lsg8. The t-test was used for significance analysis. ** means significant difference at P < 0.01.
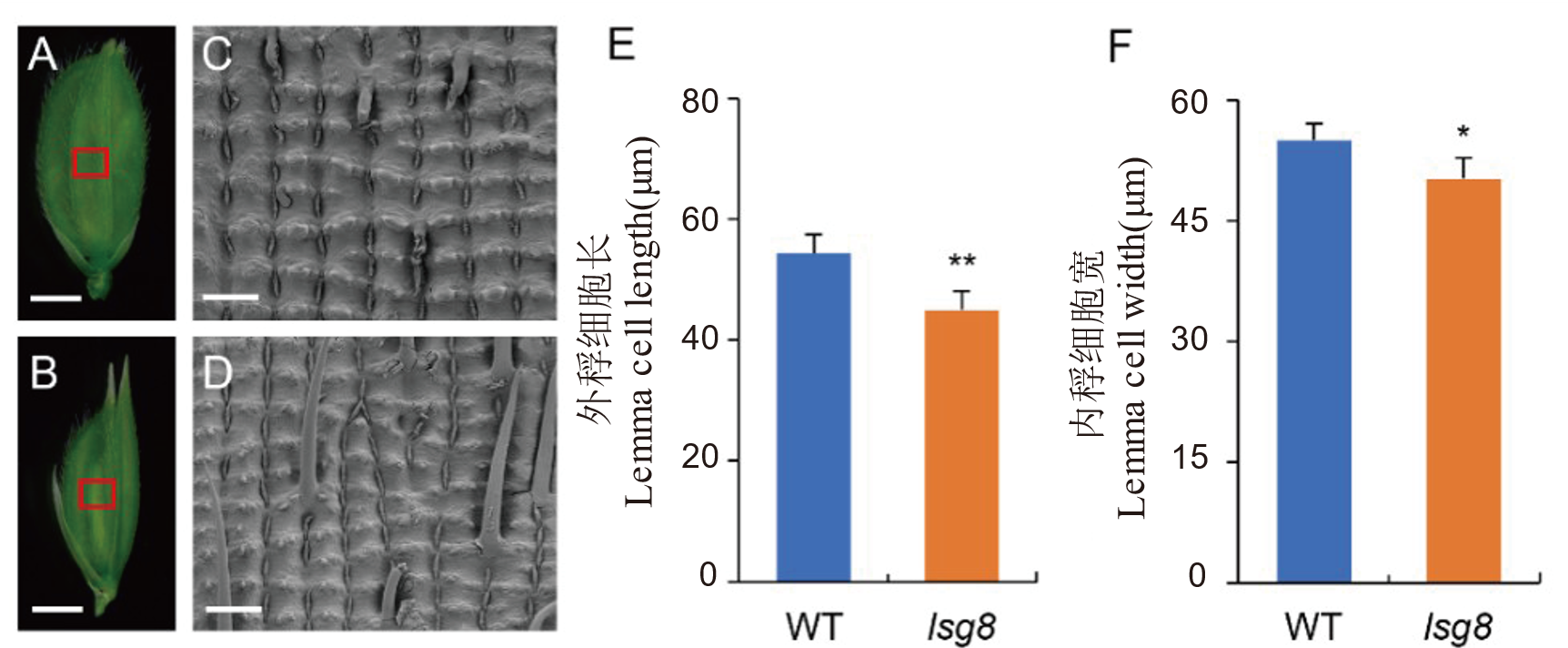
图3 野生型WYJ27与突变体lsg8颖壳外表皮细胞扫描电镜观察 A: 野生型WYJ27颖壳,标尺=2 mm。B: 突变体lsg8颖壳,标尺=2 mm。C: 野生型WYJ27颖壳外表皮细胞,标尺=100 μm。D: 突变体lsg8颖壳外表皮细胞,标尺=100 μm。E: 野生型WYJ27和突变体lsg8外稃细胞长度比较。F: 野生型WYJ27和突变体lsg8外稃细胞宽度比较。用t检验进行显著性分析。*, **分别表示在P<0.05和P<0.01水平上差异显著。
Fig. 3. Scanning electron microscopy of epidermal cells of the glume in wild type WYJ27 and the lsg8 mutant A, Glume of wild type WYJ27, scale bar=2 mm. B, Glume of mutant lsg8, scale bar= 2 mm. C, Outer epidermal cells of wild type WYJ27 glume, scale bar=100 μm. D, Outer epidermal cells of mutant lsg8 glume, scale bar=100 μm. E, Comparison of lemma cell length between wild type WYJ27 and lsg8. F, Comparison of lemma cell width between wild type WYJ27 and lsg8. Significance analysis was performed using t-test. * P<0.05; ** P<0.01.
| 杂交组合 Cross | F1 | F2 | χ2 (3:1) | |||
|---|---|---|---|---|---|---|
| 正常表型 Wild type | 突变表型 Mutant | 正常表型 Wild type | 突变表型 Mutant | |||
| lsg8 /IR36 | 15 | 0 | 1253 | 439 | 1.1046 | |
| IR36/ lsg8 | 23 | 0 | 856 | 295 | 0.2436 | |
表5 突变体lsg8的遗传分析
Table 5. Genetic analysis of lsg8 mutant
| 杂交组合 Cross | F1 | F2 | χ2 (3:1) | |||
|---|---|---|---|---|---|---|
| 正常表型 Wild type | 突变表型 Mutant | 正常表型 Wild type | 突变表型 Mutant | |||
| lsg8 /IR36 | 15 | 0 | 1253 | 439 | 1.1046 | |
| IR36/ lsg8 | 23 | 0 | 856 | 295 | 0.2436 | |
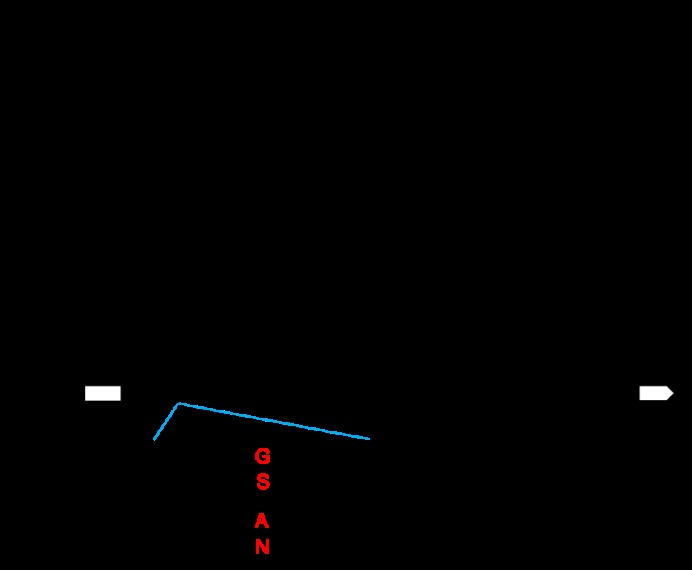
图4 LSG8基因的定位 A: LSG8在8号染色体上的定位;B: 候选基因LSG8。红色字母表示野生型和突变体在候选基因LSG8上碱基和编码氨基酸差异。
Fig. 4. Gene mapping of LSG8 A, Mapping of LSG8 on chromosome 8. B, Candidate gene LSG8. The red letters indicate the differences in bases and encoded amino acids between the wild type and the mutant on the candidate gene LSG8.
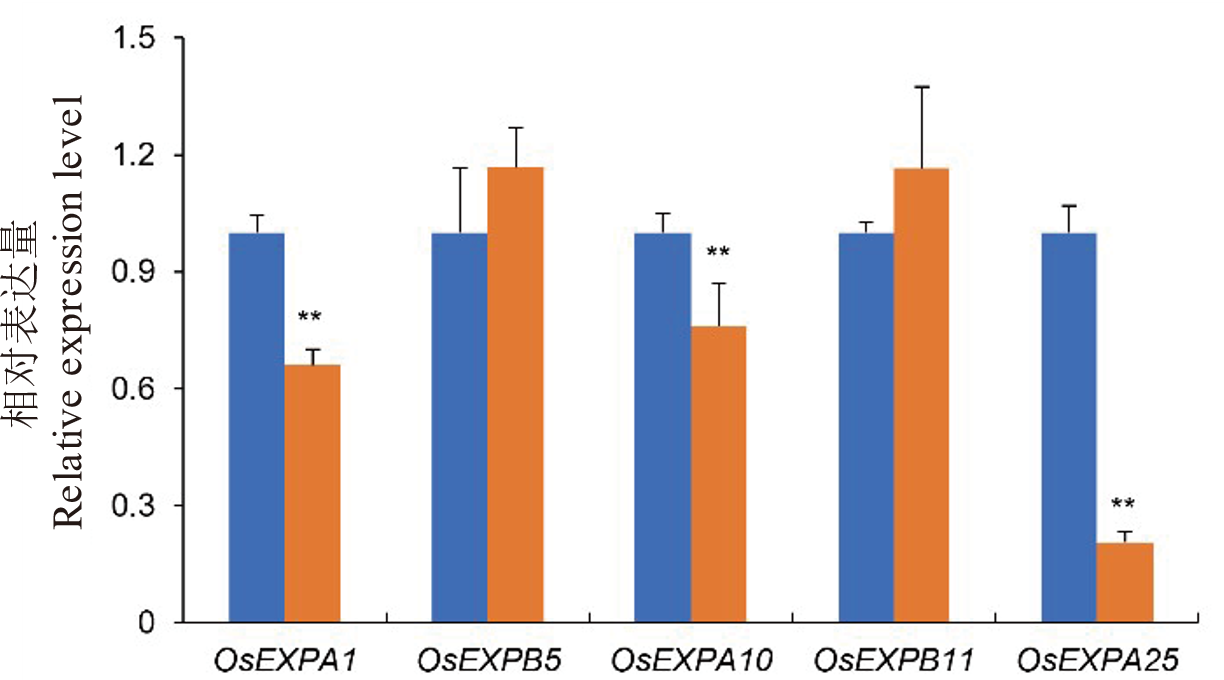
图6 水稻细胞扩展相关基因在野生型WYJ27和lsg8突变体中的相对表达水平 每个样品检测3个生物学重复。**表示P<0.01。
Fig. 6. Relative expression levels of cell expansins related genes in wild type WYJ27 and lsg8 mutant Each sample was analyzed in three independent biological replicates. ** Significant difference at P<0.01 by t-test, respectively.
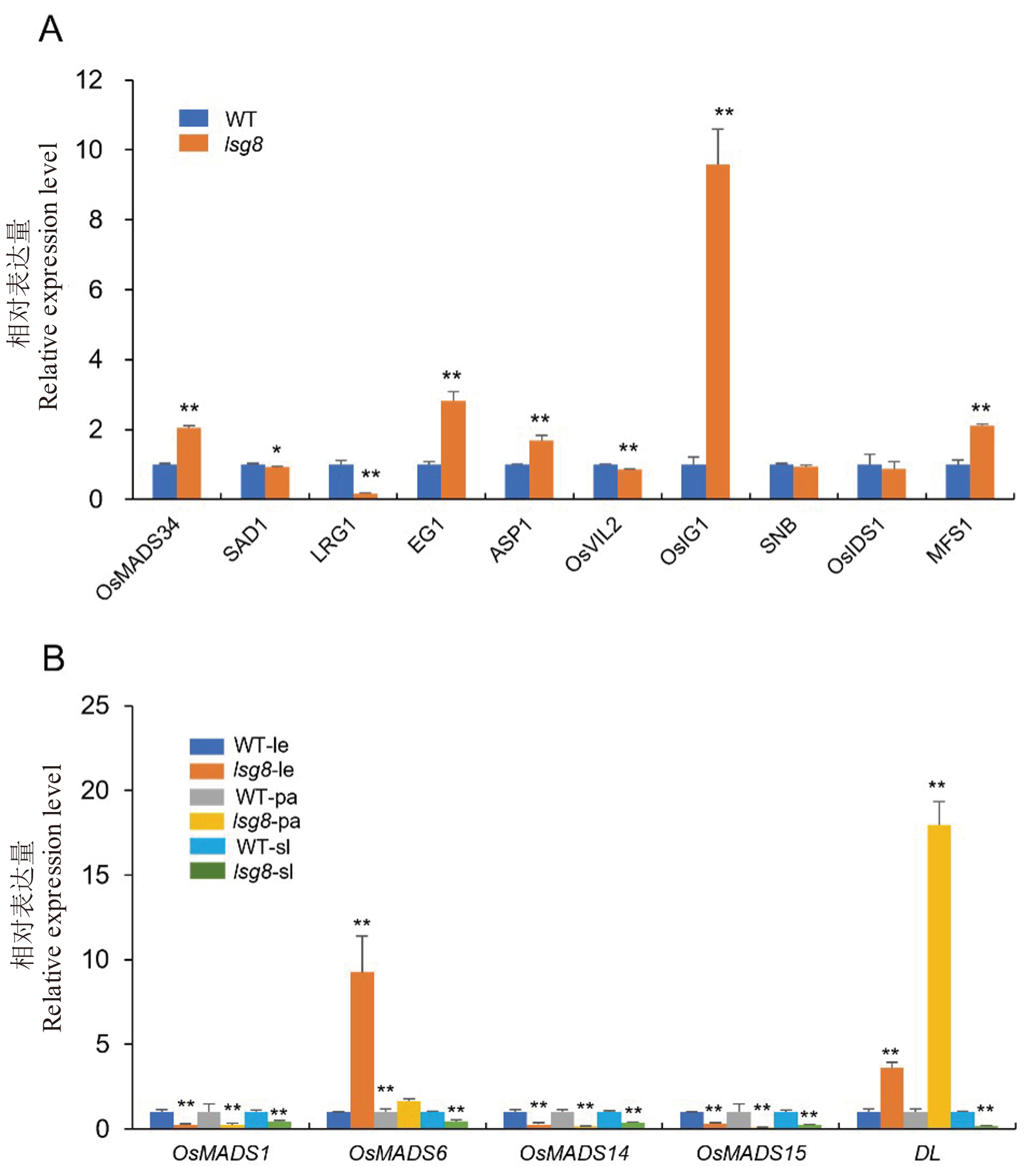
图7 水稻护颖发育调控基因和颖壳特征基因在野生型WYJ27和lsg8突变体中的相对表达水平 A: 水稻护颖发育调控基因在野生型WYJ27和lsg8突变体中的相对表达水平; B: 水稻颖壳特征基因在野生型WYJ27和lsg8突变体中的相对表达水平; le: 外稃 pa: 内稃; sl: 护颖;每个样品检测3个生物学重复, *和**表示P<0.05 和P<0.01。
Fig. 7. Relative expression levels of genes regulating sterile lemma development and hull characteristics in wild type WYJ27 and the lsg8 mutant A, qPCR analysis of sterile lemma development regulatory genes (OsMADS34, SAD1, LRG1, EG1, ASP1, OsVIL2, OsIG1, SNB, OsIDS1 and MFS’) in the wild type and lsg8 mutant. B, qPCR analysis of floral organ identity genes (OsMADS1, OsMADS6, OsMADS14, OsMADS15, and DL) in the wild type and lsg8 mutant. le, Lemma; pa, Palea; sl, Sterile lemma. Each sample was analyzed in three independent biological replicates. * and ** : Significant difference at P<0.05 and P<0.01 by t-test, respectively.
| [1] | 朱玲, 陈晓琼, 杜康兮, 韩保林, 冉秀华, 张红宇, 徐培洲, 吴先军. 水稻长护颖突变体基因的克隆与表达分析[J]. 中国农业科学, 2015, 48(11): 2085-2095. |
| Zhu L, Chen X Q, Du K X, Han B L, Ran X H, Zhang H Y, Xu P Z, Wu X J. Gene cloning and expression analysis of long empty glumes mutants in rice[J]. Scientia Agricultura Sinica, 2015, 48(11): 2085-2095. (in Chinese with English abstract) | |
| [2] | 符德保, 李燕, 肖景华, 张启发, 吴昌银. 中国水稻基因组学研究历史及现状[J]. 生命科学, 2016, 28(10): 1113-1121. |
| Fu D B, Li Y, Xiao J H, Zhang Q F, Wu C Y. The history and current status of rice genomics research in China[J]. Chinese Bulletin of Life Sciences, 2016, 28(10): 1113-1121. (in Chinese with English abstract) | |
| [3] | 张必东, 林泓, 朱思颖, 李忠成, 庄慧, 李云峰. 水稻颖壳异常突变体ah1的鉴定与候选基因分析[J]. 中国农业科学, 2024, 57(3): 429-441. |
| Zhang B D, Lin H, Zhu S Y, Li Z C, Zhuang H, Li Y F. Identification and candidate gene analysis of the ABNORMAL HULL 1 (ah1) mutant in rice (Oryza sativa L.)[J]. Scientia Agricultura Sinica, 2024, 57(3): 429-441. (in Chinese with English abstract) | |
| [4] | 罗曦, 魏林燕, 郑燕梅, 魏毅东, 连玲, 谢华安, 吴方喜. 水稻护颖发育相关基因的研究进展[J]. 南京农业大学学报2021, 44(3): 412-420. |
| Luo X, Wei L Y, Zheng Y M, Wei Y D, Lian L, Xie H A, Wu F X. Research advances on developmental genes of sterile lemmas in rice[J]. Journal of Nanjing Agricultural University, 2021, 44(3): 412-420. (in Chinese with English abstract) | |
| [5] | Ren D Y, Hu J. FZP determines grain size and sterile lemma fate in rice[J]. Journal of Experimental Botany, 2018, 69(20): 4853-4866. |
| [6] | Yoshida A, Suzaki T, Tanaka W, Hirano H Y. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet[J]. Proceedings of the National Academy of Sciences of the United States of Ameica, 2009, 106 (47): 20103-20108. |
| [7] | Liu M, Li H, Su Y, Li W, Shi C. G1/ELE functions in the development of rice lemmas in addition to determining identities of empty glumes[J]. Frontiers in Plant Science, 2016, 7: 1006. |
| [8] | Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice[J]. Plant Cell Physiology, 2010, 51(1): 47-57. |
| [9] | Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D. The SEPALLATA-Like Gene OsMADS34 is required for rice inflorescence and spikelet development[J]. Plant Physiology, 2010, 153(2): 728-740. |
| [10] | Li W Q, Yoshida A, Takahashi M, Maekawa M, Kojima M, Sakakibara H, Kyozuka J. SAD1, an RNA polymerase Ⅰ subunit A34.5 of rice, interacts with mediator and controls various aspects of plant development[J]. The Plant Jouranl, 2015, 81: 282-291. |
| [11] | Xu Q K, Yu H P, Xia S S, Cui Y J, Yu X Q, Liu H, Zeng D L, Hu J, Zhang Q, Gao Z Y, Zhang G H, Zhu L, Shen L, Guo L B, Rao Y C, Qian Q, Ren D Y. The C2H2 zinc-finger protein LACKING RUDIMENTARY GLUME 1 regulates spikelet development in rice[J]. Science Bulletin, 2020, 65: 753-764. |
| [12] | Zhuang H, Wang H L, Zhang T, Zeng X Q, Chen H, Wang Z W, Zhang J, Zheng H, Tang J, Ling Y H, Yang Z L, He G H, Li Y F. NONSTOP GLUMES1 encodes a C2H2 zinc finger protein that regulates spikelet development in rice[J]. The Plant Cell, 2020, 32: 392-413. |
| [13] | Li H G, Xue D W, Gao Z Y, Yan M X, Xu W Y, Xing Z, Huang D N, Qian Q, Xue Y B. A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice[J]. The Plant Journal, 2009, 57(4): 593-605. |
| [14] | Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano H Y. ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice[J]. The Plant Journal, 2012, 70(2): 327-339. |
| [15] | Yang J G, Lee S Y, Hang R L, Kim S R, Lee Y S, Cao X F, Amasino R, An G. OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice[J]. The Plant Journal, 2013, 73: 566-578. |
| [16] | Zhang J R, Tang W, Huang Y L, Niu X L, Zhao Y, Han Y, Liu Y S. Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice[J]. Journal of Experimental Botany, 2015, 66(1): 99-112. |
| [17] | Lee D Y, Lee J, Moon S, Park S Y, An G. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem[J]. The Plant Journal, 2007, 49(1): 64-78. |
| [18] | Lee D Y, An G. Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET1 (OsIDS1) synergistically control inflorescence architecture and floral meristem establishment in rice[J]. The Plant Journal, 2012, 69(3): 445-461. |
| [19] | Ren D Y, Li Y F, Zhao F M, Sang X C, Shi J Q, Wang N, Guo S, Ling Y H, Zhang C W, Yang Z L, He G H. MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice[J]. Plant Physiology, 2013, 162(2): 872-884. |
| [20] | Fan C C, Xing Y Z, Mao H L, Lu T T, Han B, Xu C G, Li X H, Zhang Q F. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein[J]. Theoretical and Applied Genetics, 2006, 112(6): 1164-1171. |
| [21] | Mao H L, Sun S Y, Yao J L, Wang C R, Yu S B, Xu C G, Li X H, Zhang Q F. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice[J]. Proceedings of the National Academy of Sciences of the United States of USA, 2010, 107: 19579-19584. |
| [22] | Sun S Y, Wang L, Mao H L, Shao L, Li X H, Xiao J. H, Ouyang Y D, Zhang Q F. A G-protein pathway determines grain size in rice[J]. Nature Communications, 2018, 9(1): 851. |
| [23] | Qi P, Lin Y S, Song X J, Shen J B, Huang W, Shan J X, Zhu M Z, Jiang L, Gao J P, Lin H X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3[J]. Cell Research, 2012, 22: 1666-1680. |
| [24] | Zhang X J, Wang J F, Huang J, Lan H X, Wang C L, Yin C F, Wu Y Y, Tang H J, Qian Q, Li J Y, Zhang H S. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109: 21534-21539. |
| [25] | Yu J P, Xiong H Y, Zhu X Y, Zhang H L, Li H H, Miao J L, Wang W S, Tang Z S, Zhang Z Y, Yao G X, Zhang Q, Pan Y H, Wang X, Rashid M A R, Li J J, Gao Y M, Li Z K, Yang W C, Fu X D, Li Z C. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap[J]. BMC Biology, 2017, 15: 28. |
| [26] | Ying J Z, Ma M, Bai C, Huang X H, Liu J L, Fan Y Y, Song X J. TGW3, a major QTL that negatively modulates grain length and weight in rice[J]. Molecular Plant, 2018, 11: 750-753. |
| [27] | Hu Z J, Lu S J, Wang M J, He H H, Sun L, Wang H R, Liu X H, Jiang L, Sun J L, Xin X Y, Kong W, Chu C C, Xue H W, Yang J S, Luo X J, Liu J X. A novel QTL qTGW3 encodes the GSK3/SHAGGY-Like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice[J]. Molecular Plant, 2018, 11: 736-749. |
| [28] | Song X J, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, Suzuki T, Higashiyama T, Yamasaki M, Mori H, Inukai Y, Wu J Z, Kitano H, Sakakibara H, Jacobsen S E, Ashikari M. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice[J]. Proceedings of the National Academy of Sciences USA, 2015, 112: 76-81. |
| [29] | Wang A H, Hou Q Q, Si L Z, Huang X H, Luo J H, Lu D F, Zhu J J, Shangguan Y Y, Miao J H, Xie Y F, Wang Y C, Zhao Q, Feng Q, Zhou C C, Li Y, Fan D L, Lu Y Q, Tian Q L, Wang Z X, Han B. The PLATZ transcription factor GL6 affects grain length and number in rice[J]. Plant Physiology, 2019, 180: 2077-2090. |
| [30] | Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B I, Onishi A, Miyagawa H, Katoh E. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield[J]. Nature Genetics, 2013, 45(6): 707-711. |
| [31] | Wang S K, Li S, Liu Q, Wu K, Zhang J Q, Wang S S, Wang Y, Chen X B, Zhang Y, Gao C X, Wang F, Huang H X, Fu X D. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality[J]. Nature Genetics, 2015, 47: 949-954. |
| [32] | Wang Y X, Xiong G S, Hu J, Jiang L, Yu H, Xu J, Fang Y X, Zeng L J, Xu E B, Xu J, Ye W J, Meng X B, Liu R F, Chen H Q, Jing Y H, Wang Y H, Zhu X D, Li J Y, Qian Q. Copy number variation at the GL7 locus contributes to grain size diversity in rice[J]. Nature Genetics, 2015, 47: 944-948. |
| [33] | Si L Z, Chen J Y, Huang X H, Gong H, Luo J H, Hou Q Q, Zhou T Y, Lu T T, Zhu J J, Shangguan Y Y, Chen E W, Gong C X, Zhao Q, Jing Y F, Zhao Y, Li Y, Cui L L, Fan D L, Lu Y Q, Weng Q J, Wang Y C, Zhan Q L, Liu K Y, Wei X H, An K, An G, Han B. OsSPL13 controls grain size in cultivated rice[J]. Nature Genetics, 2016, 48: 447-456. |
| [34] | Song X J, Huang W, Shi M, Zhu M Z, Lin H X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase[J]. Nature Genetics, 2007, 39: 623-630. |
| [35] | Hao J, Wang D, Wu Y, Huang K, Duan P G, Li N, Xu R, Zeng D L, Dong G J, Zhang B L, Zhang L M, Inzé D, Qian Q, Li Y H. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice[J]. Molecular Plant, 2021, 8: 1266-1280. |
| [36] | Ruan B P, Shang L G, Zhang B, Hu J, Wang Y X, Lin H, Zhang A P, Liu C L, Peng Y L, Zhu L, Ren D Y, Shen L, Dong G J, Zhang G H, Zeng D L, Guo L B, Qian Q, Gao Z Y. Natural variation in the promoter of TGW2 determines grain width and weight in rice[J]. New Phytologist, 2020, 227: 629-640. |
| [37] | Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu C J, Li X H, Xiao J H, He Y Q, Zhang Q F. Natural variation in GS5 plays an important role in regulating grain size and yield in rice[J]. Nature Genetics, 2011, 43: 1266-1269. |
| [38] | Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. Deletion in a gene associated with grain size increased yields during rice domestication[J]. Nature Genetics, 2008, 40: 1023-1028. |
| [39] | Weng J F, Gu S H, Wan X Y, Gao H, Guo T, Su N, Lei C L, Zhang X, Cheng Z J, Guo X P, Wang J L, Jiang L, Zhai H Q, Wan J M. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight[J]. Cell Research, 2008, 18: 1199-1209. |
| [40] | Duan P G, Xu J S, Zeng D L, Zhang B L, Geng M F, Zhang G Z, Huang K, Huang L J, Xu R, Ge S, Qian Q, Li Y H. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice[J]. Molecular Plant, 2017, 10(5): 685-694. |
| [41] | Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D. Control of grain size, shape and quality by OsSPL16 in rice[J]. Nature Genetics, 2012, 44: 950-954. |
| [42] | Rogers S O, Bendich A J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues[J]. Plant Molecular Biology, 1985, 5: 69-76. |
| [43] | Howarth W O. The Gramineae: A study of cereal, bamboo and grasses[J]. Nature, 1935, 136(3435): 317-319. |
| [44] | Kellogg E A. The evolutionary history of Ehrhartoideae, Oryzeae, and Oryza[J]. Rice, 2009, 2: 1-14. |
| [45] | Terrell E E, Peterson P M, Wergin W P. Epidermal features and spikelet micromorphology in Oryza and related genera (Poaceae: Oryzeae)[J]. Smithsonian Contributions to Botany, 2001, 91: 1-50. |
| [46] | Wu T K, Ali A, Wang J H, Song J H, Fang Y Q, Zhou T T, Luo Y, Zhang H Y, Chen X Q, Liao Y X, Liu Y T, Xu P Z, Wu J J. A homologous gene of OsREL2/ASP1, ASP-LSL regulates pleiotropic phenotype including long sterile lemma in rice[J]. BMC Plant Biology, 2021, 21: 390. |
| [1] | 王娟, 吴丽娟, 洪海波, 姚志文, 王磊, 鄂志国. 水稻泛素结合酶E2的生物学功能研究进展[J]. 中国水稻科学, 2025, 39(6): 744-750. |
| [2] | 陶士博, 许娜, 徐正进, 刘畅, 徐铨. 水稻发芽期耐冷基因Cold6的克隆[J]. 中国水稻科学, 2025, 39(6): 751-759. |
| [3] | 陈伟, 叶元妹, 赵剑华, 冯志明, 陈宗祥, 胡珂鸣, 左示敏. 利用CRISPR/Cas9技术改良南粳46抽穗期[J]. 中国水稻科学, 2025, 39(6): 760-770. |
| [4] | 侯桂花, 周立国, 雷建国, 陈虹, 聂元元. 水稻OsRDR5基因功能及作用机制初步解析[J]. 中国水稻科学, 2025, 39(6): 779-788. |
| [5] | 邓欢, 刘亚培, 王春连, 郭威, 陈析丰, 纪志远. 水稻抗白叶枯病新基因Xa49(t)的定位分析[J]. 中国水稻科学, 2025, 39(6): 825-831. |
| [6] | 郝雯倩, 蔡兴菁, 杨海东, 吴宇阳, 滕轩, 薛超, 龚志云. 不同类型组蛋白修饰在水稻响应非生物胁迫中的研究进展[J]. 中国水稻科学, 2025, 39(5): 575-585. |
| [7] | 王镜博, 苏畅, 冯晶, 姜思旭, 徐海, 崔志波, 赵明辉. 水稻OsAlR1基因耐铝性功能研究[J]. 中国水稻科学, 2025, 39(5): 615-623. |
| [8] | 韶也, 胡远艺, 彭彦, 毛毕刚, 刘慧敏, 唐婵娟, 雷斌, 唐丽, 余丽霞, 李文建, 罗武中, 罗治斌, 袁远涛, 李曜魁, 张丹, 周利斌, 柏连阳, 唐文帮, 赵炳然. 基于M1TDS靶向筛选技术的重离子束诱变定向改良杂交水稻卓两优1126性状的研究[J]. 中国水稻科学, 2025, 39(5): 624-634. |
| [9] | 徐群, 王珊, 袁筱萍, 金石桥, 晋芳, 郝万军, 吴小碧, 冯跃, 余汉勇, 孙燕飞, 杨窑龙, 魏兴华. 用于水稻品种真实性验证的SNP位点评价[J]. 中国水稻科学, 2025, 39(5): 635-642. |
| [10] | 张海鹏, 李莞意, 廖福兴, 马美子, 张洪程, 杨艳菊. 纳米钼对水稻根系形态生理和硝态氮吸收的影响[J]. 中国水稻科学, 2025, 39(5): 650-664. |
| [11] | 刘钰婷, 周星, 何辰延, 李秋萍, 艾小凤, 袁玉洁, 刘睿, 杨景文, 刘婷婷, 王丽, 程红, 黄蓉, 李奥运, 胡文, 胡忠, 任万军, 邓飞. 不同光照条件下减穴稳苗配置对水稻茎鞘干物质积累转运特性的影响[J]. 中国水稻科学, 2025, 39(5): 665-678. |
| [12] | 杨行洲, 崔苗苗, 魏利辉, 顾爱国, 李东霞, 乐秀虎, 冯辉. 外源miR3979处理水稻对拟禾本科根结线虫趋性、侵染和发育的影响[J]. 中国水稻科学, 2025, 39(5): 703-710. |
| [13] | 朱鹏, 凌溪铁, 王金彦, 张保龙, 杨郁文, 许轲, 裘实. 机直播条件下不同控草方式对抗除草剂水稻产量和品质差异性研究[J]. 中国水稻科学, 2025, 39(4): 501-515. |
| [14] | 董立强, 张义凯, 杨铁鑫, 冯莹莹, 马亮, 梁潇, 张玉屏, 李跃东. 北方粳稻密苗机插育秧对秧苗素质及取秧特性的影响[J]. 中国水稻科学, 2025, 39(4): 516-528. |
| [15] | 周洋, 叶凡, 刘立军. 典型促生微生物提高盐胁迫水稻抗性的研究进展[J]. 中国水稻科学, 2025, 39(4): 529-542. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||