
中国水稻科学 ›› 2025, Vol. 39 ›› Issue (4): 543-551.DOI: 10.16819/j.1001-7216.2025.240508
朱建平1,2,3, 李霞1,2,3, 李文奇1,2,3, 许扬1,2,3, 王芳权1,2,3, 陶亚军1,2,3, 蒋彦婕1,2,3, 陈智慧1,2,3, 范方军1,2,3, 杨杰1,2,3,*( )
)
收稿日期:2024-05-15
修回日期:2024-08-20
出版日期:2025-07-10
发布日期:2025-07-21
通讯作者:
*email: yangjie168@aliyun.com基金资助:
ZHU Jianping1,2,3, LI Xia1,2,3, LI Wenqi1,2,3, XU Yang1,2,3, WANG Fangquan1,2,3, TAO Yajun1,2,3, JIANG Yanjie1,2,3, CHEN Zhihui1,2,3, FAN Fangjun1,2,3, YANG Jie1,2,3,*( )
)
Received:2024-05-15
Revised:2024-08-20
Online:2025-07-10
Published:2025-07-21
Contact:
*email: yangjie168@aliyun.com
摘要:
【目的】对粉质胚乳突变体的研究有助于解析稻米品质形成的分子机制。【方法】从日本晴60Co诱变突变体库中筛选到一份粉质皱缩胚乳突变体we1(white endosperm1),对其表型、理化性质进行分析,并利用we1与9311杂交获得的F2群体对目标基因进行精细定位。【结果】we1胚乳表现为白色粉质皱缩状,淀粉颗粒排列疏松且不规则;千粒重、株高、总淀粉、直链淀粉含量显著下降,脂肪含量显著上升。遗传分析表明we1粉质皱缩胚乳性状受单个隐性基因控制。利用we1/9311 F2群体进行基因定位,WE1被定位在6号染色体短臂P16和P18之间约181 kb区间内,该区间包含23个开放阅读框(Open reading frame, ORFs)。qRT-PCR 结果显示,WE1的突变会影响淀粉合成相关基因的表达。【结论】WE1可能是一个参与淀粉合成的新基因,本研究为进一步解析WE1调控水稻淀粉合成的分子机制奠定了基础。
朱建平, 李霞, 李文奇, 许扬, 王芳权, 陶亚军, 蒋彦婕, 陈智慧, 范方军, 杨杰. 水稻粉质胚乳突变体we1的表型分析与基因定位[J]. 中国水稻科学, 2025, 39(4): 543-551.
ZHU Jianping, LI Xia, LI Wenqi, XU Yang, WANG Fangquan, TAO Yajun, JIANG Yanjie, CHEN Zhihui, FAN Fangjun, YANG Jie. Phenotypic Analysis and Gene Mapping of a Floury Endosperm Mutant we1 in Rice[J]. Chinese Journal OF Rice Science, 2025, 39(4): 543-551.
| 引物名称 Primer name | 正向引物序列 Forward primer (5’-3’) | 反向引物序列 Reverse primer(5’-3’) |
|---|---|---|
| P1 | CCCCAACTTTCAGCTTTGCT | GACAAAATCCTCCGTGGCG |
| P5 | AATGCGAAGTTTGGGGGAAC | CAATCAGACGACGAAACGGG |
| P8 | CGTGGACGGTGATGGAATTC | CCTCAGTGACATGCTCAAGG |
| P12 | ACTCAGTAGCAGTTGAGGAGA | TCAAATCCTTGCACATAACCCA |
| P16 | ACTCAGCCGATACTTGAGGG | TTTGGAGTCTATTAGGCTTGAAGC |
| P18 | TGCATGTGTTCACACTGTGA | ACTAATGATCAGCTAGGTGCCT |
表1 WE1精细定位所用引物
Table 1. Primers for fine mapping of WE1
| 引物名称 Primer name | 正向引物序列 Forward primer (5’-3’) | 反向引物序列 Reverse primer(5’-3’) |
|---|---|---|
| P1 | CCCCAACTTTCAGCTTTGCT | GACAAAATCCTCCGTGGCG |
| P5 | AATGCGAAGTTTGGGGGAAC | CAATCAGACGACGAAACGGG |
| P8 | CGTGGACGGTGATGGAATTC | CCTCAGTGACATGCTCAAGG |
| P12 | ACTCAGTAGCAGTTGAGGAGA | TCAAATCCTTGCACATAACCCA |
| P16 | ACTCAGCCGATACTTGAGGG | TTTGGAGTCTATTAGGCTTGAAGC |
| P18 | TGCATGTGTTCACACTGTGA | ACTAATGATCAGCTAGGTGCCT |
| 引物名称 | 正向引物序列 | 反向引物序列 |
|---|---|---|
| Primer name | Forward primer(5’-3’) | Reverse primer(5’-3’) |
| Actin | CATGCTATCCCTCGTCTCGACCT | CGCACTTCATGATGGAGTTGTAT |
| AGPS1 | AGAATGCTCGTATTGGAGAAAATG | GGCAGCATGGAATAAACCAC |
| AGPS2b | AACAATCGAAGCGCGAGAAA | GCCTGTAGTTGGCACCCAGA |
| AGPL1 | GGAAGACGGATGATCGAGAAAG | CACATGAGATGCACCAACGA |
| AGPL2 | AGTTCGATTCAAGACGGATAGC | CGACTTCCACAGGCAGCTTATT |
| AGPL4 | TCAACGTCGATGCAGCAAAT | ATCCCTCAGTTCCTAGCCTCATT |
| GBSSI | TCCGAGAGGTTCAGGTCATC | ATGAGCTCCTCGGCGTAGTA |
| SSI | GGGCCTTCATGGATCAACC | CCGCTTCAAGCATCCTCATC |
| SSIIa | GCTTCCGGTTTGTGTGTTCA | CTTAATACTCCCTCAACTCCACCAT |
| SSIIc | GCAAAGCGGCTATAGAGGTTCC | ATTTCCATCACCGTAGCAGTAGC |
| SSIIIa | GCCTGCCCTGGACTACATTG | GCAAACATATGTACACGGTTCTGG |
| SSIVa | GGGAGCGGCTCAAACATAAA | CCGTGCACTGACTGCAAAAT |
| BEI | TGGCCATGGAAGAGTTGGC | CAGAAGCAACTGCTCCACC |
| BEIIa | AGCGCTGAAGGCATTACCTACC | CTACTAATGCTGCAGACTGTGCTC |
| BEIIb | ATGCTAGAGTTTGACCGC | AGTGTGATGGATCCTGCC |
| ISA1 | TGCTCAGCTACTCCTCCATCATC | AGGACCGCACAACTTCAACATA |
| ISA2 | TAGAGGTCCTCTTGGAGG | AATCAGCTTCTGAGTCACCG |
| ISA3 | ACAGCTTGAGACACTGGGTTGAG | GCATCAAGAGGACAACCATCTG |
表2 实时荧光定量PCR分析所用的引物
Table 2. Primers used in real-time PCR analysis
| 引物名称 | 正向引物序列 | 反向引物序列 |
|---|---|---|
| Primer name | Forward primer(5’-3’) | Reverse primer(5’-3’) |
| Actin | CATGCTATCCCTCGTCTCGACCT | CGCACTTCATGATGGAGTTGTAT |
| AGPS1 | AGAATGCTCGTATTGGAGAAAATG | GGCAGCATGGAATAAACCAC |
| AGPS2b | AACAATCGAAGCGCGAGAAA | GCCTGTAGTTGGCACCCAGA |
| AGPL1 | GGAAGACGGATGATCGAGAAAG | CACATGAGATGCACCAACGA |
| AGPL2 | AGTTCGATTCAAGACGGATAGC | CGACTTCCACAGGCAGCTTATT |
| AGPL4 | TCAACGTCGATGCAGCAAAT | ATCCCTCAGTTCCTAGCCTCATT |
| GBSSI | TCCGAGAGGTTCAGGTCATC | ATGAGCTCCTCGGCGTAGTA |
| SSI | GGGCCTTCATGGATCAACC | CCGCTTCAAGCATCCTCATC |
| SSIIa | GCTTCCGGTTTGTGTGTTCA | CTTAATACTCCCTCAACTCCACCAT |
| SSIIc | GCAAAGCGGCTATAGAGGTTCC | ATTTCCATCACCGTAGCAGTAGC |
| SSIIIa | GCCTGCCCTGGACTACATTG | GCAAACATATGTACACGGTTCTGG |
| SSIVa | GGGAGCGGCTCAAACATAAA | CCGTGCACTGACTGCAAAAT |
| BEI | TGGCCATGGAAGAGTTGGC | CAGAAGCAACTGCTCCACC |
| BEIIa | AGCGCTGAAGGCATTACCTACC | CTACTAATGCTGCAGACTGTGCTC |
| BEIIb | ATGCTAGAGTTTGACCGC | AGTGTGATGGATCCTGCC |
| ISA1 | TGCTCAGCTACTCCTCCATCATC | AGGACCGCACAACTTCAACATA |
| ISA2 | TAGAGGTCCTCTTGGAGG | AATCAGCTTCTGAGTCACCG |
| ISA3 | ACAGCTTGAGACACTGGGTTGAG | GCATCAAGAGGACAACCATCTG |

图1 野生型与突变体we1表性分析 A,B:野生型(A)与突变体we1(B)成熟种子外观表型,标尺为1 mm;C,D:野生型(C)与突变体we1(D)成熟种子横截面,标尺为1 mm。
Fig. 1. Phenotypic analysis of wild type and the we1 mutant A, B, Mature seeds of WT and we1. Bars = 1 mm. C, D, Cross-sections of mature seeds of WT and we1. Bars = 1 mm.
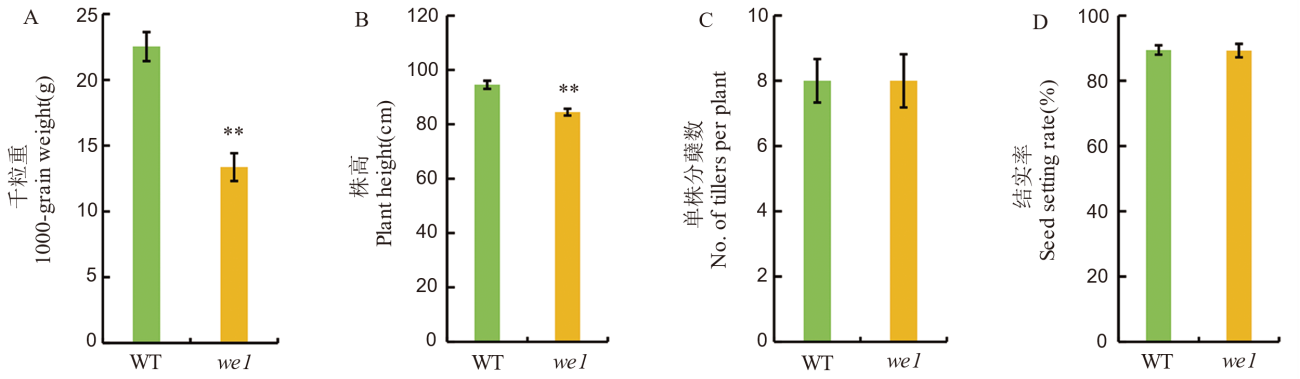
图2 野生型与突变体we1农艺性状比较 数值为平均值±标准差,n=20。采用t检验,**表示在 P<0.01 水平上差异显著。
Fig. 2. Agronomic traits comparison between wild type and the we1 mutant All values are means±SD (n=20). **indicates significant difference at P<0.01 level (Student’s t-test).
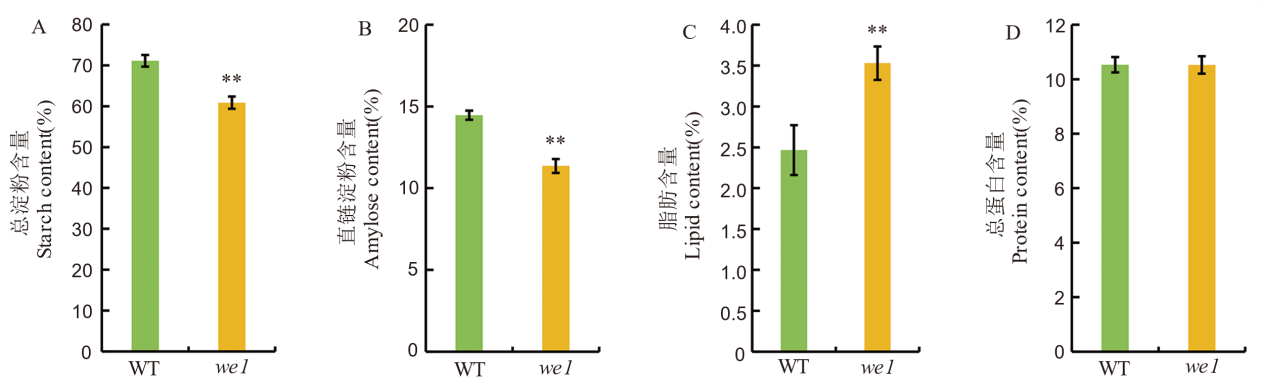
图3 野生型与突变体we1理化性质比较 所有数值为平均值±标准差,n=3。采用t检验,**表示在 P<0.01 水平上差异显著。
Fig. 3. Physicochemical properties comparison between wild type and the we1 mutant All values are means ± SD (n=3). **indicates significant difference at P<0.01 level (Student’s t-test).
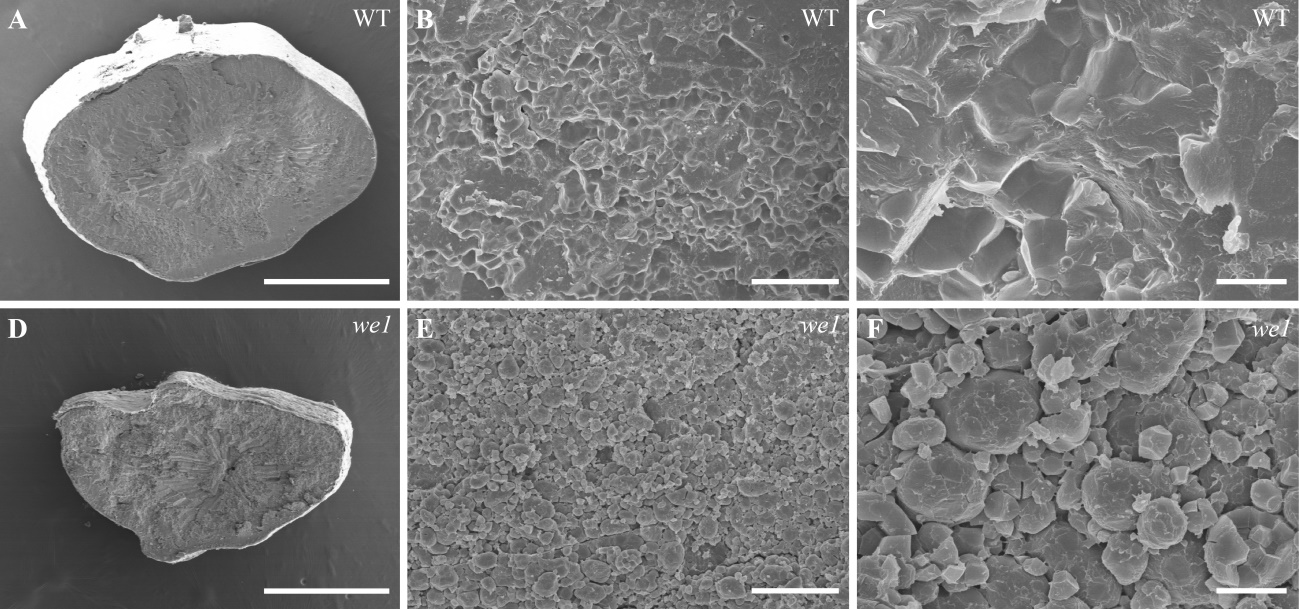
图4 野生型与突变体we1成熟种子扫描电镜观察 WT(A~C)和we1(D~F)成熟种子横切面的 SEM 观察。A,D标尺为1 mm;B,E标尺为50 μm;C,F标尺为10 μm。
Fig. 4. Scanning electron microscopic analysis of mature seeds of WT and we1 Scanning electron microscopic (SEM) analysis of cross-sections of mature seeds of WT (A~C) and we1 (D~F). A, D, Bars = 1 mm; B, E, Bars = 50 μm; C, F, Bars = 10 μm.
| 杂交组合 Cross | 正常透明种子数 No. of wild type seeds | 突变粉质种子数 No. of mutant seeds | χ2(3:1) |
|---|---|---|---|
| we1/野生型F2 F2 of we1/wild type | 132 | 50 | 0.469 |
| 野生型/we1 F2 F2 of wild type/we1 | 151 | 47 | 0.108 |
表3 突变体we1遗传分析
Table 3. Genetic analysis of the we1 mutant
| 杂交组合 Cross | 正常透明种子数 No. of wild type seeds | 突变粉质种子数 No. of mutant seeds | χ2(3:1) |
|---|---|---|---|
| we1/野生型F2 F2 of we1/wild type | 132 | 50 | 0.469 |
| 野生型/we1 F2 F2 of wild type/we1 | 151 | 47 | 0.108 |
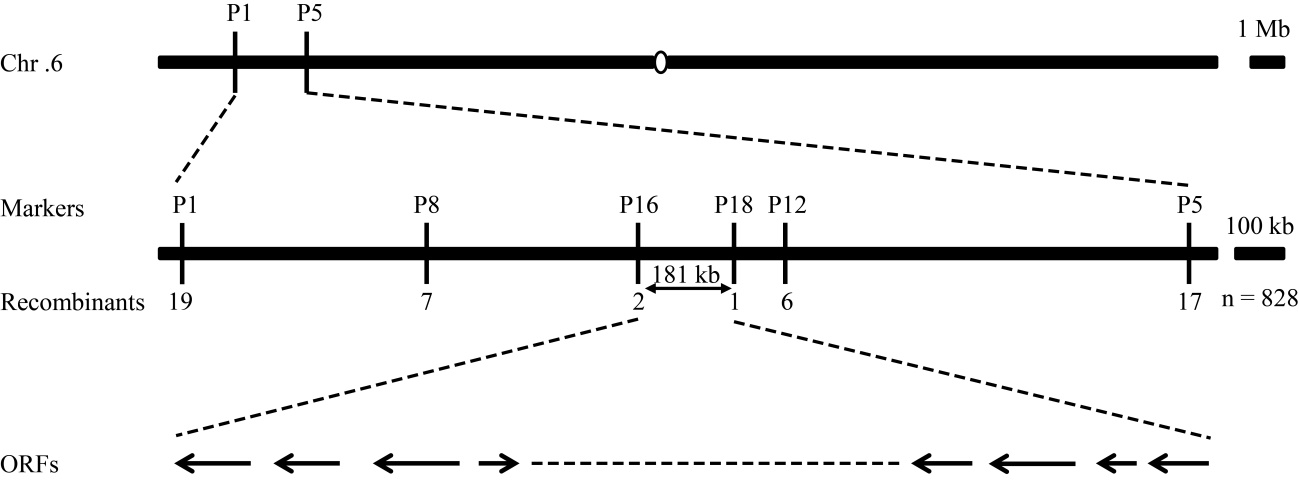
图5 WE1基因的精细定位 WE1定位在6号染色体短臂标记P16和P18之间约181 kb区间内,包含23个预测基因。
Fig. 5. Fine mapping of WE1 gene The WE1 locus was mapped to a 181 kb region between markers P16 and P18 on the long arm of chromosome 12, which contains 23 predicted open reading frames (ORFs).
| 开放阅读框 | 基因登录号 | 功能注释 |
|---|---|---|
| ORFs | Locus ID | Functional description |
| ORF1 | Os06g0165200 | 含有仙茅凝集素结构域的蛋白质Curculin-like lectin domain containing protein |
| ORF2 | Os06g0165300 | 保守假定蛋白Conserved hypothetical protein |
| ORF3 | Os06g0165500 | 含有仙茅凝集素结构域的蛋白质Curculin-like lectin domain containing protein |
| ORF4 | Os06g0165600 | 含有AP2结构域的蛋白质AP2 domain containing protein |
| ORF5 | Os06g0165800 | 咖啡酰辅酶A氧甲基转移酶Caffeoyl-CoA 3-O-methyltransferase |
| ORF6 | Os06g0166000 | 含有F-box结构域的蛋白质Cyclin-like F-box domain containing protein |
| ORF7 | Os06g0166100 | 含有FAR1结构域的蛋白质FAR1 domain containing protein |
| ORF8 | Os06g0166200 | 含有C2H2型锌指结构域的蛋白质Zinc finger, C2H2-type domain containing protein |
| ORF9 | Os06g0166400 | TINY 类蛋白质TINY-like protein |
| ORF10 | Os06g0166500 | 生长素响应蛋白IAA20 Auxin-responsive protein IAA20 |
| ORF11 | Os06g0166900 | 含有蛋白激酶结构域的蛋白质Protein kinase-like domain containing protein |
| ORF12 | Os06g0167000 | PRP8 蛋白质PRP8 protein |
| ORF13 | Os06g0167100 | 含有Armadillo类螺旋结构域的蛋白质Armadillo-like helical domain containing protein |
| ORF14 | Os06g0167200 | RING-H2型锌指蛋白RING-H2 finger protein ATL1R |
| ORF15 | Os06g0167400 | DTC家族蛋白Di-trans-poly-cis-decaprenylcistransferase family protein |
| ORF16 | Os06g0167500 | 含有LRR结构域的蛋白质Leucine-rich repeat, plant specific containing protein |
| ORF17 | Os06g0167600 | 蛋白酶体亚基-3 Proteasome subunit alpha-3 |
| ORF18 | Os06g0168000 | 谷胱甘肽硫转移酶Glutathione S-transferase, C-terminal-like domain containing protein |
| ORF19 | Os06g0168400 | 保守假定蛋白Conserved hypothetical protein |
| ORF20 | Os06g0168500 | Syntaxin类蛋白 Syntaxin-like protein |
| ORF21 | Os06g0168600 | 核糖核苷酸还原酶Ribonucleotide reductase |
| ORF22 | Os06g0168700 | 富含脯氨酸蛋白Prolin rich protein |
| ORF23 | Os06g0168800 | 蛋白激酶Protein kinase |
表4 WE1候选基因分析
Table 4. Candidate genes for WE1
| 开放阅读框 | 基因登录号 | 功能注释 |
|---|---|---|
| ORFs | Locus ID | Functional description |
| ORF1 | Os06g0165200 | 含有仙茅凝集素结构域的蛋白质Curculin-like lectin domain containing protein |
| ORF2 | Os06g0165300 | 保守假定蛋白Conserved hypothetical protein |
| ORF3 | Os06g0165500 | 含有仙茅凝集素结构域的蛋白质Curculin-like lectin domain containing protein |
| ORF4 | Os06g0165600 | 含有AP2结构域的蛋白质AP2 domain containing protein |
| ORF5 | Os06g0165800 | 咖啡酰辅酶A氧甲基转移酶Caffeoyl-CoA 3-O-methyltransferase |
| ORF6 | Os06g0166000 | 含有F-box结构域的蛋白质Cyclin-like F-box domain containing protein |
| ORF7 | Os06g0166100 | 含有FAR1结构域的蛋白质FAR1 domain containing protein |
| ORF8 | Os06g0166200 | 含有C2H2型锌指结构域的蛋白质Zinc finger, C2H2-type domain containing protein |
| ORF9 | Os06g0166400 | TINY 类蛋白质TINY-like protein |
| ORF10 | Os06g0166500 | 生长素响应蛋白IAA20 Auxin-responsive protein IAA20 |
| ORF11 | Os06g0166900 | 含有蛋白激酶结构域的蛋白质Protein kinase-like domain containing protein |
| ORF12 | Os06g0167000 | PRP8 蛋白质PRP8 protein |
| ORF13 | Os06g0167100 | 含有Armadillo类螺旋结构域的蛋白质Armadillo-like helical domain containing protein |
| ORF14 | Os06g0167200 | RING-H2型锌指蛋白RING-H2 finger protein ATL1R |
| ORF15 | Os06g0167400 | DTC家族蛋白Di-trans-poly-cis-decaprenylcistransferase family protein |
| ORF16 | Os06g0167500 | 含有LRR结构域的蛋白质Leucine-rich repeat, plant specific containing protein |
| ORF17 | Os06g0167600 | 蛋白酶体亚基-3 Proteasome subunit alpha-3 |
| ORF18 | Os06g0168000 | 谷胱甘肽硫转移酶Glutathione S-transferase, C-terminal-like domain containing protein |
| ORF19 | Os06g0168400 | 保守假定蛋白Conserved hypothetical protein |
| ORF20 | Os06g0168500 | Syntaxin类蛋白 Syntaxin-like protein |
| ORF21 | Os06g0168600 | 核糖核苷酸还原酶Ribonucleotide reductase |
| ORF22 | Os06g0168700 | 富含脯氨酸蛋白Prolin rich protein |
| ORF23 | Os06g0168800 | 蛋白激酶Protein kinase |
图6 WE1候选基因表达分析 开花后12 d胚乳的WE1候选基因表达分析(qRT-PCR),以Actin作为内参。所有数值为平均值±标准差(n = 3)。
Fig. 6. Expression of WE1 candidate genes qRT-PCR assay of the expression of WE1 candidate genes in 12 DAF endosperm. Actin was used as control. All values are means ± SD(n = 3).
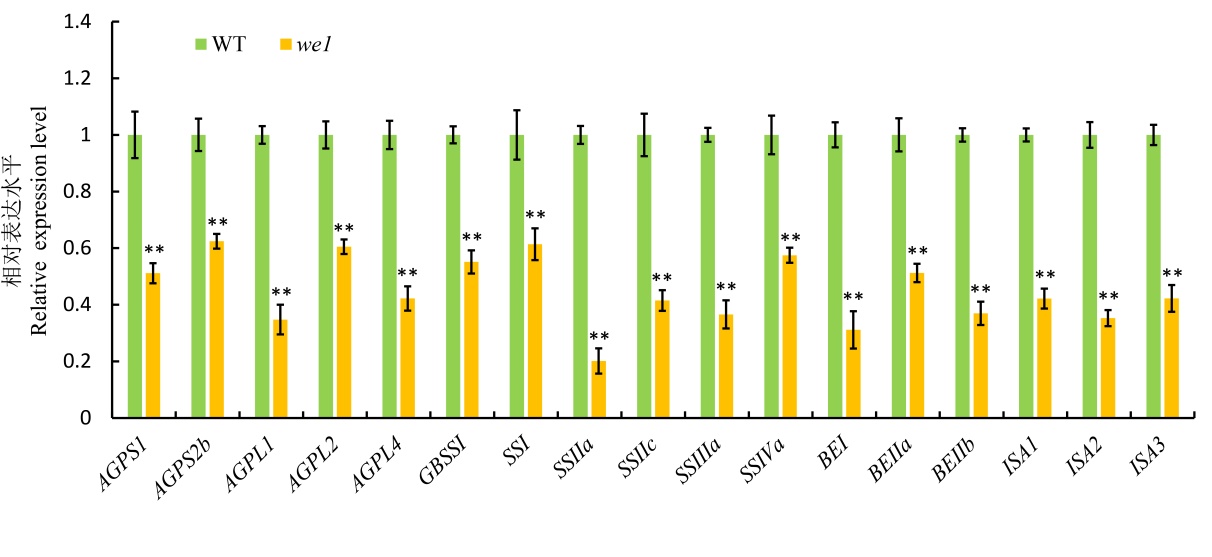
图7 淀粉合成相关基因表达分析 开花后12 d胚乳的淀粉合成相关基因表达分析(qRT-PCR),以Actin作为内参。所有数值为平均值±标准差(n = 3)。 **P<0.01 (t检验)。
Fig. 7. Expression analysis of representative genes coding starch synthesis qRT-PCR assay of the expression of representative genes coding starch synthesis in 12 DAF endosperm. Actin was used as control. All values are means ± SD(n=3). **P<0.01(Student’s t-test).
| [1] | Smith A M, Zeeman S C. Starch: A flexible, adaptable carbon store coupled to plant growth[J]. Annual Review of Plant Biology, 2020, 71: 217-245. |
| [2] | 朱霁晖, 张昌泉, 顾铭洪, 刘巧泉. 水稻Wx基因的等位变异及育种利用研究进展[J]. 中国水稻科学, 2015, 29(4): 431-438. |
| Zhu J H, Zhang C Q, Gu M H, Liu Q Q. Progress in the allelic variation of Wx gene and its application in rice breeding[J]. Chinese Journal of Rice Science, 2015, 29(4): 431-438. (in Chinese with English abstract) | |
| [3] | Nakamura Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue[J]. Plant and Cell Physiology, 2002, 43(7): 718-725. |
| [4] | Hirose T, Terao T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.)[J]. Planta, 2004, 220(1): 9-16. |
| [5] | Ohdan T, Francisco P B Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice[J]. Journal of Experimental Botany, 2005, 56(422): 3229-3244. |
| [6] | Utsumi Y, Utsumi C, Sawada T, Fujita N, Nakamura Y. Functional diversity of isoamylase oligomers: The ISA1 Homo-oligomer is essential for amylopectin biosynthesis in rice endosperm[J]. Plant Physiology, 2011, 156(1): 61-77. |
| [7] | Li P, Chen Y H, Lu J, Zhang C Q, Liu Q Q, Li Q F. Genes and their molecular functions determining seed structure, components, and quality of rice[J]. Rice, 2022, 15(1): 18. |
| [8] | 张习春, 鲁菲菲, 吕育松, 罗荣剑, 焦桂爱, 邬亚文, 唐绍清, 胡培松, 魏祥进. 两个垩白突变体的鉴定及突变基因的图位克隆[J]. 中国水稻科学, 2017, 31(6): 568-579. |
| Zhang D C, Lu F F, Lü Y S, Luo R J, Jiao G A, Wu Y W, Tang S Q, Hu P S, Wei X J. Identification and gene mapping-based clone of two chalkiness mutants in rice[J]. Chinese Journal of Rice Science, 2017, 31(6): 568-579. (in Chinese with English abstract) | |
| [9] | 杜溢墨, 潘天, 田云录, 刘世家, 刘喜, 江玲, 张文伟, 王益华, 万建民. 水稻粉质皱缩胚乳突变体fse4的表型分析与基因克隆[J]. 中国水稻科学2019, 33(6): 499-512. |
| Du Y M, Pan T, Tian Y L, Liu S J, Liu X, Jiang L, Zhang W W, Wang Y H, Wan J M. Phenotypic analysis and gene cloning of rice floury endosperm mutant fse4[J]. Chinese Journal of Rice Science, 2019, 33(6): 499-512. (in Chinese with English abstract | |
| [10] | 唐小涵, 刘世家, 刘喜, 田云录, 王云龙, 滕烜, 段二超, 张元燕, 江玲, 张文伟, 王益华, 万建民. 色氨酰-tRNA合成酶基因WRS1调控水稻种子发育[J]. 中国水稻科学, 2020, 34(5): 383-396. |
| Tang X H, Liu S J, Liu X, Tian Y L, Wang Y L, Teng X, Duan E C, Zhang Y Y, Jiang L, Zhang W W, Wang Y H, Wan J M. Tryptophanyl-tRNA synthetase gene WRS1 regulates rice seed development[J]. Chinese Journal of Rice Science, 2020, 34(5): 383-396. (in Chinese with English abstract) | |
| [11] | She K C, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M, Tsuge T, Matsumoto K, Kudoh M, Itoh E, Kikuchi S, Kishimoto N, Yazaki J, Ando T, Yano M, Aoyama T, Sasaki T, Satoh H, Shimada H. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality[J]. The Plant Cell, 2010, 22(10): 3280-3294. |
| [12] | Kang H G, Park S, Matsuoka M, An G H. White-core endosperm in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB)[J]. Plant Journal, 2005, 42(6): 901-911. |
| [13] | Ryoo N, Yu C, Park C S, Baik M Y, Park I M, Cho M H, Bhoo S H, An G, Hahn T R, Jeon J S. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice[J]. Plant Cell Reports, 2007, 26(7): 1083-1095. |
| [14] | Peng C, Wang Y H, Liu F, Ren Y L, Zhou K N, Lü J, Zheng M, Zhao S L, Zhang L, Wang C M, Jiang L, Zhang X, Guo X P, Bao Y Q, Wan J M. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm[J]. Plant Journal, 2014, 77(6): 917-930. |
| [15] | Zhang L, Ren Y L, Lu B Y, Yang C Y, Feng Z M, Liu Z, Chen J, Ma W W, Wang Y, Yu X W, Wang Y L, Zhang W W, Wang Y H, Liu S J, Wu F Q, Zhang X, Guo X P, Bao Y Q, Jiang L, Wan J M. FLOURY ENDOSPERM7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice[J]. Journal of Experimental Botany, 2016, 67(3): 633-647. |
| [16] | Yan H G, Zhang W W, Wang Y H, Jin J, Xu H C, Fu Y S, Shan Z Z, Wang X, Teng X, Li X, Wang Y X, Hu X Q, Zhang W X, Zhu C Y, Zhang X, Zhang Y, Wang R Q, Zhang J, Cai Y, You X M, Chen J, Ge X Y, Wang L, Xu J H, Jiang L, Liu S J, Lei C L, Zhang X, Wang H Y, Ren Y L, Wan J M. Rice LIKE EARLY STARVATION1 cooperates with FLOURY ENDOSPERM 6 to modulate starch biosynthesis and endosperm development[J]. The Plant Cell, 2024, 36(5): 1892-1912. |
| [17] | You X M, Zhang W W, Hu J L, Jing R N, Cai Y, Feng Z M, Kong F, Zhang J, Yan H G, Chen W W, Chen X G, Ma J, Tang X J, Wang P, Zhu S S, Liu L L, Jiang L, Wan J M. FLOURY ENDOSPERM15 encodes a glyoxalase I involved in compound granule formation and starch synthesis in rice endosperm[J]. Plant Cell Reports, 2019, 38(3): 345-359. |
| [18] | Teng X, Zhong M S, Zhu X P, Wang C M, Ren Y L, Wang Y L, Zhang H, Jiang L, Wang D, Hao Y Y, Wu M M, Zhu J P, Zhang X, Guo X P, Wang Y H, Wan J M. FLOURY ENDOSPERM16 encoding a NAD-dependent cytosolic malate dehydrogenase plays an important role in starch synthesis and seed development in rice[J]. Plant Biotechnology Journal, 2019, 17(10): 1914-1927. |
| [19] | Lei J, Teng X, Wang Y F, Jiang X K, Zhao H H, Zheng X M, Ren Y L, Dong H, Wang Y L, Duan E C, Zhang Y Y, Zhang W W, Yang H, Chen X L, Chen R B, Zhang Y, Yu M Z, Xu S B, Bao X H, Zhang P C, Liu S J, Liu X, Tian Y L, Jiang L, Wang Y H, Wan J M. Plastidic pyruvate dehydrogenase complex E1 component subunit Alpha1 is involved in galactolipid biosynthesis required for amyloplast development in rice[J]. Plant Biotechnology Journal, 2022, 20(3): 437-453. |
| [20] | Wei X J, Jiao G A, Lin H Y, Sheng Z H, Shao G N, Xie L H, Tang S Q, Xu Q G, Hu P S. GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development[J]. Journal of Integrative Plant Biology, 2017, 59(2): 134-153. |
| [21] | 方鹏飞, 李三峰, 焦桂爱, 谢黎虹, 胡培松, 魏祥进, 唐绍清. 水稻粉质胚乳突变体flo7的理化性质及基因定位[J]. 中国水稻科学, 2014, 28(5): 447-457. |
| Fang P F, Li S F, Jiao G A, Xie L H, Hu P S, Wei X J, Tang S Q. Physicochemical property analysis and gene mapping of a floury endosperm mutant flo7 in rice[J]. Chinese Journal of Rice Science, 2014, 28(5): 447-457. (in Chinese with English abstract) | |
| [22] | 李景芳, 田云录, 刘喜, 刘世家, 陈亮明, 江玲, 张文伟, 徐大勇, 王益华, 万建民. 鸟苷酸激酶 OsGK1 对水稻种子发育至关重要[J]. 中国水稻科学, 2018, 32(5): 415-426. |
| Li J F, Tian Y L, Liu X, Liu S J, Chen L M, Jiang L, Zhang W W, Xu D Y, Wang Y H, Wan J M. The guanylate kinase OsGK1 is essential for seed development in rice[J]. Chinese Journal of Rice Science, 2018, 32(5): 415-426. (in Chinese with English abstract) | |
| [23] | 于艳芳, 刘喜, 田云录, 刘世家, 陈亮明, 朱建平, 王云龙, 江玲, 张文伟, 王益华, 万建民. 水稻粉质胚乳fse3突变体的表型分析及基因定位[J]. 中国农业科学, 2018, 51(11): 2023-2037. |
| Yu Y F, Liu X, Tian Y L, Liu S J, Chen L M, Zhu J P, Wang Y L, Jiang L, Zhang W W, Wang Y H, Wan J M. Phenotypic analysis and gene mapping of a floury and shrunken endosperm mutant fse3 in rice[J]. Scientia Agricultura Sinica, 2018, 51(11): 2023-2037. (in Chinese with English abstract) | |
| [24] | 潘鹏屹, 朱建平, 王云龙, 郝媛媛, 蔡跃, 张文伟, 江玲, 王益华, 万建民. 水稻粉质胚乳突变体ws 的表型分析及基因克隆[J]. 中国水稻科学, 2016, 30(5): 447-457. |
| Pan P Y, Zhu J P, Wang Y L, Hao Y Y, Cai Y, Zhang W W, Jiang L, Wang Y H, Wan J M. Phenotyping and gene cloning of a floury endosperm mutant ws in rice[J]. Chinese Journal of Rice Science, 2016, 30(5): 447-457. (in Chinese with English abstract) | |
| [25] | Hu T T, Tian Y L, Zhu J P, Wang Y L, Jing R N, Lei J, Sun Y L, Yu Y F, Li J F, Chen X L, Zhu X P, Hao Y Y, Liu L L, Wang Y H, Wan J M. OsNDUFA9 encoding a mitochondrial complex I subunit is essential for embryo development and starch synthesis in rice[J]. Plant Cell Reports, 2018, 37(12): 1667-1679. |
| [26] | Wang J C, Xu H, Zhu Y, Liu Q Q, Cai X L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm[J]. Journal of Experimental Botany, 2013, 64(11): 3453-3466. |
| [27] | Fu F F, Xue H W. Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator[J]. Plant Physiology, 2010, 154(2): 927-938. |
| [28] | Xiong Y F, Ren Y, Li W, Wu F S, Yang W J, Huang X L, Yao J L. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice[J]. Journal of Experimental Botany, 2019, 70(15): 3765-3780. |
| [29] | Bello B K, Hou Y X, Zhao J, Jiao G A, Wu Y W, Li Z Y, Wang Y F, Tong X H, Wang W, Yuan W Y, Wei X J, Zhang J. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.)[J]. Plant Biotechnology Journal, 2019, 17(7): 1222-1235. |
| [30] | Wang J, Chen Z C, Zhang Q, Meng S S, Wei C X. The NAC Transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis[J]. Plant Physiology, 2020, 184(4): 1775-1791. |
| [31] | Wu M W, Liu J X, Bai X, Chen W Q, Ren Y L, Liu J L, Chen M M, Zhao H, Yao X F, Zhang J D, Wan J M, Liu C M. Transcription factors NAC20 and NAC26 interact with RPBF to activate albumin accumulations in rice endosperm[J]. Plant Biotechnology Journal, 2023, 21(5): 890-892. |
| [1] | 朱鹏, 凌溪铁, 王金彦, 张保龙, 杨郁文, 许轲, 裘实. 机直播条件下不同控草方式对抗除草剂水稻产量和品质差异性研究[J]. 中国水稻科学, 2025, 39(4): 501-515. |
| [2] | 董立强, 张义凯, 杨铁鑫, 冯莹莹, 马亮, 梁潇, 张玉屏, 李跃东. 北方粳稻密苗机插育秧对秧苗素质及取秧特性的影响[J]. 中国水稻科学, 2025, 39(4): 516-528. |
| [3] | 周洋, 叶凡, 刘立军. 典型促生微生物提高盐胁迫水稻抗性的研究进展[J]. 中国水稻科学, 2025, 39(4): 529-542. |
| [4] | 黄福灯, 吴春艳, 郝媛媛, 韩一飞, 张小斌, 孙会锋, 潘刚. 不同氮肥水平下水稻倒二叶叶鞘的转录组分析[J]. 中国水稻科学, 2025, 39(4): 563-574. |
| [5] | 卢椰子, 邱结华, 蒋楠, 寇艳君, 时焕斌. 稻瘟病菌效应子研究进展[J]. 中国水稻科学, 2025, 39(3): 287-294. |
| [6] | 王超瑞, 周宇琨, 温雅, 张瑛, 法晓彤, 肖治林, 张耗. 秸秆还田方式对稻田土壤特性和温室气体排放的影响及其水肥互作调控[J]. 中国水稻科学, 2025, 39(3): 295-305. |
| [7] | 王雅宣, 王新峰, 杨后红, 刘芳, 肖晶, 蔡玉彪, 魏琪, 傅强, 万品俊. 稻飞虱适应水稻抗性机制的研究进展[J]. 中国水稻科学, 2025, 39(3): 306-321. |
| [8] | 黄涛, 魏兆根, 陈玘, 程泽, 刘欣, 王广达, 胡珂鸣, 谢文亚, 陈宗祥, 冯志明, 左示敏. 水稻类病斑突变体lm52的基因克隆及其广谱抗病性分析[J]. 中国水稻科学, 2025, 39(3): 322-330. |
| [9] | 马顺婷, 胡运高, 高方远, 刘利平, 牟昌铃, 吕建群, 苏相文, 刘松, 梁毓玉, 任光俊, 郭鸿鸣. 水稻真核翻译起始因子OseIF6.2调控粒型的功能研究[J]. 中国水稻科学, 2025, 39(3): 331-342. |
| [10] | 张彬涛, 刘聪聪, 郭明亮, 杨绍华, 吴世强, 郭龙彪, 朱义旺. 水稻OsDR8基因的稻瘟病抗性评价及优异单倍型鉴定[J]. 中国水稻科学, 2025, 39(3): 343-351. |
| [11] | 韦新宇, 曾跃辉, 肖长春, 黄建鸿, 阮宏椿, 杨旺兴, 邹文广, 许旭明. 水稻康丰B抗稻瘟病基因Pi-kf2(t)的克隆与功能验证[J]. 中国水稻科学, 2025, 39(3): 352-364. |
| [12] | 李文奇, 许扬, 王芳权, 朱建平, 陶亚军, 李霞, 范方军, 蒋彦婕, 陈智慧, 杨杰. 广谱抗稻瘟病基因PigmR的KASP标记开发及应用[J]. 中国水稻科学, 2025, 39(3): 365-372. |
| [13] | 韦还和, 汪璐璐, 马唯一, 张翔, 左博源, 耿孝宇, 朱旺, 朱济邹, 孟天瑶, 陈英龙, 高平磊, 许轲, 戴其根. 盐−旱复合胁迫下粳稻品种南粳9108籽粒灌浆特性及其与产量形成的关系[J]. 中国水稻科学, 2025, 39(3): 373-386. |
| [14] | 沈智达, 余秋华, 张斌, 曹玉东, 王少华, 王红飞, 伍永清, 戴志刚, 李小坤. 磷肥施用量对湖北省直播水稻产量、磷素积累及利用率的影响[J]. 中国水稻科学, 2025, 39(3): 399-411. |
| [15] | 何勇, 张诗骞, 王志成, 詹逍康, 丁一可, 刘晓瑞, 马素素, 田志宏. 印度梨形孢与复合肥组合施用对水稻机插秧秧苗素质的影响[J]. 中国水稻科学, 2025, 39(3): 412-422. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||