
中国水稻科学 ›› 2025, Vol. 39 ›› Issue (4): 552-562.DOI: 10.16819/j.1001-7216.2025.240713
陈嘉乐1,5, 于清涛2, 郑琛凡1,5, 汪庆3, 谭瑗瑗4, 陈百翠2, 李承欣2, 蒋萌1,5, 舒庆尧1,5,*( )
)
收稿日期:2024-07-17
修回日期:2024-08-17
出版日期:2025-07-10
发布日期:2025-07-21
通讯作者:
*email: qyshu@zju.edu.cn基金资助:
CHEN Jiale1,5, YU Qingtao2, ZHENG Chenfan1,5, WANG Qing3, TAN Yuanyuan4, CHEN Baicui2, LI Chengxin2, JIANG Meng1,5, SHU Qingyao1,5,*( )
)
Received:2024-07-17
Revised:2024-08-17
Online:2025-07-10
Published:2025-07-21
Contact:
*email: qyshu@zju.edu.cn
摘要:
【目的】水稻谷粒宽度是粒型和粒重的重要决定因子,可影响水稻产量和稻米外观品质。迄今已发现大量基因突变可影响粒宽,但因突变存在其他显著负效应,真正具有育种价值的基因不多,挖掘新的粒宽基因特别是对品质和产量没有负效应的自然变异具有重要意义。【方法】通过小粒型籼稻(DR610)和大粒型粳稻(哈粳稻7号,HG7)的F2群体极端粒重个体的QTL-seq和已知影响粒型基因的联合分析,发现DR610的OsNF-YC10存在一个无义突变,这可能是其谷粒比HG7窄的原因。为探明OsNF-YC10变异与水稻粒宽的关系,我们对4000余份水稻种质中该基因的自然变异类型及其与粒宽的相关性进行分析。【结果】OsNF-YC10存在10种主要的单倍型,其中,以HG7为代表的单倍型(Hap1)为粳稻中的高频单倍型(1464/1509);以DR610为代表的单倍型(Hap5)在第1048位核苷酸发生了一个C(HapC)→T(HapT)的无义突变,HapC型品种的粒宽(3.036 mm,n=1596)显著大于HapT型品种的粒宽(2.938 mm,n=309),表明该基因的自然变异显著影响水稻粒宽。进一步分析发现,Hap1和Hap5单倍型存在明显的籼粳分化,后者主要分布在低纬度地区。最后,利用四引物ARMS-PCR,开发了一个区分C→T变异的分子标记,可用于粒型的分子标记辅助选择。【结论】本研究确定了OsNF-YC10的自然变异及其单倍型与粒宽的相关性,为粒宽的设计育种提供了重要参考。
陈嘉乐, 于清涛, 郑琛凡, 汪庆, 谭瑗瑗, 陈百翠, 李承欣, 蒋萌, 舒庆尧. 水稻OsNF-YC10自然变异及其与谷粒宽度的相关性[J]. 中国水稻科学, 2025, 39(4): 552-562.
CHEN Jiale, YU Qingtao, ZHENG Chenfan, WANG Qing, TAN Yuanyuan, CHEN Baicui, LI Chengxin, JIANG Meng, SHU Qingyao. Natural Variation of OsNF-YC10 and Its Correlation with Grain Width in Rice[J]. Chinese Journal OF Rice Science, 2025, 39(4): 552-562.
| 引物类型 Type of primer | 引物序列(5’ to 3’) Primer sequence (5’ to 3’) | 等位基因 Allele | 产物长度 Size (bp) |
|---|---|---|---|
| 扩增OsNF-YC10全长部分 | |||
| Forward primer | CTGTGGGACTGTAAGATCATCTATG | 1466 | |
| Reverse primer | CTTGTTGCCATTACTGGTGCTTGG | ||
| Tetra-primer ARMS-PCR | |||
| Forward outer primer | TAGCCCTGCGGTTCTTGCAAGTATGA | ||
| Reverse outer primer | AATCCTGGTGTCCGAAACATCATCCT | ||
| Forward inner primer | TGCTGCTACAAAGGTTTTcAGTGcCt | T | 241 |
| Reverse inner primer | GTTTCTTTCTCATGTTGTTCTTCaTg | C | 361 |
表1 用于本研究的PCR引物
Table 1. PCR primers used in the present study
| 引物类型 Type of primer | 引物序列(5’ to 3’) Primer sequence (5’ to 3’) | 等位基因 Allele | 产物长度 Size (bp) |
|---|---|---|---|
| 扩增OsNF-YC10全长部分 | |||
| Forward primer | CTGTGGGACTGTAAGATCATCTATG | 1466 | |
| Reverse primer | CTTGTTGCCATTACTGGTGCTTGG | ||
| Tetra-primer ARMS-PCR | |||
| Forward outer primer | TAGCCCTGCGGTTCTTGCAAGTATGA | ||
| Reverse outer primer | AATCCTGGTGTCCGAAACATCATCCT | ||
| Forward inner primer | TGCTGCTACAAAGGTTTTcAGTGcCt | T | 241 |
| Reverse inner primer | GTTTCTTTCTCATGTTGTTCTTCaTg | C | 361 |
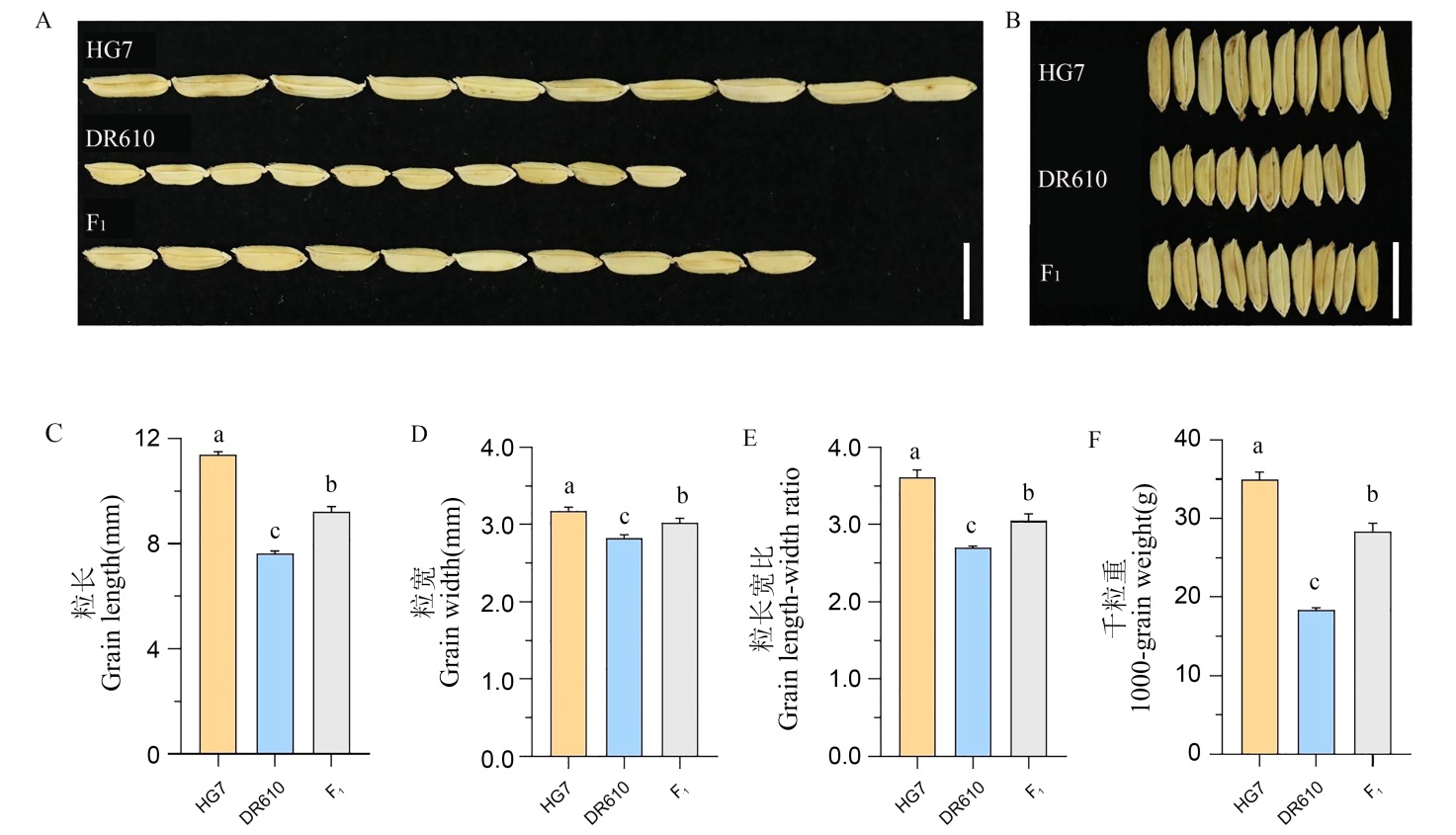
图1 HG7、DR610及其杂种 F1籽粒性状 A和B: 籽粒外观,比例尺为1 cm; C~F: 粒长、粒宽、长宽比和千粒重(均数±标准差,n=3),采用单因素方差和Tukey’s多重比较检验分析,不同字母表示P < 0.05。
Fig. 1. Grain traits of HG7, DR610 and their F1 A and B, Grain phenotype, bar = 1 cm; C-F, Grain length, grain width, grain length-to-width ratio, and 1000-grain weight. Values are mean ± SD (n = 3). Different letters indicate significant differences at P < 0.05(analyzed by one-way ANOVA followed by Tukey’s multiple comparison test).
| 样品名称 Sample | 总读序数 Total reads | 比对率 Alignment (%) | 基因组覆盖率 Genome coverage (%) | 测序深度 Average depth (×) |
|---|---|---|---|---|
| HG7 | 85 448 290 | 90.70 | 97.47 | 30 |
| DR610 | 113 535 356 | 98.54 | 92.73 | 43 |
| LGMP | 99 534 914 | 96.99 | 99.00 | 37 |
表2 DR610、HG7及其F2大粒混池的测序数据及统计
Table 2. Statistics of whole-genome sequencing results of DR610, HG7 and their F2 large grain mixing pool (LGMP)
| 样品名称 Sample | 总读序数 Total reads | 比对率 Alignment (%) | 基因组覆盖率 Genome coverage (%) | 测序深度 Average depth (×) |
|---|---|---|---|---|
| HG7 | 85 448 290 | 90.70 | 97.47 | 30 |
| DR610 | 113 535 356 | 98.54 | 92.73 | 43 |
| LGMP | 99 534 914 | 96.99 | 99.00 | 37 |
| 染色体 Chr | 开始 Start | 结束 End | 大小 Size (Mb) | 基因数目 Number of genes |
|---|---|---|---|---|
| 1 | 12 931 694 | 13 481 021 | 0.55 | 28 |
| 1 | 13 645 077 | 13 688 772 | 0.04 | 0 |
| 1 | 13 705 131 | 13 820 507 | 0.12 | 8 |
| 3 | 23 567 245 | 24 695 746 | 1.13 | 86 |
| Total | - | - | 1.84 | 122 |
表3 DR610×HG7 F2大粒混池QTL-seq确定的4个粒重QTL基因组区间及其包含的注释基因数
Table 3. Genomic regions and number of annotated genes, in which four QTLs of grain weight were mapped using QTL-seq of large grain F2 plants of DR610×HG7
| 染色体 Chr | 开始 Start | 结束 End | 大小 Size (Mb) | 基因数目 Number of genes |
|---|---|---|---|---|
| 1 | 12 931 694 | 13 481 021 | 0.55 | 28 |
| 1 | 13 645 077 | 13 688 772 | 0.04 | 0 |
| 1 | 13 705 131 | 13 820 507 | 0.12 | 8 |
| 3 | 23 567 245 | 24 695 746 | 1.13 | 86 |
| Total | - | - | 1.84 | 122 |
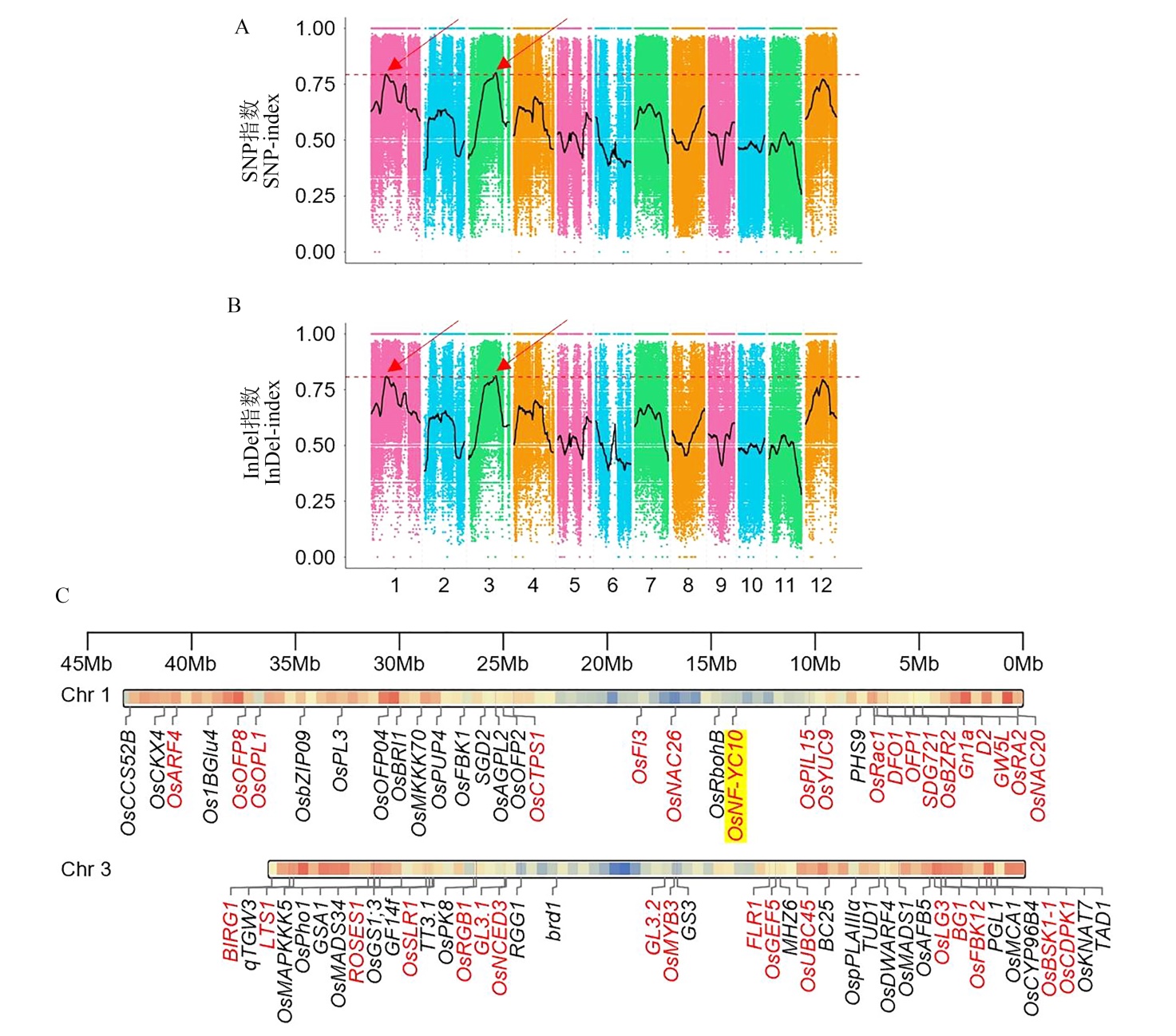
图3 水稻粒重QTL-seq与候选基因鉴定 QTL-seq分析的SNP指数(A)和InDel指数图(B)。红色箭头代表候选区域。(C)为1、3号染色体上已报道影响粒型的注释基因,红色表示的是双亲在CDS上存在差异的基因,黄色背景基因为粒重QTL所在区域与1号染色体上已知粒型基因存在交集的基因。
Fig. 3. QTL-seq for rice grain QTL mapping and candidate gene identification The SNP-index plot (A) and InDel-index plot (B) revealed by QTL-seq, the red arrows indicate the candidate regions of grain QTLs. (C) The reported grain shape genes located on chromosomes 1 and 3. Genes highlighted in red indicated coding sequence(CDS) variation between DR610 and HG7. The gene (OsNF-YC10) marked with yellow background coincided with the QTL regions on panels A and B.
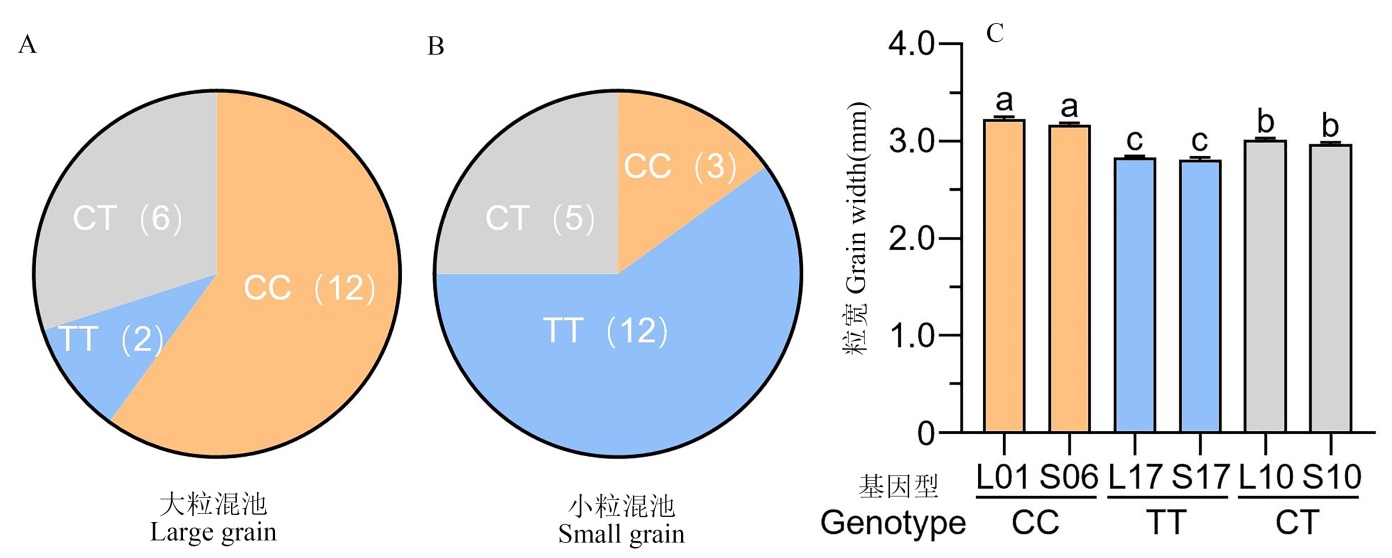
图4 DR610×HG7 的F2大粒(A)和小粒(B)单株OsNF-YC10第13789514位SNP的基因型及其单株数,以及不同OsNF-YC10基因型单株的粒宽(C)
Fig. 4. Number of plants with different OsNF-YC10 genotypes (based on the 13789514 SNP) having extreme large (A) and extreme small grains (B) in the DR610×HG7 F2 population, and grain width of F2 plants with different OsNF-YC10 genotypes (C)
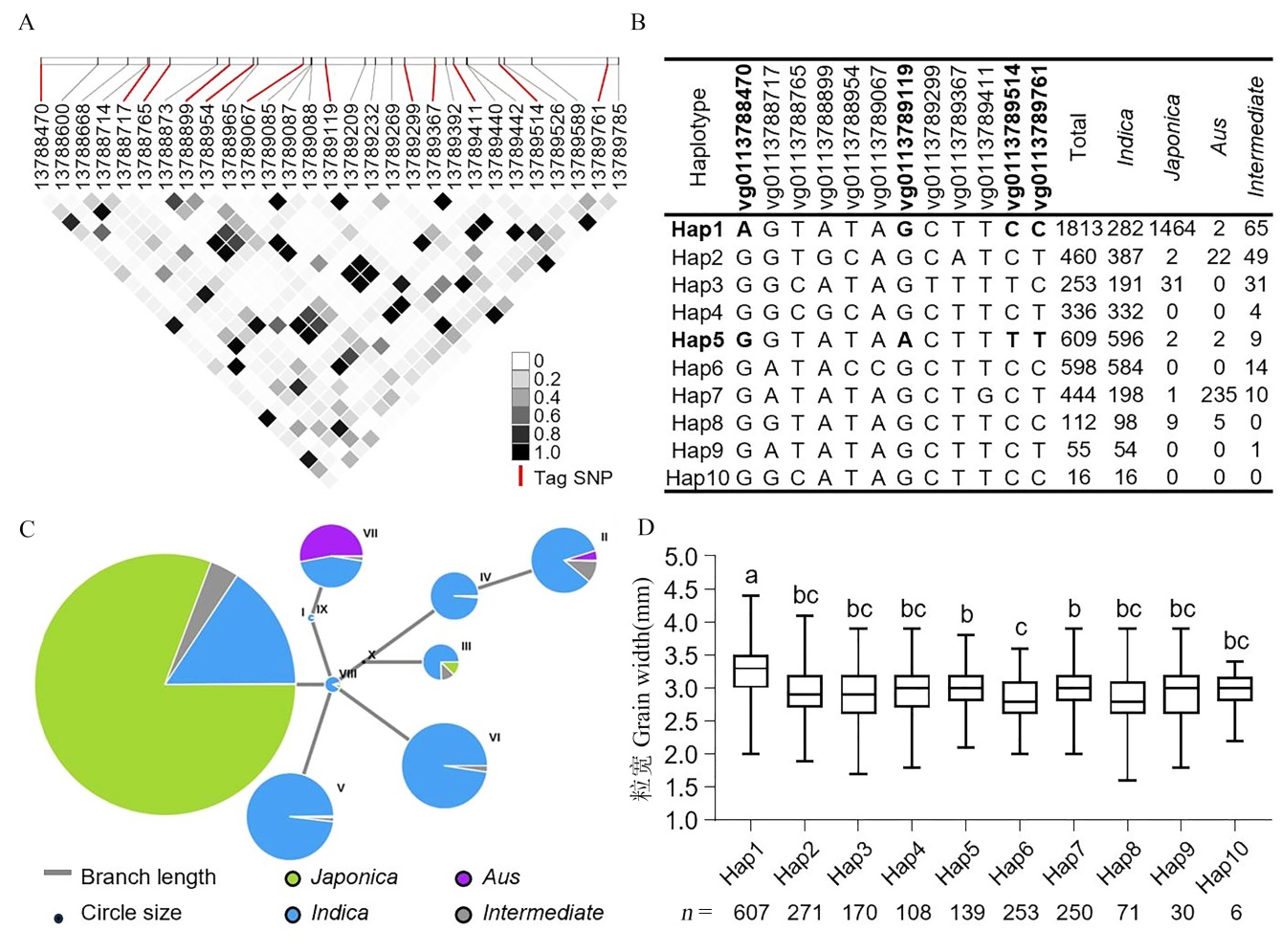
图5 水稻种质资源中OsNF-YC10的单倍型分析 A: OsNF-YC10的编码序列(coding sequence,CDS)中所有多态性位点的成对r2值(一种连锁不平衡的度量)。每个格子颜色的暗度对应r2值,红线对应的SNP为标签SNP (Tag SNP)。B: 4726份水稻材料中OsNF-YC10 CDS区域的单倍型(Hap1-10)及其分布。C: OsNF-YC10的单倍型网络分析。D: 约2000份水稻种质中不同OsNF-YC10单倍型材料的粒宽比较。采用单因素方差分析和Tukey多重比较检验进行分析;不同字母表示P < 0.05。
Fig. 5. Haplotype analysis of OsNF-YC10 in rice germplasm collections A, Pairwise r2 values (a measure of linkage disequilibrium) among all polymorphic sites in OsNF-YC10 coding sequence (CDS). The darkness of the color of each box corresponds to the r2 value, as shown in the legend. B, Haplotypes (Hap) of the OsNF-YC10 CDS region among 4726 rice accessions. C, Haplotype network analysis of OsNF-YC10. D, Grain width comparison of different OsNF-YC10 haplotypes from about 2000 rice germplasm accessions. Different letters indicate significant differences at P < 0.05(analyzed by one-way ANOVA followed by Tukey’s multiple comparison test).
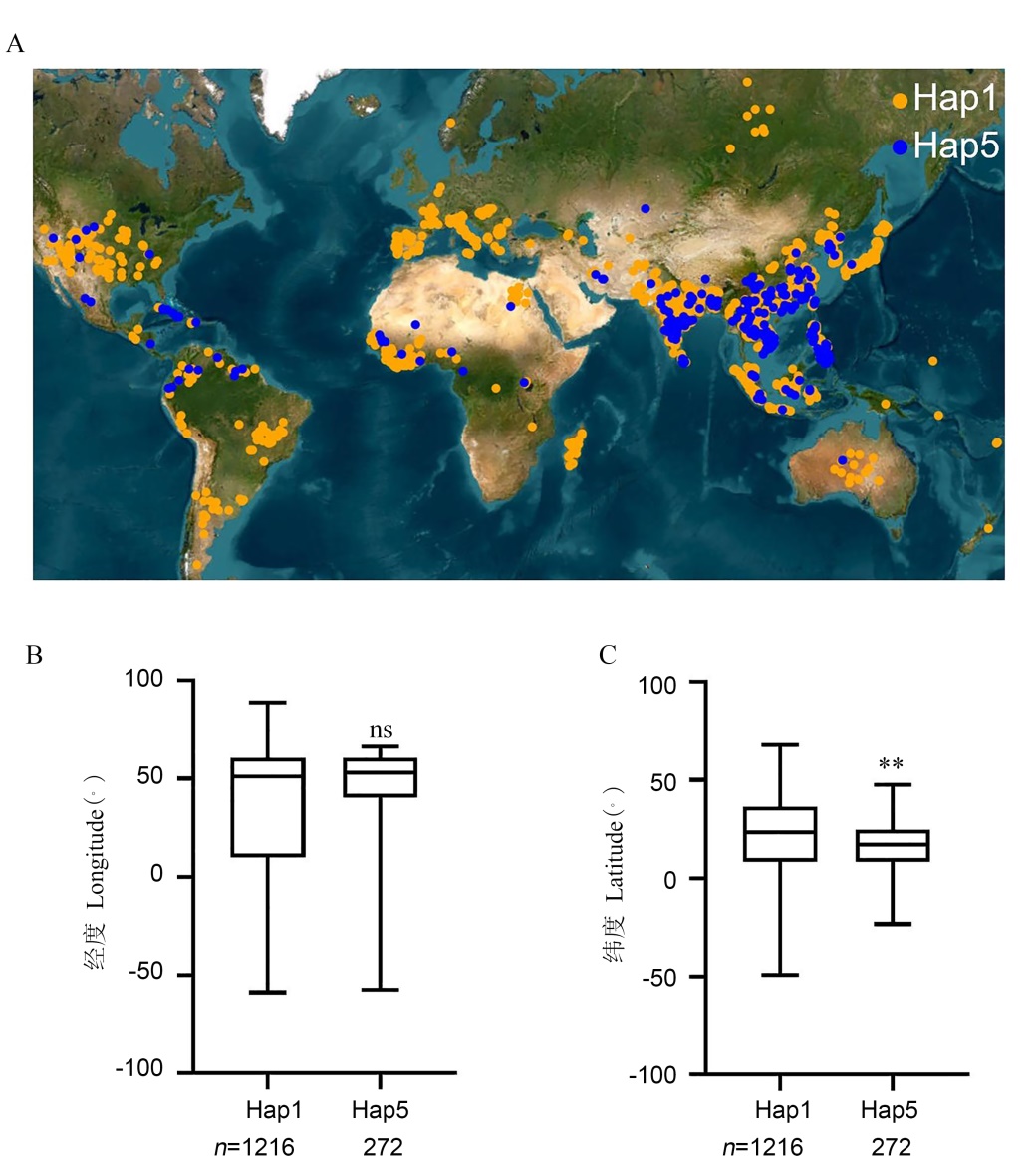
图6 OsNF-YC10的两种主要单倍型品种的全球分布 A: 两种主要OsNF-YC10单倍型品种的地理分布。两种单倍型水稻品系按经度(B)和纬度(C)分布的箱线图。**表示P < 0.01(Mann-Whitney U检验)。
Fig. 6. Global distribution of rice accessions of two major OsNF-YC10 haplotypes A, Geographical distribution of rice accessions with different OsNF-YC10 haplotypes. B and C, Box plot showing the distribution of rice accessions according to longitude (B) and latitude (C), respectively. ** indicates significant differences at P < 0.01(by Mann-Whitney U test).
| [1] | Xing Y Z, Zhang Q. Genetic and molecular bases of rice yield[J]. Annual Review of Plant Biology, 2010, 61(1): 421-442. |
| [2] | Yang W F, Zhan P L, Lin S J, Gou Y J, Zhang G Q, Wang S K. Research progress of grain shape genetics in rice[J]. Journal of South China Agricultural University, 2019, 40(5): 203-210. |
| [3] | Li G M, Tang J Y, Zheng J K, Chu C C. Exploration of rice yield potential: Decoding agronomic and physiological traits[J]. The Crop Journal, 2021, 9(3): 577-589. |
| [4] | 李堂, 曹华盛, 熊亮, 王福军, 李曙光, 顾海永, 罗文永, 何高, 梁世胡. 水稻粒型调控基因功能研究进展[J]. 广东农业科学, 2023, 50(12): 12-28. |
| Li T, Cao H S, Xiong L, Wang F J, Li S G, Gu H Y, Luo W Y, He G, Liang S H. Research progress on the function of rice grain type genes[J]. Guangdong Agricultural Sciences, 2023, 50(12): 12-28. (in Chinese with English abstract) | |
| [5] | Zuo J R, Li J Y. Molecular genetic dissection of quantitative trait loci regulating rice grain size[J]. Annual Review of Genetics, 2014, 48: 99-118. |
| [6] | Li N, Xu R, Li Y H. Molecular networks of seed size control in plants[J]. Annual Review of Plant Biology, 2019, 70: 435-463. |
| [7] | Ren D Y, Ding C Q, Qian Q. Molecular bases of rice grain size and quality for optimized productivity[J]. Science Bulletin, 2023, 68(3): 314-350. |
| [8] | Jia S Z, Xiong Y F, Xiao P P, Wang X, Yao J L. OsNF-YC10, a seed preferentially expressed gene regulates grain width by affecting cell proliferation in rice[J]. Plant Science, 2019, 280: 219-227. |
| [9] | Si L Z, Chen J Y, Huang X H, Gong H, Luo J H, Hou Q Q, Zhou T Y, Lu T Q, Zhu J J, Shangguan Y Y, Chen E W, Gong C X, Zhao Q, Jing Y F, Zhao Y, Li Y, Cui L L, Fan D L, Lu Y Q, Weng Q J, Wang Y C, Zhan Q L, Liu K Y, Wei X H, An K, An G, Han B. OsSPL13 controls grain size in cultivated rice[J]. Nature Genetics, 2016, 48(4): 447-456. |
| [10] | Yuan H, Qin P, Hu L, Zhan S J, Wang S F, Gao P, Li J, Jin M Y, Xu Z Y, Gao Q, Du A P, Tu B, Chen W L, Ma B T, Wang Y P, Li S G. OsSPL18 controls grain weight and grain number in rice[J]. Journal of Genetics and Genomics, 2019, 46(1): 41-51. |
| [11] | Huang J P, Chen Z M, Lin J J, Chen J W, Wei M H, Liu L, Yu F, Zhang Z S, Chen F Y, Jiang L R, Zheng J S, Wang T S, Chen H Y, Xie W Y, Huang S H, Wang H C, Huang Y M, Huang R Y. Natural variation of the BRD2 allele affects plant height and grain size in rice[J]. Planta, 2022, 256(2): 27. |
| [12] | Yan Y, Wei M X, Li Y, Tao H, Wu H Y, Chen Z F, Li C, Xu J H. MiR529a controls plant height, tiller number, panicle architecture and grain size by regulating SPL target genes in rice (Oryza sativa L.)[J]. Plant Science, 2021, 302: 110728. |
| [13] | Li R S, Li Z, Ye J, Yang Y Y, Ye J H, Xu S L, Liu J R, Yuan X P, Wang Y P, Zhang M C, Yu H Y, Xu Q, Wang S, Yang Y L, Wang S, Wei X H, Feng Y. Identification of SMG3, a QTL coordinately controls grain size, grain number per panicle, and grain weight in rice[J]. Frontiers in Plant Science, 2022, 13: 880919. |
| [14] | Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano L M, Kamoun S, Terauchi R. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations[J]. The Plant Journal, 2013, 74(1): 174-183. |
| [15] | Bommisetty R, Chakravartty N, Bodanapu R, Naik J B, Panda S K, Lekkala S P, Lalam K, Thomas G, Mallikarjuna S J, Eswar G R, Kadambari G M, Bollineni S N, Issa K, Akkareddy S, Srilakshmi C, Hariprasadreddy K, Rameshbabu P, Sudhakar P, Gupta S, Lachagari V B R, Vemireddy L R. Discovery of genomic regions and candidate genes for grain weight employing next generation sequencing based QTL-seq approach in rice (Oryza sativa L.)[J]. Molecular Biology Reports, 2020, 47(11): 8615-8627. |
| [16] | Bommisetty R, Chakravartty N, Hariprasad K R, Rameshbabu P, Sudhakar P, Bodanapu R, Naik J B, Bhaskar Reddy B V, Lekkala S P, Gupta S, Tanti B, Lachagari V B R, Vemireddy L R. Identification of a novel QTL for grain number per panicle employing NGS-based QTL-seq approach in rice (Oryza sativa L.)[J]. Plant Biotechnology Reports, 2023, 17(2): 191-201. |
| [17] | 王豪, 张健, 王加峰, 杨瑰丽, 郭涛, 陈志强, 王慧. 基于QTL-seq的水稻粒质量QTL定位及候选基因分析[J]. 华北农学报, 2020, 35(2): 18-28. |
| Wang H, Zhang J, Wang J F, Yang G L, Guo T, Chen Z Q, Wang H. QTL Mapping and candidate gene analysis of rice grain weight based on QTL-seq. Acta Agriculturae Boreali-Sinica, 2020, 35(2): 18-28. (in Chinese with English abstract) | |
| [18] | Sun H Z, Yuan Z K, Li F H, Zhang Q Q, Peng T, Li J Z, Du Y X. Mapping of qChalk1 controlling grain chalkiness in japonica rice[J]. Molecular Biology Reports, 2023, 50(7): 5879-5887. |
| [19] | Yuan H, Xu Z Y, Tan X Q, Gao P, Jin M Y, Song W C, Wang S G, Kang Y H, Liu P X, Tu B, Wang Y P, Qin P, Li S G, Ma B T, Chen W L. A natural allele of TAW1 contributes to high grain number and grain yield in rice[J]. The Crop Journal, 2021, 9(5): 1060-1069. |
| [20] | Alexandrov N, Tai S, Wang W, Mansueto L, Palis K, Fuentes R R, Ulat V J, Chebotarov D, Zhang G, Li Z, Mauleon R, Hamilton R S, McNally K L. SNP-Seek database of SNPs derived from 3000 rice genomes[J]. Nucleic Acids Research, 2015, 43(D1): D1023-D1027. |
| [21] | Zhao H, Yao W, Ouyang Y D, Yang W N, Wang G W, Lian X M, Xing Y Z, Chen L L, Xie W B. RiceVarMap: A comprehensive database of rice genomic variations[J]. Nucleic Acids Research, 2015, 43(D1): D1018-D1022. |
| [22] | Chen S, Zhou Y, Chen Y, Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): i884-i890. |
| [23] | Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009, 25(14): 1754-1760. |
| [24] | Danecek P, Bonfield J K, Liddle J, Marshall J, Ohan V, Pollard M O, Whitwham A, Keane T, McCarthy S A, Davies R M, Li H. Twelve years of SAMtools and BCFtools[J]. GigaScience, 2021, 10(2): giab008. |
| [25] | McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo M A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Research, 2010, 20(9): 1297-1303. |
| [26] | Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R. Genome sequencing reveals agronomically important loci in rice using MutMap[J]. Nature Biotechnology, 2012, 30(2): 174-178. |
| [27] | Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs[J]. Nucleic Acids Research, 1997, 25(17): 3389-3402. |
| [28] | Medrano R F V, de Oliveira C A. Guidelines for the Tetra-primer ARMS-PCR technique development[J]. Molecular Biotechnology, 2014, 56(7): 599-608. |
| [29] | Zhang K, Calabrese P, Nordborg M, Sun F Z. Haplotype block structure and its applications to association studies: power and study designs[J]. The American Journal of Human Genetics, 2002, 71(6): 1386-1394. |
| [1] | 马顺婷, 胡运高, 高方远, 刘利平, 牟昌铃, 吕建群, 苏相文, 刘松, 梁毓玉, 任光俊, 郭鸿鸣. 水稻真核翻译起始因子OseIF6.2调控粒型的功能研究[J]. 中国水稻科学, 2025, 39(3): 331-342. |
| [2] | 邵雅芳, 朱大伟, 郑欣, 牟仁祥, 章林平, 陈铭学. 2002−2022长三角地区粳稻品质发展状况和地域差异性分析[J]. 中国水稻科学, 2025, 39(2): 264-276. |
| [3] | 杜彦修, 孙文玉, 袁泽科, 张倩倩, 李富豪, 李俊周, 孙红正. 利用QTL-Seq结合分子标记定位粳稻垩白粒率控制位点qChalk8[J]. 中国水稻科学, 2024, 38(6): 665-671. |
| [4] | 何勇, 刘耀威, 熊翔, 祝丹晨, 王爱群, 马拉娜, 王廷宝, 张健, 李建雄, 田志宏. 利用CRISPR/Cas9技术编辑OsOFP30基因创制水稻粒型突变体[J]. 中国水稻科学, 2024, 38(5): 507-515. |
| [5] | 丁正权, 潘月云, 施扬, 黄海祥. 基于基因芯片的嘉禾系列长粒优质食味粳稻综合评价与比较[J]. 中国水稻科学, 2024, 38(4): 397-408. |
| [6] | 吕宙, 易秉怀, 陈平平, 周文新, 唐文帮, 易镇邪. 施氮量与移栽密度对小粒型杂交水稻产量形成的影响[J]. 中国水稻科学, 2024, 38(4): 422-436. |
| [7] | 刘慧敏, 周杰强, 胡远艺, 田妍, 雷斌, 李建武, 魏中伟, 唐文帮. 水稻小粒不育系新组合卓两优1126的高产特征[J]. 中国水稻科学, 2024, 38(2): 160-171. |
| [8] | 兰金松, 庄慧. 水稻株型的分子机理研究进展[J]. 中国水稻科学, 2023, 37(5): 449-458. |
| [9] | 韦敏益, 马增凤, 黄大辉, 秦媛媛, 刘驰, 卢颖萍, 罗同平, 李振经, 张月雄, 秦钢. 基于QTL-Seq的水稻抗细菌性条斑病QTL定位[J]. 中国水稻科学, 2023, 37(2): 133-141. |
| [10] | 黄涛, 王燕宁, 钟奇, 程琴, 杨朦朦, 王鹏, 吴光亮, 黄诗颖, 李才敬, 余剑峰, 贺浩华, 边建民. 利用染色体片段置换系群体定位和分析水稻粒重和粒型QTL[J]. 中国水稻科学, 2022, 36(2): 159-170. |
| [11] | 龚柯, 薛炮, 温小霞, 廖飞飞, 孙滨, 彭泽群, 程式华, 曹立勇, 张迎信, 吴玮勋, 孙廉平, 占小登. 水稻特大粒种质BG1和优质恢复系华占的粒形基因研究及相关功能标记开发[J]. 中国水稻科学, 2021, 35(6): 543-553. |
| [12] | 闫浩亮, 王松, 王雪艳, 党程成, 周梦, 郝蓉蓉, 田小海. 不同水稻品种在高温逼熟下的表现及其与气象因子的关系[J]. 中国水稻科学, 2021, 35(6): 617-628. |
| [13] | 朱玉君, 左紫薇, 张振华, 樊叶杨. 一种水稻微效QTL精细定位和克隆新途径[J]. 中国水稻科学, 2021, 35(4): 407-414. |
| [14] | 邓雪梅, 胡鹏, 王月影, 文艺, 谭义青, 伍豪, 吴凯雄, 王俊格, 侯琳琳, 朱黎欣, 朱丽, 陈光, 曾大力, 张光恒, 郭龙彪, 高振宇, 任德勇, 钱前, 胡江. 水稻粒宽突变体gw4的鉴定与基因定位[J]. 中国水稻科学, 2021, 35(3): 259-268. |
| [15] | 康艺维, 陈玉宇, 张迎信. 水稻粒型基因克隆研究进展及育种应用展望[J]. 中国水稻科学, 2020, 34(6): 479-490. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||