
中国水稻科学 ›› 2025, Vol. 39 ›› Issue (5): 624-634.DOI: 10.16819/j.1001-7216.2025.240106
韶也1,2,#, 胡远艺1,2,#, 彭彦1,2,#, 毛毕刚1,2, 刘慧敏1,2, 唐婵娟1, 雷斌1,2, 唐丽1,2, 余丽霞3, 李文建3, 罗武中3, 罗治斌1,2, 袁远涛1,2, 李曜魁1,2, 张丹1,2, 周利斌3, 柏连阳2,4,*( ), 唐文帮1,2,*(
), 唐文帮1,2,*( ), 赵炳然1,2,*(
), 赵炳然1,2,*( )
)
收稿日期:2024-01-09
修回日期:2024-04-17
出版日期:2025-09-10
发布日期:2025-09-10
通讯作者:
*email:lybai@hunaas.cn,作者简介:第一联系人:#共同第一作者
基金资助:
SHAO Ye1,2,#, HU Yuanyi1,2,#, PENG Yan1,2,#, MAO Bigang1,2, LIU Huimin1,2, TANG Chanjuan1, LEI Bin1,2, TANG Li1,2, YU Lixia3, LI Wenjian3, LUO Wuzhong3, LUO Zhibin1,2, YUAN Yuantao1,2, LI Yaokui1,2, ZHANG Dan1,2, ZHOU Libin3, BAI Lianyang2,4,*( ), TANG Wenbang1,2,*(
), TANG Wenbang1,2,*( ), ZHAO Bingran1,2,*(
), ZHAO Bingran1,2,*( )
)
Received:2024-01-09
Revised:2024-04-17
Online:2025-09-10
Published:2025-09-10
About author:First author contact:#These authors contributed equally to this work
摘要:
【目的】以两系杂交水稻卓两优1126为“底盘品种”,快速创制具有镉低积累、香味、耐储藏、耐淹、低升糖指数等性状的改良品种。【方法】12C6+碳离子束诱变卓两优1126亲本卓234S和湘农恢1126,利用M1TDS技术从诱变M1群体中鉴定OsNRAMP5、OsBADH2、OsLOX3、OsPAO5、OsSSIIIa和OsBEIIb基因嵌合突变株,通过KASP分型从诱变M2群体中鉴定分离目标基因纯合(杂合)突变株,利用OsNRAMP5突变亲本配制OsNRAMP5突变组合。【结果】从重离子束诱变M1群体中鉴定到13株为5个目标基因突变的嵌合突变株,其中7个M2株系中鉴定到目标基因的纯合或杂合突变;OsNRAMP5纯合突变株镉含量较原始亲本显著降低,OsBADH2纯合突变株香味特征化合物2-乙酰基吡咯啉(2-AP)含量较原始亲本显著上升;OsNRAMP5基因突变亲本配制的组合,在镉污染土(土壤有效镉含量0.677 mg/kg,pH 5.6)盆栽种植,其籽粒镉含量为0.05 mg/kg,而原始组合为0.91 mg/kg。【结论】利用“重离子束诱变+M1TDS技术”实现了卓两优1126镉低积累等性状的快速改良,为生物传统诱变育种进化到定向诱变育种提拱了成功案例和共性技术参考。
韶也, 胡远艺, 彭彦, 毛毕刚, 刘慧敏, 唐婵娟, 雷斌, 唐丽, 余丽霞, 李文建, 罗武中, 罗治斌, 袁远涛, 李曜魁, 张丹, 周利斌, 柏连阳, 唐文帮, 赵炳然. 基于M1TDS靶向筛选技术的重离子束诱变定向改良杂交水稻卓两优1126性状的研究[J]. 中国水稻科学, 2025, 39(5): 624-634.
SHAO Ye, HU Yuanyi, PENG Yan, MAO Bigang, LIU Huimin, TANG Chanjuan, LEI Bin, TANG Li, YU Lixia, LI Wenjian, LUO Wuzhong, LUO Zhibin, YUAN Yuantao, LI Yaokui, ZHANG Dan, ZHOU Libin, BAI Lianyang, TANG Wenbang, ZHAO Bingran. Directed Improvement of Hybrid Rice Zhuoliangyou 1126 by Heavy Ion Beam Mutagenesis Based on M1TDS Targeted Screening Technology[J]. Chinese Journal OF Rice Science, 2025, 39(5): 624-634.
| 名称 Name | 序列 Sequence |
|---|---|
| OsNRAMP5-246-8-F | GAAGGTGACCAAGTTCATGCTTGTGGTTGGCTCCGGCTTAA |
| OsNRAMP5-246-8-H | GAAGGTCGGAGTCAACGGATTTGTGGTTGGCTCCGGCTTGC |
| OsNRAMP5-246-8-C | CACAAAATGAAACAGTGGAAACCGATC |
| OsNRAMP5-234-6-F | GAAGGTGACCAAGTTCATGCTCACAAAATGAAACAGTGGAAACCGAT |
| OsNRAMP5-234-6-H | GAAGGTCGGAGTCAACGGATTCACAAAATGAAACAGTGGAAACCGAC |
| OsNRAMP5-234-6-C | CGTACCTCATATCTGTGGTTGGCTC |
| OsNRAMP5-204-6-F | GAAGGTGACCAAGTTCATGCTCACAAAATGAAACAGTGGAAACCGA |
| OsNRAMP5-204-6-H | GAAGGTCGGAGTCAACGGATTCACAAAATGAAACAGTGGAAACCGC |
| OsNRAMP5-204-6-C | CGTACCTCATATCTGTGGTTGGCTC |
| OsNRAMP5-37-7-F | GAAGGTGACCAAGTTCATGCTTGAAGAAGCTAAGAGAGGAAGCAACAA |
| OsNRAMP5-37-7-H | GAAGGTCGGAGTCAACGGATTTGAAGAAGCTAAGAGAGGAAGCAACAG |
| OsNRAMP5-37-7-C | ACTAGCTCTCCAGCTGATGCTC |
| OsBADH2-168-1-F | GAAGGTGACCAAGTTCATGCTGTGTAGTTGGGTTGATCACACCTT |
| OsBADH2-168-1-H | GAAGGTCGGAGTCAACGGATTGTGTAGTTGGGTTGATCACACCTC |
| OsBADH2-168-1-C | TGGGTCATAAATAAATATAAGCGCAGG |
| OsBADH2-224-8-F | GAAGGTGACCAAGTTCATGCTTTCGTCTTTTCTTGACAGCCTGTT |
| OsBADH2-224-8-H | GAAGGTCGGAGTCAACGGATTTTCGTCTTTTCTTGACAGCCTGTC |
| OsBADH2-224-8-C | AGGACTTTTTCCACCAAGTTCCAG |
| OsLOX3-760-7-F | GAAGGTGACCAAGTTCATGCTTATCTTCACCTATGCCACAAGGC |
| OsLOX3-760-7-H | GAAGGTCGGAGTCAACGGATTTATCTTCACCTATGCCACAAGGA |
| OsLOX3-760-7-C | CAGGGTATCATCATCACGTAAGAAC |
| OsLOX3-115-5-F | GAAGGTGACCAAGTTCATGCTGAGAGCAAAGTGTTGAACATGAAC |
| OsLOX3-115-5-H | GAAGGTCGGAGTCAACGGATTGAGAGCAAAGTGTTGAACATGAAG |
| OsLOX3-115-5-C | GGACCGACCCGGTTCTTGAG |
| OsPAO5-549-5-F | GAAGGTGACCAAGTTCATGCTTGCTTGACAGGAATCCACACCTA |
| OsPAO5-549-5-H | GAAGGTCGGAGTCAACGGATTTGCTTGACAGGAATCCACACCTG |
| OsPAO5-549-5-C | GCACCACCATTCCAGATATAGGTG |
| OsPAO5-1028-4-F | GAAGGTGACCAAGTTCATGCTAACTTTCCTCCAGGGTTACCAAAAT |
| OsPAO5-1028-4-H | GAAGGTCGGAGTCAACGGATTAACTTTCCTCCAGGGTTACCAAATC |
| OsPAO5-1028-4-C | GCTTGTCCCATCTTCAACACATAC |
| OsPAO5-84-2-F | GAAGGTGACCAAGTTCATGCTCTCAGCTCAAGAAAATGCTACCAGG |
| OsPAO5-84-2-H | GAAGGTCGGAGTCAACGGATTCTCAGCTCAAGAAAATGCTACCACT |
| OsPAO5-84-2-C | GACAGTACTCAACTGTACATGGTCTG |
| OsSSIIIa-598-7-F | GAAGGTGACCAAGTTCATGCTCCATTTGGTTCAAGGCCTAGAACTC |
| OsSSIIIa-598-7-H | GAAGGTCGGAGTCAACGGATTCCATTTGGTTCAAGGCCTAGAACTG |
| OsSSIIIa-598-7-C | CACTGTTTTCGACGTAGACCATG |
| OsBEIIb-931-5-F | GAAGGTGACCAAGTTCATGCTGTGTGATTGTTATTAAATCTCACCAGGA |
| OsBEIIb-931-5-H | GAAGGTCGGAGTCAACGGATTGTGTGATTGTTATTAAATCTCACCAGGC |
| OsBEIIb-931-5-C | GGCTATCTTAACTTTATGGGAAATGAGTTC |
表1 KASP基因分型引物序列
Table 1. Primer sequence for KASP genotyping
| 名称 Name | 序列 Sequence |
|---|---|
| OsNRAMP5-246-8-F | GAAGGTGACCAAGTTCATGCTTGTGGTTGGCTCCGGCTTAA |
| OsNRAMP5-246-8-H | GAAGGTCGGAGTCAACGGATTTGTGGTTGGCTCCGGCTTGC |
| OsNRAMP5-246-8-C | CACAAAATGAAACAGTGGAAACCGATC |
| OsNRAMP5-234-6-F | GAAGGTGACCAAGTTCATGCTCACAAAATGAAACAGTGGAAACCGAT |
| OsNRAMP5-234-6-H | GAAGGTCGGAGTCAACGGATTCACAAAATGAAACAGTGGAAACCGAC |
| OsNRAMP5-234-6-C | CGTACCTCATATCTGTGGTTGGCTC |
| OsNRAMP5-204-6-F | GAAGGTGACCAAGTTCATGCTCACAAAATGAAACAGTGGAAACCGA |
| OsNRAMP5-204-6-H | GAAGGTCGGAGTCAACGGATTCACAAAATGAAACAGTGGAAACCGC |
| OsNRAMP5-204-6-C | CGTACCTCATATCTGTGGTTGGCTC |
| OsNRAMP5-37-7-F | GAAGGTGACCAAGTTCATGCTTGAAGAAGCTAAGAGAGGAAGCAACAA |
| OsNRAMP5-37-7-H | GAAGGTCGGAGTCAACGGATTTGAAGAAGCTAAGAGAGGAAGCAACAG |
| OsNRAMP5-37-7-C | ACTAGCTCTCCAGCTGATGCTC |
| OsBADH2-168-1-F | GAAGGTGACCAAGTTCATGCTGTGTAGTTGGGTTGATCACACCTT |
| OsBADH2-168-1-H | GAAGGTCGGAGTCAACGGATTGTGTAGTTGGGTTGATCACACCTC |
| OsBADH2-168-1-C | TGGGTCATAAATAAATATAAGCGCAGG |
| OsBADH2-224-8-F | GAAGGTGACCAAGTTCATGCTTTCGTCTTTTCTTGACAGCCTGTT |
| OsBADH2-224-8-H | GAAGGTCGGAGTCAACGGATTTTCGTCTTTTCTTGACAGCCTGTC |
| OsBADH2-224-8-C | AGGACTTTTTCCACCAAGTTCCAG |
| OsLOX3-760-7-F | GAAGGTGACCAAGTTCATGCTTATCTTCACCTATGCCACAAGGC |
| OsLOX3-760-7-H | GAAGGTCGGAGTCAACGGATTTATCTTCACCTATGCCACAAGGA |
| OsLOX3-760-7-C | CAGGGTATCATCATCACGTAAGAAC |
| OsLOX3-115-5-F | GAAGGTGACCAAGTTCATGCTGAGAGCAAAGTGTTGAACATGAAC |
| OsLOX3-115-5-H | GAAGGTCGGAGTCAACGGATTGAGAGCAAAGTGTTGAACATGAAG |
| OsLOX3-115-5-C | GGACCGACCCGGTTCTTGAG |
| OsPAO5-549-5-F | GAAGGTGACCAAGTTCATGCTTGCTTGACAGGAATCCACACCTA |
| OsPAO5-549-5-H | GAAGGTCGGAGTCAACGGATTTGCTTGACAGGAATCCACACCTG |
| OsPAO5-549-5-C | GCACCACCATTCCAGATATAGGTG |
| OsPAO5-1028-4-F | GAAGGTGACCAAGTTCATGCTAACTTTCCTCCAGGGTTACCAAAAT |
| OsPAO5-1028-4-H | GAAGGTCGGAGTCAACGGATTAACTTTCCTCCAGGGTTACCAAATC |
| OsPAO5-1028-4-C | GCTTGTCCCATCTTCAACACATAC |
| OsPAO5-84-2-F | GAAGGTGACCAAGTTCATGCTCTCAGCTCAAGAAAATGCTACCAGG |
| OsPAO5-84-2-H | GAAGGTCGGAGTCAACGGATTCTCAGCTCAAGAAAATGCTACCACT |
| OsPAO5-84-2-C | GACAGTACTCAACTGTACATGGTCTG |
| OsSSIIIa-598-7-F | GAAGGTGACCAAGTTCATGCTCCATTTGGTTCAAGGCCTAGAACTC |
| OsSSIIIa-598-7-H | GAAGGTCGGAGTCAACGGATTCCATTTGGTTCAAGGCCTAGAACTG |
| OsSSIIIa-598-7-C | CACTGTTTTCGACGTAGACCATG |
| OsBEIIb-931-5-F | GAAGGTGACCAAGTTCATGCTGTGTGATTGTTATTAAATCTCACCAGGA |
| OsBEIIb-931-5-H | GAAGGTCGGAGTCAACGGATTGTGTGATTGTTATTAAATCTCACCAGGC |
| OsBEIIb-931-5-C | GGCTATCTTAACTTTATGGGAAATGAGTTC |
| 名称 Name | 序列 Sequence |
|---|---|
| OsNRAMP5-3E-540F | GTTGGTCCTGGATTCATGGTGTC |
| OsNRAMP5-3E-540R | TCCAATCAGAATCACCCAGAGCAG |
| OsNRAMP5-1E-265F | TCTTCGTCTACTTCCAGCTAGCC |
| OsNRAMP5-1E-265R | ACAGTGAACAACTGTCCATGGTG |
| OsBADH-4-471F | TTGATGAAGCAGCATGGGACATG |
| OsBADH-4-471R | GCTGCTAGGTACAATTTGTGAGAC |
| OsBADH-8-357F | CTTCAGCTGCTCCTATGGTTAAGG |
| OsBADH-8-357R | TCTAGCATCCAGCTCAGTTAAGTGC |
| OsLOX3-8-535F | GAGAAGAACCTCGAAGGCCTCAG |
| OsLOX3-8-535R | CCATCACAGCATGTGTGTTGAGC |
| OsPAO-6/7-563F | GCTCTCTTTGATAAGGATGGTCGTC |
| OsPAO-6/7-563R | ATGGCCACCAGTAAGAACATGTTC |
| OsBEIIb-17-359F | GGCATGAGAACCTCACATACTG |
| OsBEIIb-17-359R | GGAAGTACTTGTGGAGCTCTTGG |
表2 Sanger测序引物序列
Table 2. Primers for Sanger sequencing
| 名称 Name | 序列 Sequence |
|---|---|
| OsNRAMP5-3E-540F | GTTGGTCCTGGATTCATGGTGTC |
| OsNRAMP5-3E-540R | TCCAATCAGAATCACCCAGAGCAG |
| OsNRAMP5-1E-265F | TCTTCGTCTACTTCCAGCTAGCC |
| OsNRAMP5-1E-265R | ACAGTGAACAACTGTCCATGGTG |
| OsBADH-4-471F | TTGATGAAGCAGCATGGGACATG |
| OsBADH-4-471R | GCTGCTAGGTACAATTTGTGAGAC |
| OsBADH-8-357F | CTTCAGCTGCTCCTATGGTTAAGG |
| OsBADH-8-357R | TCTAGCATCCAGCTCAGTTAAGTGC |
| OsLOX3-8-535F | GAGAAGAACCTCGAAGGCCTCAG |
| OsLOX3-8-535R | CCATCACAGCATGTGTGTTGAGC |
| OsPAO-6/7-563F | GCTCTCTTTGATAAGGATGGTCGTC |
| OsPAO-6/7-563R | ATGGCCACCAGTAAGAACATGTTC |
| OsBEIIb-17-359F | GGCATGAGAACCTCACATACTG |
| OsBEIIb-17-359R | GGAAGTACTTGTGGAGCTCTTGG |
| 目标基因 Target gene | 基因编号 Gene ID | 目标表型 Target phenotype | 外显子数 No. of exons | 目标区域覆盖度 Coverage of target area(%) | 平均测序深度 Average sequencing depth |
|---|---|---|---|---|---|
| OsNRAMP5 | Os07g0257200 | 镉低积累 Low cadmium accumulation | 13 | 100 | 53398× |
| OsBADH2 | Os08g0424500 | 香味Aroma | 15 | 100 | 53086× |
| OsLOX3 | Os03g0699700 | 耐储藏Storage-tolerance | 9 | 100 | 50202× |
| OsPAO5 | Os04g0671300 | 耐淹 Submergence-tolerance | 10 | 100 | 54263× |
| OsSSIIIa | Os08g0191433 | 低升糖指数 Low glycemic index (GI) | 14 | 100 | 54510× |
| OsBEIIb | Os02g0528200 | 低升糖指数 Low glycemic index (GI) | 22 | 100 | 52192× |
表3 目标基因及靶向测序分析情况
Table 3. Target gene and targeted sequencing analysis
| 目标基因 Target gene | 基因编号 Gene ID | 目标表型 Target phenotype | 外显子数 No. of exons | 目标区域覆盖度 Coverage of target area(%) | 平均测序深度 Average sequencing depth |
|---|---|---|---|---|---|
| OsNRAMP5 | Os07g0257200 | 镉低积累 Low cadmium accumulation | 13 | 100 | 53398× |
| OsBADH2 | Os08g0424500 | 香味Aroma | 15 | 100 | 53086× |
| OsLOX3 | Os03g0699700 | 耐储藏Storage-tolerance | 9 | 100 | 50202× |
| OsPAO5 | Os04g0671300 | 耐淹 Submergence-tolerance | 10 | 100 | 54263× |
| OsSSIIIa | Os08g0191433 | 低升糖指数 Low glycemic index (GI) | 14 | 100 | 54510× |
| OsBEIIb | Os02g0528200 | 低升糖指数 Low glycemic index (GI) | 22 | 100 | 52192× |
| 目标基因 Target gene | 亲本 Parent | 混池编号 No. of the pool | 突变位置 Mutation site | 野生/突变基因型 Wild/mutant genotype |
|---|---|---|---|---|
| OsNRAMP5 | 卓234S Zhuo 234S | 217-288E | 第3外显子Exon 3 | C/CTTAA |
| 卓234S Zhuo 234S | 217-288W | 第3外显子Exon 3 | C/CTTAA | |
| 卓234S Zhuo 234S | 217-288E | 第3外显子Exon 3 | GA/G | |
| 湘农恢1126 Xiangnonghui 1126 | 145-216S | 第3外显子Exon 3 | GCAGAT/G | |
| 湘农恢1126 Xiangnonghui 1126 | 1-72N | 第1外显子Exon 1 | CTCAATCTCCAT/C | |
| OsBADH2 | 卓234S Zhuo 234S | 145-216E | 第4外显子和第4内含子Exon 4/Intron 4 | CCTTGGTATTTCACATTTTTCT/C |
| 湘农恢1126 Xiangnonghui 1126 | 217-238S | 第8外显子Exon 8 | GTT/G | |
| OsLOX3 | 卓234S Zhuo 234S | 718-789W | 第7外显子Exon 7 | G/GC |
| 湘农恢1126 Xiangnonghui 1126 | 73-144S | 第9外显子Exon 9 | GAAC/G | |
| 湘农恢1126 Xiangnonghui 1126 | 73-144E | 第9外显子Exon 9 | GAAC/G | |
| OsPAO5 | 卓234S Zhuo 234S | 504-575W | 第6外显子和第6内含子Exon 6/Intron 6 | CACTTACTTT/C |
| 卓234S Zhuo 234S | 1004-1040E | 第9外显子Exon 9 | ATGAT/A | |
| 卓234S Zhuo 234S | 991-1040W | 第9外显子Exon 9 | ATGAT/A | |
| 湘农恢1126 Xiangnonghui 1126 | 73-144E | 第9外显子Exon 9 | GTAGCTCC/G | |
| OsSSIIIa | 卓234S Zhuo 234S | 576-647E | 第13外显子Exon 13 | CTCG/C |
| OsBEIIb | 卓234S Zhuo 234S | 863-932W | 第17外显子Exon 17 | GATGT/G |
表4 混池目标基因靶向测序捕获突变情况
Table 4. Mutations were captured by mixed-pool targeted gene sequencing
| 目标基因 Target gene | 亲本 Parent | 混池编号 No. of the pool | 突变位置 Mutation site | 野生/突变基因型 Wild/mutant genotype |
|---|---|---|---|---|
| OsNRAMP5 | 卓234S Zhuo 234S | 217-288E | 第3外显子Exon 3 | C/CTTAA |
| 卓234S Zhuo 234S | 217-288W | 第3外显子Exon 3 | C/CTTAA | |
| 卓234S Zhuo 234S | 217-288E | 第3外显子Exon 3 | GA/G | |
| 湘农恢1126 Xiangnonghui 1126 | 145-216S | 第3外显子Exon 3 | GCAGAT/G | |
| 湘农恢1126 Xiangnonghui 1126 | 1-72N | 第1外显子Exon 1 | CTCAATCTCCAT/C | |
| OsBADH2 | 卓234S Zhuo 234S | 145-216E | 第4外显子和第4内含子Exon 4/Intron 4 | CCTTGGTATTTCACATTTTTCT/C |
| 湘农恢1126 Xiangnonghui 1126 | 217-238S | 第8外显子Exon 8 | GTT/G | |
| OsLOX3 | 卓234S Zhuo 234S | 718-789W | 第7外显子Exon 7 | G/GC |
| 湘农恢1126 Xiangnonghui 1126 | 73-144S | 第9外显子Exon 9 | GAAC/G | |
| 湘农恢1126 Xiangnonghui 1126 | 73-144E | 第9外显子Exon 9 | GAAC/G | |
| OsPAO5 | 卓234S Zhuo 234S | 504-575W | 第6外显子和第6内含子Exon 6/Intron 6 | CACTTACTTT/C |
| 卓234S Zhuo 234S | 1004-1040E | 第9外显子Exon 9 | ATGAT/A | |
| 卓234S Zhuo 234S | 991-1040W | 第9外显子Exon 9 | ATGAT/A | |
| 湘农恢1126 Xiangnonghui 1126 | 73-144E | 第9外显子Exon 9 | GTAGCTCC/G | |
| OsSSIIIa | 卓234S Zhuo 234S | 576-647E | 第13外显子Exon 13 | CTCG/C |
| OsBEIIb | 卓234S Zhuo 234S | 863-932W | 第17外显子Exon 17 | GATGT/G |
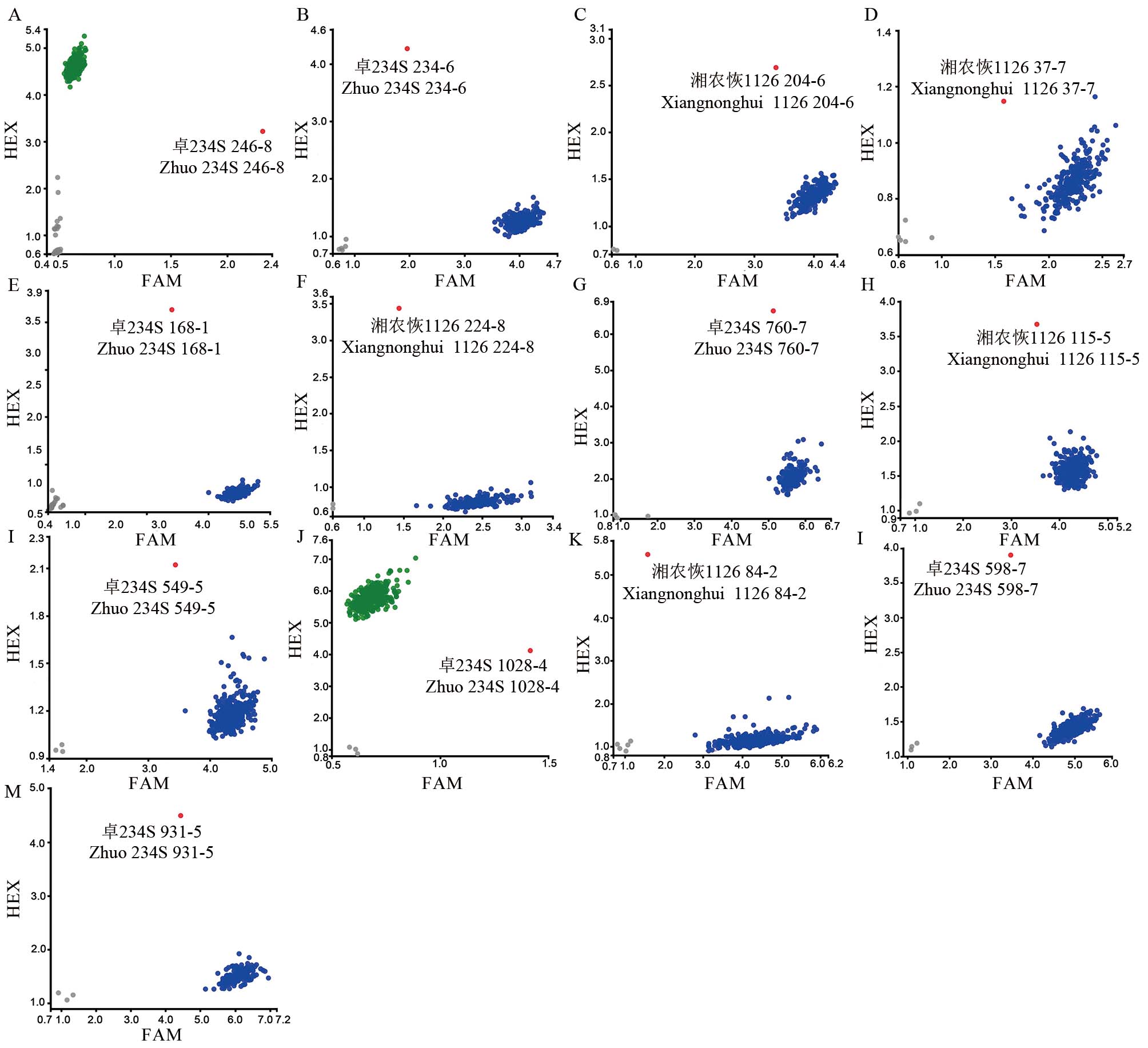
图1 目标基因突变混池的KASP基因分型 A~D分别代表OsNRAMP5基因不同突变位点基因分型;E, F 分别代表OsBADH2基因不同突变位点基因分型;G, H分别代表OsLOX3基因不同突变位点基因分型;I~K分别代表OsPAO5基因不同突变位点基因分型;L, M分别代表OsSSIIIa基因和OsBEIIb基因突变位点基因分型;A. 图中246-8代表诱变M1群体246行第8株(下同);蓝点/绿点代表野生基因型,红点代表突变基因型,灰点代表阴性对照;FAM和HEX分别代表两种不同颜色的荧光修饰标签。
Fig. 1. KASP genotyping in the mixed pool of target gene mutations A-D represent the genotyping of different mutation sites of OsNRAMP5 gene; E and F represent the genotyping of different mutation sites of OsBADH2 gene, respectively; G and H represent genotyping of different mutation sites of OsLOX3 gene, respectively; I-K represent genotyping of different mutation sites of OsPAO5 gene; L, M represent the genotyping of mutation sites in OsSSIIIa gene and OsBEIIb gene, respectively; In panel A, 246-8 represents the 8th plant in 246 rows of M1 population (the same below); Blue/green dots represent wild genotypes, red dots represent mutant genotypes, and gray dots represent negative controls; FAM and HEX represent fluorescently modified labels of two different colors, respectively.
| 基因 Gene | 品种 Variety | 突变株定位(行-株) Location of mutants (row-plant) | 突变分蘖占比 Proportion of tillers containing mutations |
|---|---|---|---|
| OsNRAMP5 | 卓234S Zhuo 234S | 246-8 | 18/18 |
| OsNRAMP5 | 卓234S Zhuo 234S | 234-6 | 4/18 |
| OsNRAMP5 | 湘农恢1126 Xiangnonghui 1126 | 204-6 | 3/6 |
| OsNRAMP5 | 湘农恢1126 Xiangnonghui 1126 | 37-7 | 1/8 |
| OsBADH2 | 卓234S Zhuo 234S | 168-1 | 3/18 |
| OsBADH2 | 湘农恢1126 Xiangnonghui 1126 | 224-8 | 1/6 |
| OsLOX3 | 卓234S Zhuo 234S | 760-7 | 1/18 |
| OsLOX3 | 湘农恢1126 Xiangnonghui 1126 | 115-5 | 2/6 |
| OsPAO5 | 卓234S Zhuo 234S | 549-5 | 1/18 |
| OsPAO5 | 卓234S Zhuo 234S | 1028-4 | 5/18 |
| OsPAO5 | 湘农恢1126 Xiangnonghui 1126 | 84-2 | 1/6 |
| OsSSIIIa | 卓234S Zhuo 234S | 598-7 | 1/18 |
| OsBEIIb | 卓234S Zhuo 234S | 931-5 | 4/18 |
表5 诱变M1突变株分蘖比例
Table 5. Tiller ratio of mutagenic M1 mutants
| 基因 Gene | 品种 Variety | 突变株定位(行-株) Location of mutants (row-plant) | 突变分蘖占比 Proportion of tillers containing mutations |
|---|---|---|---|
| OsNRAMP5 | 卓234S Zhuo 234S | 246-8 | 18/18 |
| OsNRAMP5 | 卓234S Zhuo 234S | 234-6 | 4/18 |
| OsNRAMP5 | 湘农恢1126 Xiangnonghui 1126 | 204-6 | 3/6 |
| OsNRAMP5 | 湘农恢1126 Xiangnonghui 1126 | 37-7 | 1/8 |
| OsBADH2 | 卓234S Zhuo 234S | 168-1 | 3/18 |
| OsBADH2 | 湘农恢1126 Xiangnonghui 1126 | 224-8 | 1/6 |
| OsLOX3 | 卓234S Zhuo 234S | 760-7 | 1/18 |
| OsLOX3 | 湘农恢1126 Xiangnonghui 1126 | 115-5 | 2/6 |
| OsPAO5 | 卓234S Zhuo 234S | 549-5 | 1/18 |
| OsPAO5 | 卓234S Zhuo 234S | 1028-4 | 5/18 |
| OsPAO5 | 湘农恢1126 Xiangnonghui 1126 | 84-2 | 1/6 |
| OsSSIIIa | 卓234S Zhuo 234S | 598-7 | 1/18 |
| OsBEIIb | 卓234S Zhuo 234S | 931-5 | 4/18 |
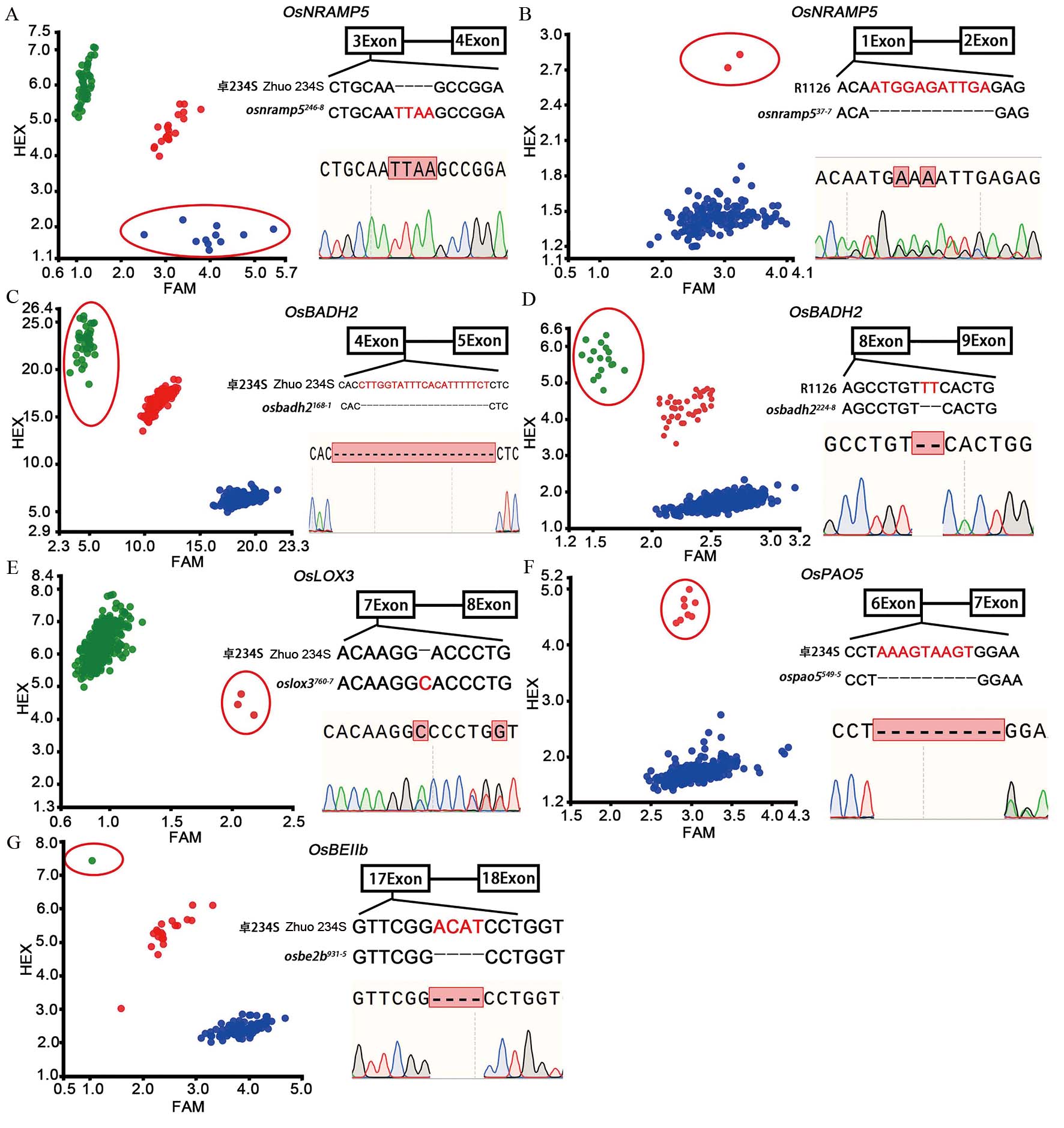
图2 M2群体的KASP鉴定及突变单株的Sanger测序验证 A, B:卓234S和湘农恢1126(R1126) M2群体OsNRAMP5基因KASP鉴定及突变单株Sanger测序验证,osnramp5246-8和osnramp537-7分别代表卓234S和湘农恢1126的OsNRAMP5基因突变单株; C, D:卓234S和湘农恢1126 M2群体OsBADH2基因KASP鉴定及Sanger测序验证,osbadh2168-1和 osbadh2224-8分别代表卓234S和湘农恢1126的OsBADH2基因突变单株;E~G:卓234S M2群体OsLOX3、OsPAO5和OsBEⅡb基因KASP鉴定及Sanger测序验证,oslox3760-7、ospao5549-5和osbe2b931-5分别代表卓234S的OsLOX3、OsPAO5和OsBEⅡb基因突变单株。图A中红点代表杂合基因型,蓝点代表纯合突变基因型,绿点代表野生基因型;图B~G中红点代表杂合基因型,绿点代表纯合突变基因型,蓝点代表野生基因型;红色椭圆形框内圆点代表目标基因纯合/杂合突变基因型。
Fig. 2. KASP identification of M2 generation and Sanger sequencing of mutant plants A and B, KASP identification of OsNRAMP5 gene in Zhuo 234S and Xiangnonghui 1126 M2 population and Sanger sequencing of mutants. osnramp5246-8 and osnramp537-7 represent mutant plant of OsNRAMP5 gene in Zhuo 234S and Xiangnonghui 1126, respectively; C and D, KASP identification of OsBADH2 gene in Zhuo 234S and Xiangnonghui 1126 M2 population and Sanger sequencing of mutants; osbadh2168-1and osbadh2224-8 represent mutant plant of OsBADH2 gene in Zhuo234S and Xiang Nonghui 1126, respectively. E-G, KASP identification of OsLOX3, OsPAO5 and OsBEⅡb gene in Zhuo 234S M2 population and Sanger sequencing of mutants. oslox3760-7, ospao5549-5 and osbe2b931-5 represented mutant plant of OsLOX3, OsPAO5 and OsBEⅡb gene of Zhuo 234S, respectively. In panel A, red dots represent heterozygous genotype, blue dots represent homozygous mutant genotype, green dots represent wild genotype. In panels B-G, red dots represent heterozygous genotype, green dots represent homozygous mutant genotype, blue dots represent wild genotype. The dots in the red oval box represent the homozygous/heterozygous genotype of target genes.
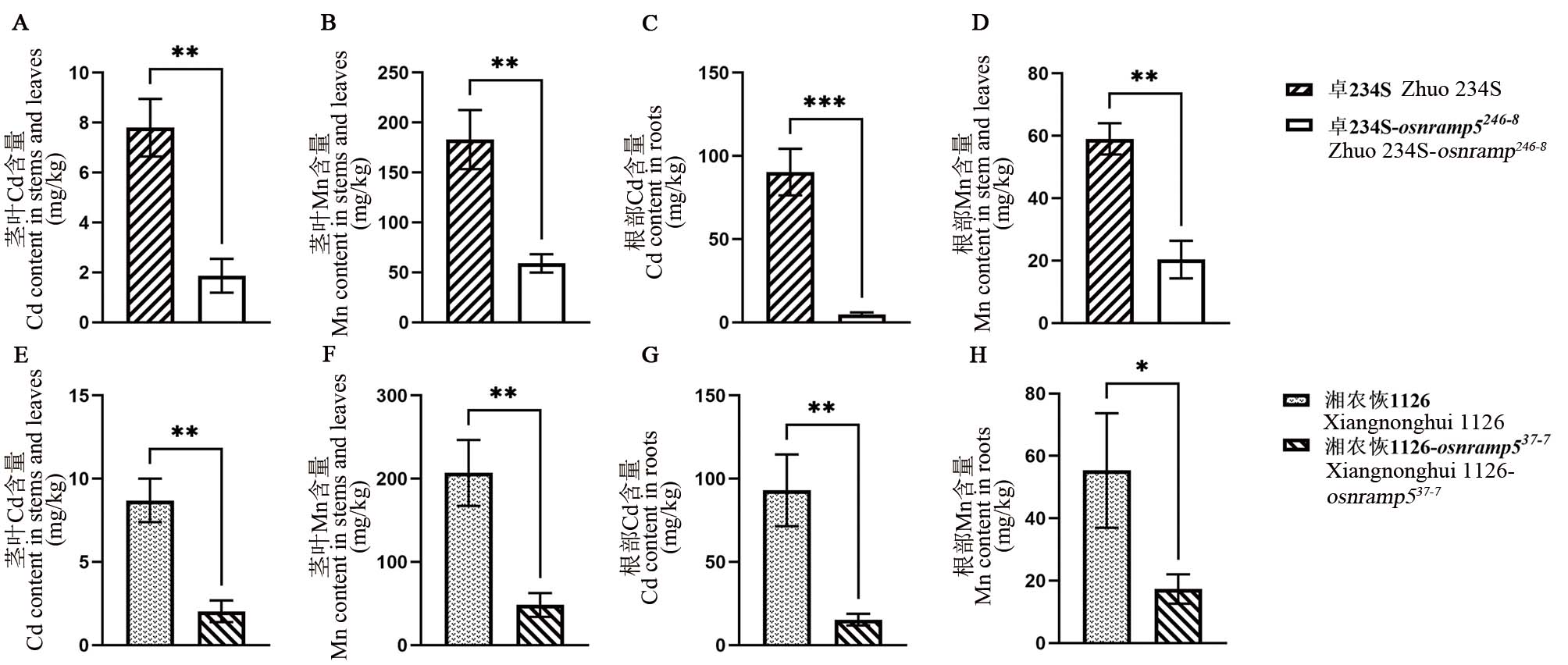
图3 OsNRAMP5纯合突变株及对照Cd、Mn含量 A-D: 纯合突变不育系卓234S-osnramp5246-8及原始对照材料Cd、Mn含量;E-H:纯合突变恢复系湘农恢1126-osnramp537-7及原始对照材料Cd、Mn含量;*、**和***分别表示在0.05、0.01和0.001水平上的差异显著(t检验)。
Fig. 3. Contents of Cd and Mn in homozygous mutant of OsNRAMP5 and control A-D, Contents of Cd and Mn in homozygous mutant Zhuo 234S-osnramp5246-8 and its original control material; E-H, Contents of Cd and Mn in the homozygous mutant Xiangnonghui 1126-osnramp537-7and its original control material; *, **, and *** represent difference was significant at 0.05, 0.01, and 0.001 levels, respectively (t-test).
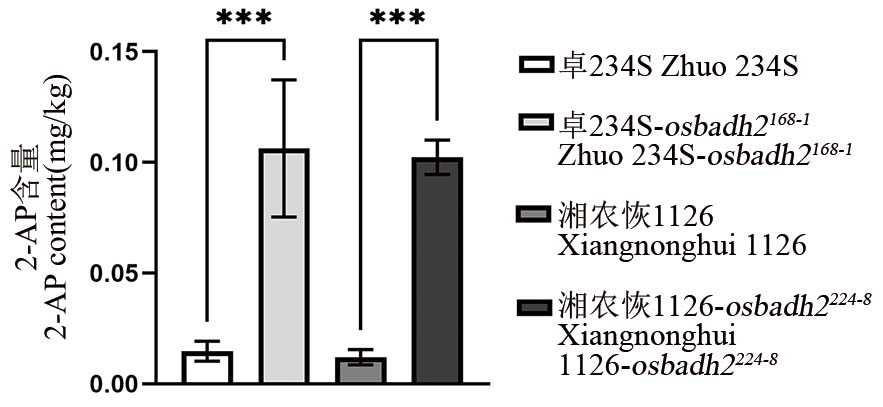
图4 OsBADH2纯合突变株及对照香味物质2-AP含量 ***表示在0.001水平上的差异(t检验)。
Fig. 4. 2-AP content in M2 homozygous mutant of OsBADH2 and control *** represent difference at the level of 0.001 (t-test).
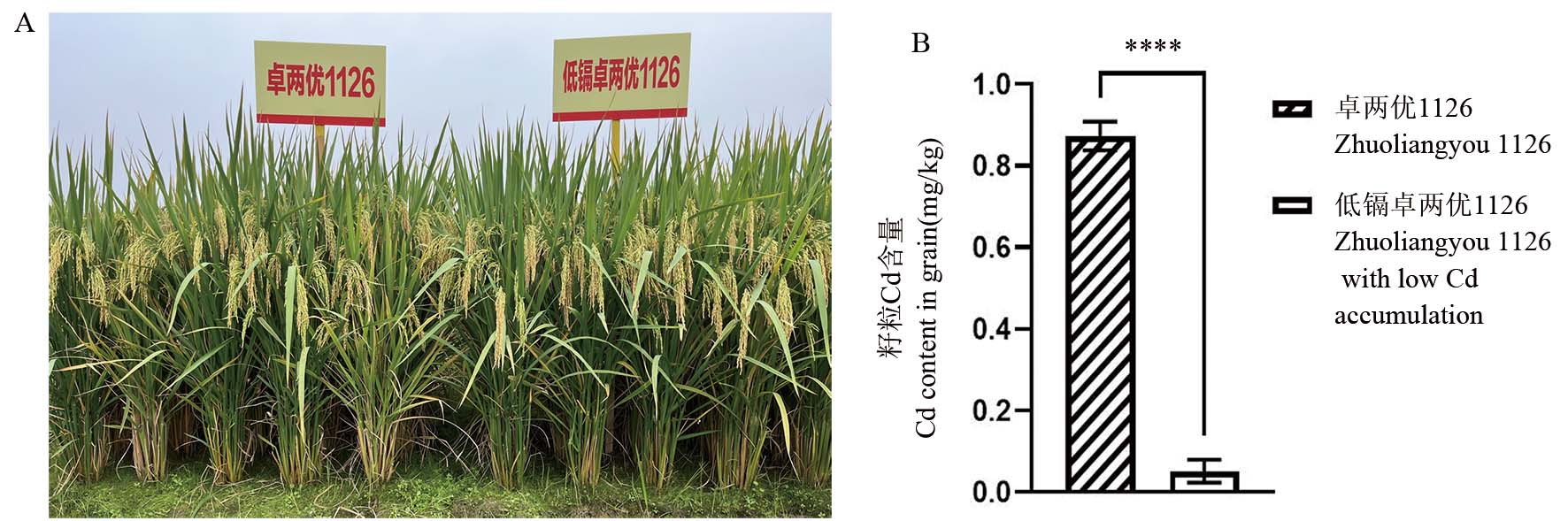
图5 镉低积累卓两优1126与原始对照品种田间表型及盆栽籽粒镉含量
Fig. 5. Field phenotype and cadmium content in grain of Zhuoliangyou 1126 with low cadmium accumulation compared with its original control
| 材料 Material | 株高 Plant height (cm) | 有效分蘖数 No. of effective tillers | 主穗长 Panicle length(cm) | 一次枝梗数 No. of primary branches | 二次枝梗数 No. of secondary branches |
|---|---|---|---|---|---|
| 卓两优1126(对照) Zhuoliangyou 1126(CK) | 124.6±3.3 a | 13.2±1.3 a | 28.9±1.4 a | 18.2±1.3 a | 99.4±6.4 a |
| 镉低积累卓两优1126 Zhuoliangyou 1126 with low cadmium accumulation | 124.8±2.7 a | 13.6±1.1 a | 27.9±0.9 a | 19.0±0.7 a | 104.2±5.7 a |
| 材料 Material | 总粒数 Spikelets per panicle | 实粒数 Grains per panicle | 结实率 Seed setting rate(%) | 千粒重 1000-grain weight(g) | |
| 卓两优1126(对照) Zhuoliangyou 1126(CK) | 347.4±39.1 a | 301.6±28.9 a | 87.0±2.0 a | 22.9±0.4 a | |
| 镉低积累卓两优1126 Zhuoliangyou 1126 with low cadmium accumulation | 360.8±24.1 a | 316.8±19.6 a | 88.0±2.0 a | 22.9±0.5 a |
表6 镉低积累卓两优1126及其原始对照品种农艺性状比较
Table 6. Comparison of agronomic traits of Zhuoliangyou 1126 with low cadmium accumulation and its original control varieties
| 材料 Material | 株高 Plant height (cm) | 有效分蘖数 No. of effective tillers | 主穗长 Panicle length(cm) | 一次枝梗数 No. of primary branches | 二次枝梗数 No. of secondary branches |
|---|---|---|---|---|---|
| 卓两优1126(对照) Zhuoliangyou 1126(CK) | 124.6±3.3 a | 13.2±1.3 a | 28.9±1.4 a | 18.2±1.3 a | 99.4±6.4 a |
| 镉低积累卓两优1126 Zhuoliangyou 1126 with low cadmium accumulation | 124.8±2.7 a | 13.6±1.1 a | 27.9±0.9 a | 19.0±0.7 a | 104.2±5.7 a |
| 材料 Material | 总粒数 Spikelets per panicle | 实粒数 Grains per panicle | 结实率 Seed setting rate(%) | 千粒重 1000-grain weight(g) | |
| 卓两优1126(对照) Zhuoliangyou 1126(CK) | 347.4±39.1 a | 301.6±28.9 a | 87.0±2.0 a | 22.9±0.4 a | |
| 镉低积累卓两优1126 Zhuoliangyou 1126 with low cadmium accumulation | 360.8±24.1 a | 316.8±19.6 a | 88.0±2.0 a | 22.9±0.5 a |
| [1] | Oladosu Y, Rafii M, Abdullah N, Hussin G, Ramli A, Rahim H, Miah G, Usman M. Principle and application of plant mutagenesis in crop improvement: A review[J]. Biotechnology & Biotechnological Equipment, 2016, 30(1): 1-16. |
| [2] | Sahu P, Sao R, Mondal S, Vishwakarma G, Gupta S, Kumar V, Singh S, Sharma D, Das B. Next generation sequencing based forward genetic approaches for identification and mapping of causal mutations in crop plants: A comprehensive review[J]. Plants (Basel), 2020, 9(10): 1355. |
| [3] | Bagher A, Nahid A, Mohsen M, Vahid M. Nuclear techniques in agriculture and genetics[J]. American Journal of Bioscience, 2014, 2(3): 102-105. |
| [4] | Jankowicz-Cieslak J, Tai T, Kumlehn J, Till B. Biotechnologies for plant mutation breeding: Protocols[M]. Springer Nature, 2017. |
| [5] | Spencer-Lopes M, Forster B, Jankuloski L. Manual on mutation breeding[M]. Food and Agriculture Organization of the United Nations (FAO), 2018. |
| [6] | Tokuyama Y, Furusawa Y, Ide H, Yasui A, Terato H. Role of isolated and clustered DNA damage and the post-irradiating repair process in the effects of heavy ion beam irradiation[J]. Journal of Radiation Research, 2015, 56(3): 446-455. |
| [7] | Hamada N, Imaoka T, Masunaga S, Ogata T, Okayasu R, Takahashi A, Kato T A, Kobayashi Y, Ohnishi T, Ono K, Shimada Y, Teshima T. Recent advances in the biology of heavy-ion cancer therapy[J]. Journal of Radiation Research, 2010, 51(4): 365-383. |
| [8] | Tanaka A, Shikazono N, Hase Y. Studies on biological effects of ion beams on lethality, molecular nature of mutation, mutation rate, and spectrum of mutation phenotype for mutation breeding in higher plants[J]. Journal of Radiation Research, 2010, 51(3): 223-233. |
| [9] | Hirano T, Kazama Y, Ishii K, Ohbu S, Shirakawa Y, Abe T. Comprehensive identification of mutations induced by heavy-ion beam irradiation in Arabidopsis thaliana[J]. Plant Journal, 2015, 82(1): 93-104. |
| [10] | Dong X, Yan X, Li W. Plant mutation breeding with heavy ion irradiation at IMP[J]. Journal of Agricultural Science, 2016, 8(5): 34-41. |
| [11] | Abe T, Kazama Y, Hirano T. Ion beam breeding and gene discovery for function analyses using mutants[J]. Nuclear Physics News, 2015, 25(4): 30-34. |
| [12] | 韶也, 彭彦, 毛毕刚, 余丽霞, 唐丽, 李曜魁, 胡远艺, 张丹, 袁智成, 罗武中, 彭选明, 李文建, 周利斌, 柏连阳, 赵炳然. M1TDS技术及镉低积累杂交水稻亲本创制与组合选育[J]. 杂交水稻, 2022, 37(1): 1-11. |
| Shao Y, Peng Y, Mao B G, Yu L X, Tang L, Li Y K, Hu Y Y, Zhang D, Yuan Z C, Luo W Z, Peng X M, Li W J, Zhou L B, Bai L Y, Zhao B R. M1TDS technology and breeding of Cd low-accumulating hybrid rice parents and combinations[J]. Hybrid Rice, 2022, 37(1): 1-11. (in Chinese with English abstract) | |
| [13] | 赵连芝, 王浩瀚, 王勇, 李雁民, 甄东升, 颉红梅. 重离子辐照选育春小麦新品种初探[J]. 西北农业学报, 2006(3): 17-19. |
| Zhao L Z, Wang H H, Wang Y, Li Y M, Zhen D S, Xie H M. Preliminary study on breeding new spring wheat varieties by heavy-ion irradiation[J]. Acta Agriculturae Boreali-occidentalis Sinica, 2006(3): 17-19. (in Chinese with English abstract) | |
| [14] | Melsen K, van de Wouw M, Contreras R. Mutation breeding in ornamentals[J]. HortScience, 2021, 56(10): 1154-1165. |
| [15] | Hu W, Li W, Chen J. Recent advances of microbial breeding via heavy-ion mutagenesis at IMP[J]. Letters in Applied Microbiology, 2017, 65(4): 274-280. |
| [16] | 陆栋, 王颖, 刘青芳, 吴鑫, 王菊芳, 马爽, 李文建. 12C离子束辐照对酵母发酵能力的影响[J]. 核技术, 2010, 33(5): 350-353. |
| Lu D, Wang Y, Liu Q F, Wu X, Wang J F, Ma S, Li W J. Effects of 12C ion beam irradiation on yeast fermentation capacity[J]. Nuclear Techniques, 2010, 33(5): 350-353. (in Chinese with English abstract) | |
| [17] | McCallum C M, Comai L, Greene E A, Henikoff S. Targeted screening for induced mutations[J]. Nature Biotechnology, 2000, 18(4): 455-7. |
| [18] | Colbert T, Till B, Tompa R, Reynolds S, Steine M, Yeung A, McCallum C, Comai L, Henikoff S. High-throughput screening for induced point mutations[J]. Plant Physiology, 2001, 126(2): 480-4. |
| [19] | Taheri S, Abdullah T, Jain S, Sahebi M, Azizi P. TILLING, high-resolution melting (HRM), and next-generation sequencing (NGS) techniques in plant mutation breeding[J]. Molecular Breeding, 2017, 37: 1-23. |
| [20] | Till B J, Datta S, Jankowicz-Cieslak J. TILLING: The next generation[J]. Advances in Biochemical Engineering-biotechnology, 2018, 164: 139-160. |
| [21] | Kumar A, McKeown P, Boualem A, Ryder P, Brychkova G, Bendahmane A, Sarkar A, Chatterjee M, Spillane C. TILLING by Sequencing (TbyS) for targeted genome mutagenesis in crops[J]. Molecular Breeding, 2017, 37: 1-12. |
| [22] | Kashtwari M, Wani A, Rather R. TILLING: An alternative path for crop improvement[J]. Journal of Crop Improvement, 2019, 33(1): 83-109. |
| [23] | Dai P, Wu L R, Chen S X, Wang M X, Cheng L Y, Zhang J X, Hao P, Yao W, Zarka J, Issa G C, Kwong L, Zhang D Y. Calibration-free NGS quantitation of mutations below 0.01% VAF[J]. Nature Communications, 2021, 12(1): 6123. |
| [24] | Song P, Chen S X, Yan Y H, Pinto A, Cheng L Y, Dai P, Patel A A, Zhang D Y. Selective multiplexed enrichment for the detection and quantitation of low-fraction DNA variants via low-depth sequencing[J]. Nature Biomedical Engineering, 2021, 5(7): 690-701. |
| [25] | Tang L, Mao B, Li Y, Lü Q, Zhang L, Chen C, He H, Wang W, Zeng X, Shao Y, Pan Y, Hu Y, Peng Y, Fu X, Li H, Xia S, Zhao B. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield[J]. Scientific Reports, 2017, 7(1): 14438. |
| [26] | Hui S, Li H, Mawia AM, Zhou L, Cai J, Ahmad S, Lai C, Wang J, Jiao G, Xie L, Shao G, Sheng Z, Tang S, Wang J, Wei X, Hu S, Hu P. Production of aromatic three-line hybrid rice using novel alleles of BADH2[J]. Plant Biotechnology Journal, 2022, 20(1): 59-74. |
| [27] | Xu H, Wei Y, Zhu Y, Lian L, Xie H, Cai Q, Chen Q, Lin Z, Wang Z, Xie H, Zhang J. Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity[J]. Plant Biotechnology Journal, 2015, 13(4): 526-39. |
| [28] | Lü Y, Shao G, Jiao G, Sheng Z, Xie L, Hu S, Tang S, Wei X, Hu P. Targeted mutagenesis of POLYAMINE OXIDASE 5 that negatively regulates mesocotyl elongation enables the generation of direct-seeding rice with improved grain yield[J]. Molecular Plant, 2021, 14(2): 344-351. |
| [29] | Sun Y, Jiao G, Liu Z, Zhang X, Li J, Guo X, Du W, Du J, Francis F, Zhao Y, Xia L. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes[J]. Frontiers in Plant Science, 2017, 8: 298. |
| [30] | Wang A, Jing Y, Cheng Q, Zhou H, Wang L, Gong W, Kou L, Liu G, Meng X, Chen M, Ma H, Shu X, Yu H, Wu D, Li J. Loss of function of SSIIIa and SSIIIb coordinately confers high RS content in cooked rice[J]. Proceedings of the National Academy of Sciences of the United States America, 2023, 120(19): e2220622120. |
| [31] | Kitamura S, Satoh K, Oono Y. Detection and characterization of genome-wide mutations in M1 vegetative cells of gamma-irradiated Arabidopsis[J]. PLoS Genetics, 2022, 18(1): e1009979. |
| [32] | Sasikala R, Kalaiyarasi R. Sensitivity of rice varieties to gamma irradiation[J]. Electronic Journal of Plant Breeding, 2010, 1(4): 845-889. |
| [33] | Gowthami R, Vanniarajan C, Souframanien J, Arumugam M. Comparison of radiosensitivity of two rice (Oryza sativa L.) varieties to gamma rays and electron beam in M1 generation[J]. Electronic Journal of Plant Breeding, 2017, 8(3): 732-741. |
| [34] | Yamaguchi H. Characteristics of ion beams as mutagens for mutation breeding in rice and chrysanthemums[J]. Japan Agricultural Research Quarterly, 2013, 47(4): 339-346. |
| [1] | 郝雯倩, 蔡兴菁, 杨海东, 吴宇阳, 滕轩, 薛超, 龚志云. 不同类型组蛋白修饰在水稻响应非生物胁迫中的研究进展[J]. 中国水稻科学, 2025, 39(5): 575-685. |
| [2] | 王镜博, 苏畅, 冯晶, 姜思旭, 徐海, 崔志波, 赵明辉. 水稻OsAlR1基因耐铝性功能研究[J]. 中国水稻科学, 2025, 39(5): 615-623. |
| [3] | 杨佳欣, 管玉圣, 杜润, 李贤勇, 蔡座坤, 王楚桃, 阳启样, 何永歆, 朱子超, 张毅. 利用CRISPR/Cas9技术创制无芽鞘紫线的香型环境非敏感隐性核雄性不育种质[J]. 中国水稻科学, 2025, 39(5): 643-649. |
| [4] | 张海鹏, 李莞意, 廖福兴, 马美子, 张洪程, 杨艳菊. 纳米钼对水稻根系形态生理和硝态氮吸收的影响[J]. 中国水稻科学, 2025, 39(5): 650-664. |
| [5] | 刘钰婷, 周星, 何辰延, 李秋萍, 艾小凤, 袁玉洁, 刘睿, 杨景文, 刘婷婷, 王丽, 程红, 黄蓉, 李奥运, 胡文, 胡忠, 任万军, 邓飞. 不同光照条件下减穴稳苗配置对水稻茎鞘干物质积累转运 特性的影响[J]. 中国水稻科学, 2025, 39(5): 665-678. |
| [6] | 杨行洲, 崔苗苗, 魏利辉, 顾爱国, 李东霞, 乐秀虎, 冯辉. 外源miR3979处理水稻对拟禾本科根结线虫趋性、侵染和发育的影响[J]. 中国水稻科学, 2025, 39(5): 703-710. |
| [7] | 朱鹏, 凌溪铁, 王金彦, 张保龙, 杨郁文, 许轲, 裘实. 机直播条件下不同控草方式对抗除草剂水稻产量和品质差异性研究[J]. 中国水稻科学, 2025, 39(4): 501-515. |
| [8] | 董立强, 张义凯, 杨铁鑫, 冯莹莹, 马亮, 梁潇, 张玉屏, 李跃东. 北方粳稻密苗机插育秧对秧苗素质及取秧特性的影响[J]. 中国水稻科学, 2025, 39(4): 516-528. |
| [9] | 周洋, 叶凡, 刘立军. 典型促生微生物提高盐胁迫水稻抗性的研究进展[J]. 中国水稻科学, 2025, 39(4): 529-542. |
| [10] | 朱建平, 李霞, 李文奇, 许扬, 王芳权, 陶亚军, 蒋彦婕, 陈智慧, 范方军, 杨杰. 水稻粉质胚乳突变体we1的表型分析与基因定位[J]. 中国水稻科学, 2025, 39(4): 543-551. |
| [11] | 黄福灯, 吴春艳, 郝媛媛, 韩一飞, 张小斌, 孙会锋, 潘刚. 不同氮肥水平下水稻倒二叶叶鞘的转录组分析[J]. 中国水稻科学, 2025, 39(4): 563-574. |
| [12] | 卢椰子, 邱结华, 蒋楠, 寇艳君, 时焕斌. 稻瘟病菌效应子研究进展[J]. 中国水稻科学, 2025, 39(3): 287-294. |
| [13] | 王超瑞, 周宇琨, 温雅, 张瑛, 法晓彤, 肖治林, 张耗. 秸秆还田方式对稻田土壤特性和温室气体排放的影响及其水肥互作调控[J]. 中国水稻科学, 2025, 39(3): 295-305. |
| [14] | 王雅宣, 王新峰, 杨后红, 刘芳, 肖晶, 蔡玉彪, 魏琪, 傅强, 万品俊. 稻飞虱适应水稻抗性机制的研究进展[J]. 中国水稻科学, 2025, 39(3): 306-321. |
| [15] | 黄涛, 魏兆根, 陈玘, 程泽, 刘欣, 王广达, 胡珂鸣, 谢文亚, 陈宗祥, 冯志明, 左示敏. 水稻类病斑突变体lm52的基因克隆及其广谱抗病性分析[J]. 中国水稻科学, 2025, 39(3): 322-330. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||