
中国水稻科学 ›› 2025, Vol. 39 ›› Issue (3): 331-342.DOI: 10.16819/j.1001-7216.2025.2401010
马顺婷1, 胡运高1, 高方远2, 刘利平2, 牟昌铃2, 吕建群2, 苏相文2, 刘松2, 梁毓玉1, 任光俊2,*( ), 郭鸿鸣2,*(
), 郭鸿鸣2,*( )
)
收稿日期:2024-10-25
修回日期:2025-03-27
出版日期:2025-05-10
发布日期:2025-05-21
通讯作者:
*email: guangjun61@sina.com;hongmingguo552@163.com基金资助:
MA Shunting1, HU Yungao1, GAO Fangyuan2, LIU Liping2, MOU Changling2, LÜ Jianqun2, SU Xiangwen2, LIU Song2, LIANG Yuyu1, REN Guangjun2,*( ), GUO Hongming2,*(
), GUO Hongming2,*( )
)
Received:2024-10-25
Revised:2025-03-27
Online:2025-05-10
Published:2025-05-21
Contact:
*email: guangjun61@sina.com;hongmingguo552@163.com摘要:
【目的】本研究旨在通过反向遗传学策略,深入探究水稻真核翻译起始因子OseIF6.2在水稻生长发育中的具体功能,以期为OseIF6.2功能的进一步解析奠定基础。【方法】首先,利用生物信息学方法进行多重序列比对及进化树的构建,以分析OseIF6.2与其他真核生物中eIF6蛋白的系统发育关系;运用CRISPR/Cas9基因编辑技术,对水稻(日本晴)中的OseIF6.2基因进行精准编辑。其次,通过qRT-PCR技术,检测OseIF6.2在水稻不同组织中的表达水平;借助激光共聚焦显微镜,观察OseIF6.2在细胞内的精确定位。然后对野生型与oseif6.2突变体的粒长、粒宽、粒厚及千粒重等关键农艺性状进行比较分析。最后,通过酵母双杂交(Y2H)及双分子荧光互补(BiFC)实验,验证OseIF6.2与其潜在互作蛋白OseIF6.1的相互作用。【结果】系统发育分析结果表明,OseIF6.2与其他真核生物中的eIF6蛋白具有高度保守性,凸显了其在真核生物翻译调控中的普遍重要性。qRT-PCR结果显示,OseIF6.2在水稻茎中的表达水平较高。通过将OseIF6.2蛋白与GFP融合后注入本氏烟叶片细胞,并在激光共聚焦显微镜下观察,发现OseIF6.2在细胞核及细胞质区域均有荧光信号分布,明确了其亚细胞定位。此外,与野生型相比,oseif6.2突变体在粒长、粒厚及千粒重等性状上均表现为显著降低。Y2H及BiFC实验均成功验证了OseIF6.2与OseIF6.1蛋白之间的直接相互作用。【结论】综上所述,OseIF6.2作为水稻真核翻译起始因子,其功能异常显著影响水稻的粒型特征,进一步揭示了该因子在水稻生长发育及粒型形成中的关键作用。
马顺婷, 胡运高, 高方远, 刘利平, 牟昌铃, 吕建群, 苏相文, 刘松, 梁毓玉, 任光俊, 郭鸿鸣. 水稻真核翻译起始因子OseIF6.2调控粒型的功能研究[J]. 中国水稻科学, 2025, 39(3): 331-342.
MA Shunting, HU Yungao, GAO Fangyuan, LIU Liping, MOU Changling, LÜ Jianqun, SU Xiangwen, LIU Song, LIANG Yuyu, REN Guangjun, GUO Hongming. Functional Study of Rice Eukaryotic Translation Initiation Factor OseIF6.2 in Grain Size Regulation[J]. Chinese Journal OF Rice Science, 2025, 39(3): 331-342.
| 引物名称 Primer name | 引物序列 Sequence (5'-3') | 用途 Purpose |
|---|---|---|
| sgRNA序列 | GCGAGATTGGAGTGTTCGCGAGG | OseIF6.2基因sgRNA序列 |
| eIF6.2-CDS-F | ATGATTTTTTTTTTCAAAAATCGGTTCTGCATTGCA | CDS 扩增 |
| eIF6.2-CDS-R | CTATAGGTAGCTGCTGACGACGAGCGA | CDS amplification |
| K21-F | AGTTTGTAACAGCTGCTGGGATTAC | 亚细胞定位载体 |
| K21-NOSR-R | CCATCTCATAAATAACGTCATGCATTACATGTT | Subcellular localization vector |
| eIF6.2-Cas9-F | AGGAGTTCTTGGTTGGCTGACTGCGA | 基因编辑材料鉴定 |
| eIF6.2-Cas9-R | ACGCACATCCTCCCGATGATCCT | Identification of gene editing materials |
| OsCas9-1000-F | GAGGAGACAATCACCCCCTGGAACT | Cas9蛋白鉴定 |
| OsCas9-1000-R | TTATCGGACTTGCCCCTGTTCTTGT | Identification of Cas9 protein |
| eIF6.1-2YC-767-F | ACGATAGTTAATTAACATGGCGACCCGTATTCAGTTTG | BiFC载体 |
| eIF6.1-2YC-767-R | GATCGACAGTTATGTGCACTAGTGGCGCGCCC | Bimolecular fluorescence complementation vector |
| eIF6.1-CDS-759-F | CCTTAATTAAGGATGGCGACCCGTATTCAGTTTGAGAAC | CDS 扩增 |
| eIF6.1-CDS-759-R | TTGGCGCGCCAACACATAACTGTCGATCAAAG | CDS amplification |
| eIF6.2-BD-798-F | GAGGACCTGCATATGATGATTTTTTTTTTCAAAAATCG | 酵母双杂载体 |
| eIF6.2-BD-798-R | GATCCCCGGGAATTCCTATAGGTAGCTG | Construction of yeast two-hybrid vector |
| eIF6.2-2YN-797-F | AC GATAGTTAATTAACATGATTTTTTTTTTCAAAAATCGG | BiFC载体构建 |
| eIF6.2-2YN-797-R | CACTAGTGGCGCGCCCTAGGTAGCTGC | Constructionof of bimolecular fluorescence complementation vector |
| Actin-qPCR-F | TGTATGCCAGTGGTCGTACCA | 内参基因定量引物 |
| Actin-qPCR-R | CCAGCAAGGTCGAGACGAA | Primers of internal reference gene |
| eIF6.2-qPCR-148-F | AGAGAACTTCTTCAGCGTGTTC | OseIF6.2 定量引物 |
| eIF6.2-qPCR-148-R | CTCTTGGTCTGTGGTGGTATG | qRT-PCR primers of OseIF6.2 |
| OsEXPA5-F | AAGGCTGTGGCTTGATTGACA | 细胞膨大相关基因定量引物 |
| OsEXPA5-R | TTAGGCCCAATTTTGCTATTTTG | qRT-PCR primers of genes related to cell expansion |
| OsEXPA8-F | GCGATGAGCCGCAACTG | 细胞膨大相关基因定量引物 |
| OsEXPA8-R | CTCTTCCATCCTATGCCACG | qRT-PCR primers of genes related to cell expansion |
| OsEXPA30-F | CCAAGTTCAGGGCGATGCAG | 细胞膨大相关基因定量引物 |
| OsEXPA30-R | ACGGTGATCGCCGGCGAGCC | qRT-PCR primers of genes related to cell expansion |
| OsEXPB3-F | CTTTGAGTGGTTGGAGTGGTGG | 细胞膨大相关基因定量引物 |
| OsEXPB3-R | GCAGCCTTCTTGGAGATGGAA | qRT-PCR primers of genes related to cell expansion |
表1 本研究中使用的引物
Table 1. Primers used in this study
| 引物名称 Primer name | 引物序列 Sequence (5'-3') | 用途 Purpose |
|---|---|---|
| sgRNA序列 | GCGAGATTGGAGTGTTCGCGAGG | OseIF6.2基因sgRNA序列 |
| eIF6.2-CDS-F | ATGATTTTTTTTTTCAAAAATCGGTTCTGCATTGCA | CDS 扩增 |
| eIF6.2-CDS-R | CTATAGGTAGCTGCTGACGACGAGCGA | CDS amplification |
| K21-F | AGTTTGTAACAGCTGCTGGGATTAC | 亚细胞定位载体 |
| K21-NOSR-R | CCATCTCATAAATAACGTCATGCATTACATGTT | Subcellular localization vector |
| eIF6.2-Cas9-F | AGGAGTTCTTGGTTGGCTGACTGCGA | 基因编辑材料鉴定 |
| eIF6.2-Cas9-R | ACGCACATCCTCCCGATGATCCT | Identification of gene editing materials |
| OsCas9-1000-F | GAGGAGACAATCACCCCCTGGAACT | Cas9蛋白鉴定 |
| OsCas9-1000-R | TTATCGGACTTGCCCCTGTTCTTGT | Identification of Cas9 protein |
| eIF6.1-2YC-767-F | ACGATAGTTAATTAACATGGCGACCCGTATTCAGTTTG | BiFC载体 |
| eIF6.1-2YC-767-R | GATCGACAGTTATGTGCACTAGTGGCGCGCCC | Bimolecular fluorescence complementation vector |
| eIF6.1-CDS-759-F | CCTTAATTAAGGATGGCGACCCGTATTCAGTTTGAGAAC | CDS 扩增 |
| eIF6.1-CDS-759-R | TTGGCGCGCCAACACATAACTGTCGATCAAAG | CDS amplification |
| eIF6.2-BD-798-F | GAGGACCTGCATATGATGATTTTTTTTTTCAAAAATCG | 酵母双杂载体 |
| eIF6.2-BD-798-R | GATCCCCGGGAATTCCTATAGGTAGCTG | Construction of yeast two-hybrid vector |
| eIF6.2-2YN-797-F | AC GATAGTTAATTAACATGATTTTTTTTTTCAAAAATCGG | BiFC载体构建 |
| eIF6.2-2YN-797-R | CACTAGTGGCGCGCCCTAGGTAGCTGC | Constructionof of bimolecular fluorescence complementation vector |
| Actin-qPCR-F | TGTATGCCAGTGGTCGTACCA | 内参基因定量引物 |
| Actin-qPCR-R | CCAGCAAGGTCGAGACGAA | Primers of internal reference gene |
| eIF6.2-qPCR-148-F | AGAGAACTTCTTCAGCGTGTTC | OseIF6.2 定量引物 |
| eIF6.2-qPCR-148-R | CTCTTGGTCTGTGGTGGTATG | qRT-PCR primers of OseIF6.2 |
| OsEXPA5-F | AAGGCTGTGGCTTGATTGACA | 细胞膨大相关基因定量引物 |
| OsEXPA5-R | TTAGGCCCAATTTTGCTATTTTG | qRT-PCR primers of genes related to cell expansion |
| OsEXPA8-F | GCGATGAGCCGCAACTG | 细胞膨大相关基因定量引物 |
| OsEXPA8-R | CTCTTCCATCCTATGCCACG | qRT-PCR primers of genes related to cell expansion |
| OsEXPA30-F | CCAAGTTCAGGGCGATGCAG | 细胞膨大相关基因定量引物 |
| OsEXPA30-R | ACGGTGATCGCCGGCGAGCC | qRT-PCR primers of genes related to cell expansion |
| OsEXPB3-F | CTTTGAGTGGTTGGAGTGGTGG | 细胞膨大相关基因定量引物 |
| OsEXPB3-R | GCAGCCTTCTTGGAGATGGAA | qRT-PCR primers of genes related to cell expansion |
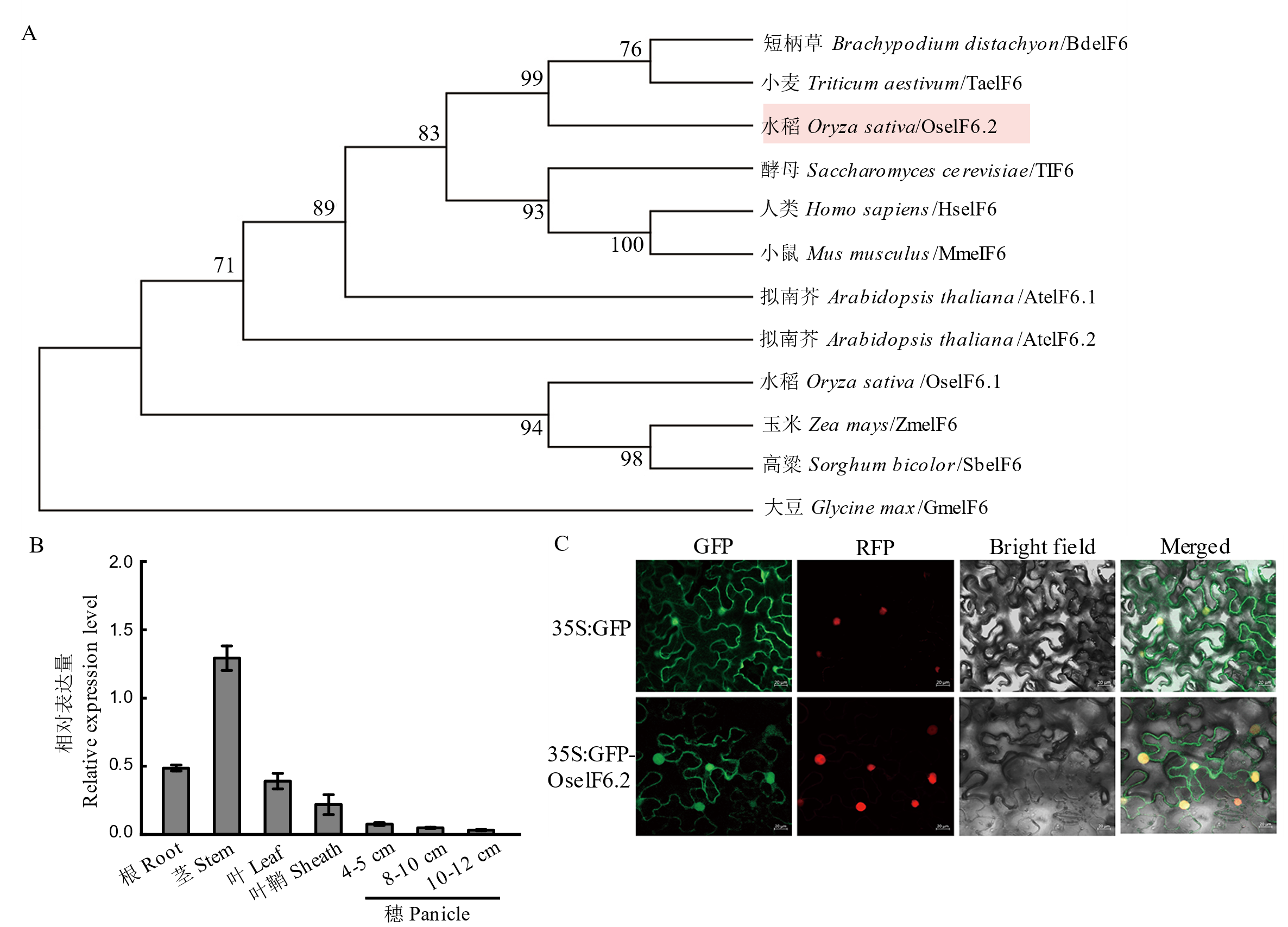
图1 OseIF6.2的表达模式及亚细胞定位 A:对不同真核生物物种中eIF6蛋白的系统发育比较分析。B:OseIF6.2在幼苗根、茎、叶、鞘和4~5、8~10、10~12 cm幼穗中的表达情况,OsActin作为内参。数据是来自三组独立实验的平均值±标准差。C:OseIF6.2在烟草叶中的亚细胞定位。GFP:绿色荧光蛋白,RFP:红色荧光蛋白。标尺=20 μm。
Fig. 1. Expression pattern and subcellular localization of OseIF6.2 A, Comparative phylogenetic analysis of eIF6 protein cross diverse eukaryotic species. B, Relative expression of OseIF6.2 in root, stem, leaf, and sheath of young seedlings and developing panicles at lengths of 4-5, 8-10, and 10-12 cm. OsActin served as the internal control. Values represent means ± SE from at least three independent experiments. C, Subcellular localization of OseIF6.2 in N. benthamiana leaves. YFP: Yellow fluorescent protein, RFP: Red fluorescent protein. Scale bars = 20 μm.

图2 OseIF6.2突变体类型及表型分析 A:OseIF6.2突变类型。B:野生型和oseif6.2突变体植株成熟期的形态和穗部结构。株高比例尺=10 cm,穗长比例尺=1 cm。C:野生型和oseif6.2突变体植株的穗长。D:野生型和oseif6.2突变体穗的一次枝梗数。E:野生型和oseif6.2突变体穗的二次枝梗数。F:野生型和oseif6.2突变体植株的结实率。数据是来自三组独立实验的平均值±标准差, t检验:*P< 0.05, **P< 0.01, ***P< 0.001。
Fig. 2. Types and phenotypic analysis of OseIF6.2 mutants A, Mutation types of OseIF6.2. B, Plants and panicles of WT and oseif6.2 mutant at the mature stage. Scale bars = 10 cm for plant height and 1 cm for panicle length. C, Panicle length of WT and oseif6.2 mutant. D, Number of primary branches per panicle of WT and oseif6.2 mutant. E, Number of secondary branches of WT and oseif6.2 mutant. F, Seed setting rate of WT and oseif6.2 mutant. Values represent means ± SE from at least three independent experiments, Student's t-test: *P< 0.05, **P< 0.01, ***P< 0.001.

图3 OseIF6.2的突变影响水稻粒型 A: 野生型和oseif6.2突变体株系的成熟水稻籽粒。比例尺=1 cm。B: 野生型和oseif6.2突变体株系的粒长;C: 粒宽;D: 粒厚;E: 千粒重;F: 每穗粒数;G: 单株穗数;H: 单株产量。数据是来自三组独立实验的平均值±标准差,t检验:* P< 0.05, **P< 0.01, ***P< 0.001。
Fig. 3. Mutation of OseIF6.2 alters grain shape A, Mature grains of WT and oseif6.2 mutants. Scale bars =1 cm. B, Grain length of WT and oseif6.2 mutants; C, Grain width; D, Grain thickness; E, 1000-grain weight; F, Filled grains per panicle; G, Panicle number per plant; H, Grain yield per plant. Values represent means ± SE from at least three independent experiments, Student's t-test: *P < 0.05, **P < 0.01, ***P< 0.001.

图4 OseIF6.2通过影响细胞膨大调节水稻粒型 A:野生型和oseif6.2突变体成熟谷粒的颖壳扫描电子显微镜图像。比例尺=100 μm。野生型和oseif6.2突变体颖壳细胞长度(B)和宽度(C)。D:野生型和oseif6.2突变体颖壳细胞面积。数据是来自三组独立实验的平均值±标准差。t检验: *P< 0.05, **P< 0.01, ***P< 0.001。
Fig. 4. OseIF6.2 regulates grain size by affecting cell expansion A, Scanning electron microscopy images of the glume outer surfaces of WT and oseif6.2 mutants. Scale bars = 100 μm. Cell length (B) and width (C) of the outer epidermal of WT and oseif6.2 mutant lemmas. D, Cell area of the outer epidermal of WT and oseif6.2 mutant lemmas. Values represent means ± SE from at least three independent experiments. Student's t-test: *P < 0.05, **P < 0.01, ***P < 0.001.
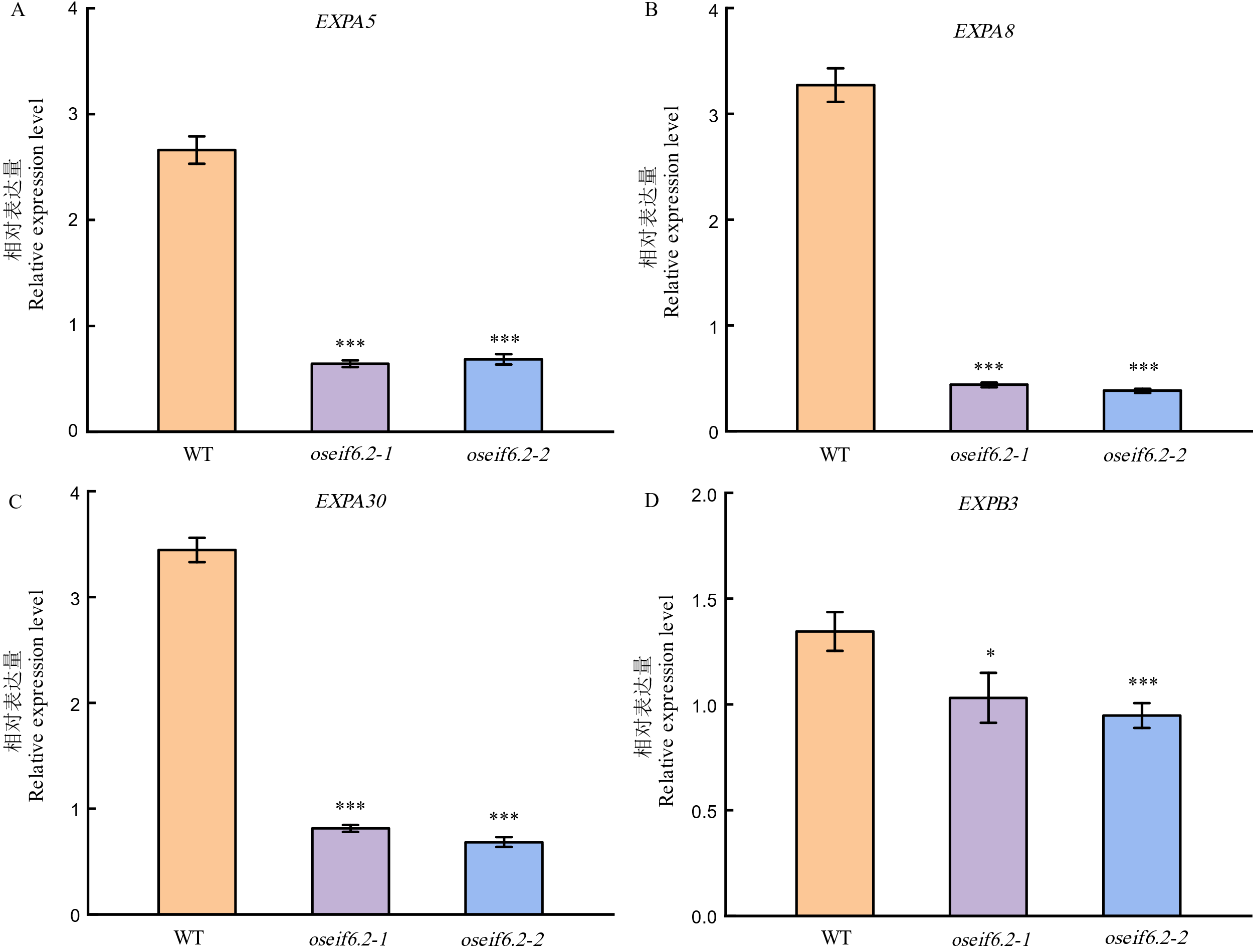
图5 野生型和oseif6.2突变体株系细胞膨大相关基因的表达水平 数据是来自三组独立实验的平均值±标准差。t检验: *P< 0.05, **P< 0.01, ***P< 0.001。
Fig. 5. Expression levels of genes related to cell expansion in WT and oseif6.2 mutant lines The values represent means ± SE derived from at least three independent experiments. Student's t-test: *P < 0.05, **P < 0.01, ***P < 0.001.

图6 OseIF6.2与OseIF6.1互作 A:OseIF6.2在酵母细胞中与OseIF6.1相互作用。转化在DDO或QDO/X/A培养基上培养。B:双分子荧光互补分析验证烟草中OseIF6.2与OseIF6.1的相互作用。OseIF6.2-nYFP与OseIF6.1-cYFP在烟草细胞中共表达。比例尺=10 μm。
Fig. 6. OseIF6.2 physically interacts with OseIF6.1 A, OseIF6.2 interacts with OseIF6.1 in yeast cells. Transformed cells were cultured on DDO or QDO/X/A media. B, Bimolecular fluorescence complementation assays verifies the interaction between OseIF6.2 and OseIF6.1 in N. benthamiana. OseIF6.2-nYFP was coexpressed with OseIF6.1-cYFP in cells of N. benthamiana. Scale bars = 10 μm.
| [1] | Su S, Hong J, Chen X, Zhang C, Chen M, Luo Z, Chang S, Bai S, Liang W, Liu Q, Zhang D. Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice[J]. Plant Biotechnology Journal, 2021, 19(11): 2304-2318. |
| [2] | Merrick W C, Pavitt G D. Protein synthesis initiation in eukaryotic cells[J]. Cold Spring Harbor Perspectives in Biology, 2018, 10(12): a033092. |
| [3] | Jackson R J, Hellen C U T, Pestova T V. The mechanism of eukaryotic translation initiation and principles of its regulation[J]. Nature Reviews Molecular Cell Biology, 2010, 11(2): 113-127. |
| [4] | Guo J, Jin Z, Yang X, Li J F, Chen J G. Eukaryotic initiation factor 6, an evolutionarily conserved regulator of ribosome biogenesis and protein translation[J]. Plant Signaling & Behavior, 2011, 6(5): 766-771. |
| [5] | Aitken C E, Lorsch J R. A mechanistic overview of translation initiation in eukaryotes[J]. Nature structural & Molecular Biology, 2012, 19(6): 568-576. |
| [6] | Raabe K, Honys D, Michailidis C. The role of eukaryotic initiation factor 3 in plant translation regulation[J]. Plant Physiology and Biochemistry, 2019, 145: 75-83. |
| [7] | Castellano M, Merchante C. Peculiarities of the regulation of translation initiation in plants[J]. Current Opinion in Plant Biology, 2021, 63: 102073. |
| [8] | Singha D L, Maharana J, Panda D, Dehury B, Modi M K, Singh S. Understanding the thermal response of rice eukaryotic transcription factor eIF4A1 towards dynamic temperature stress: insights from expression profiling and molecular dynamics simulation[J]. Journal of Biomolecular Structure and Dynamics, 2021, 39(7): 2575-2584. |
| [9] | Ma L, Yang Y, Wang Y, Cheng K, Zhou X, Li J, Zhang J, Li R, Zhang L, Wang K, Zeng N, Gong Y, Zhu D, Deng Z, Qu G, Zhu B, Fu D, Luo Y, Zhu H. SlRBP1 promotes translational efficiency via SleIF4A2 to maintain chloroplast function in tomato[J]. The Plant Cell, 2022, 34(7): 2747-2764. |
| [10] | Rausell A, Kanhonou R, Yenush L, Serrano R, Ros R. The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants[J]. Plant Journal, 2003, 34: 257-267. |
| [11] | Sahoo R K, Gill S S, Tuteja N. Pea DNA helicase 45 promotes salinity stress tolerance in IR64 rice with improved yield[J]. Plant Signaling&Behavior, 2012, 7: 1042-104. |
| [12] | Zhang L, Liu X, Gaikwad K, Kou X, Wang F, Tian X, Xin M, Ni Z, Sun Q, Peng H, Vierling E. Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in Arabidopsis thaliana[J]. The Plant Cell, 2017, 29(8): 1952-1969. |
| [13] | Zheng T C, Zang L N, Dai L J, Yang C P, Qu G Z. Two novel eukaryotic translation initiation factor 5A genes from Populus simonii×P. nigra confer tolerance to abiotic stresses in Saccharomyces cerevisiae[J]. Journal Frontiers Research, 2017, 28: 453-46. |
| [14] | Li L S, Luo C, Huang F, Liu Z L, An Z Y, Dong L, He X H. Identification and characterization of the mango eIF gene family reveals MieIF1A-a, which confers tolerance to salt stress in transgenic Arabidopsis[J]. Scientia Horticulturae, 2019, 248: 274-281. |
| [15] | Wang L, Xu C, Wang C, Wang Y. Characterization of a eukaryotic translation initiation factor 5A homolog from Tamarix androssowii involved in plant abiotic stress tolerance[J]. BMC Plant Biology, 2012, 12(1): 118. |
| [16] | 王雷, 孙尧, 李瑶, 吴琼, 夏新莉. 罗布麻eIF-5A基因功能[J]. 北方园艺, 2017(23): 190-193. |
| Wang L, Sun Y, Li Y, Wu Q, XIia X L. Function of eIF-5A gene in Apocynum venetum L[J]. Norther Horticulture, 2017(23):190-193. | |
| [17] | Kenia S D, Mayra A L, Eloísa H L, Brenda N R, Marlon A P T, Jesús H J P, Marina G R, Tzvetanka D D. Arabidopsis thaliana eIF4E1 and eIF(iso)4E participate in cold response and promote translation of some stress-related mRNAs[J]. Frontiers in Plant Science, 2021, 12(12): 698585. |
| [18] | Wang W, Xu M, Liu X, Tu J. The rice eukaryotic translation initiation factor 3 subunit e (OseIF3e) influences organ size and pollen maturation[J]. Frontiers in Plant Science, 2016, 7: 1399. |
| [19] | Huang Y, Zheng P, Liu X, Chen H, Tu J. OseIF3h regulates plant growth and pollen development at translational level presumably through interaction with OsMTA2[J]. Plants, 2021, 10(6): 1101. |
| [20] | Kim T H, Kim B H, Yahalom A, Chamovitz D A, von Arnim A G. Translational regulation via 5' mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h[J]. The Plant Cell, 2004, 16(12): 3341-3356. |
| [21] | Kim B H, Cai X, Vaughn J N, vonArnim A G. On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation[J]. Genome Biology. 2007, 8: R60. |
| [22] | Li N, Li Y. Signaling pathways of seed size control in plants[J]. Current opinion in Plant Bitology, 2016, 33: 23-32. |
| [23] | Xia C, Wang Y J, Li W Q, Chen Y R, Deng Y, Zhang X Q, Chen L Q, Ye D. The Arabidopsis eukaryotic translation initiation factor 3, subunit F (AteIF3f), is required for pollen germination and embryogenesis[J]. The Plant Journal, 2010, 63(2): 189-202. |
| [24] | Feng H, Chen Q, Feng J, Zhang J, Yang X, Zuo J. Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death[J]. Plant Physiology, 2007, 144: 1531-1545. |
| [25] | Hopkins M T, Lampi Y, Wang T W, Liu Z, Thompson J E. Eukaryotic translation initiation factor 5A is involved in pathogen-induced cell death and development of disease symptoms in Arabidopsis[J]. Plant Physiology, 2008, 148: 479-489. |
| [26] | Wang T W, Lu L, Wang D, Thompson J E. Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eucaryotic translation initiation factor 5A from tomato[J]. Journal of Biological Chemistry, 2001, 276(20): 17541. |
| [27] | Lin L, Cao J, Du A, An Q, Chen X, Yuan S, Batool W, Shabbir A, Zhang D, Wang Z, Norvienyeku J. EIF3k domain-containing protein regulates conidiogenesis, appressorium turgor, virulence, stress tolerance, and physiological and pathogenic development of Magnaporthe oryzae Oryzae[J]. Frontiers in Plant Science, 2021, 12: 748120. |
| [28] | Zhang X, Yin Y, Su Y, Jia Z, Jiang L, Lu Y, Zheng H, Peng J, Rao S, Wu G, Chen J, Yan F. eIF4A, a target of siRNA derived from rice stripe virus, negatively regulates antiviral autophagy by interacting with ATG5 in Nicotiana benthamiana[J]. PLoS Pathogens, 2021, 17(9): e1009963. |
| [29] | Roy B, von Arnim A G. Translational regulation of cytoplasmic mRNAs[J]. The Arabidopsis book/American Society of Plant Biologists, 2013, 11: e0165. |
| [30] | Si K, Maitra U. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor[J]. Molecular and Cellular Biology, 1999, 19(2): 1416-1426. |
| [31] | Basu U, Si K, Warner J R, Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis[J]. Molecular and Cellular Biology, 2001, 21(5): 1453-1462. |
| [32] | Ceci M, Gaviraghi C, Gorrini C, Sala L A, Offenhäuser N, Marchisio P C, Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly[J]. Nature, 2003, 426(6966): 579-584. |
| [33] | Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation[J]. EMBO Reports, 2009, 10(5): 459-465. |
| [34] | Gandin V, Miluzio A, Barbieri A M, Beugnet A, Kiyokawa H, Marchisio P C, Biffo S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation[J]. Nature, 2008, 455(7213): 684-688. |
| [35] | Kausik S, Jayanta C, Jorge C H. Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6[J]. Proceedings of the National Academy of Sciences, 1997, 94(26): 14285-14290. |
| [36] | Sanvito F, Piatti S, Villa A, Bossi M, Lucchini G, Marchisio PC, Biffo S. The beta4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly.[J]. The Journal of Cell Biology, 1999, 144(5): 823-837. |
| [37] | Kapp L D, Lorsch J R. The molecular mechanics of eukaryotic translation[J]. Annual Review of Biochemistry, 2004, 73(1): 657-704. |
| [38] | Sonenberg N, Hinnebusch A G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets[J]. Cell, 2009, 136(4): 731-745. |
| [39] | Russell D W, Spremulli L L. Mechanism of action of the wheat germ ribosome dissociation factor: Interaction with the 60S subunit[J]. Archives of Biochemistry and Biophysics, 1980, 201(2): 518-526. |
| [40] | Kato Y, Konishi M, Shigyo M, Yoneyama T, Yanagisawa S. Characterization of plant eukaryotic translation initiation factor 6 (eIF6) genes: The essential role in embryogenesis and their differential expression in Arabidopsis and rice[J]. Biochemical and Biophysical Research Communications, 2010, 397(4): 673-678. |
| [41] | Guo H M, Lv J Q, Su X W, Chen L, Ren J S, Liu L P, Ren M X, Liu S, Dai M L, Ren G J, Gao F Y. Rice OseIF6. 1 encodes a eukaryotic translation initiation factor and is essential for the development of grain and anther[J]. Frontiers in Plant Science, 2024, 15: 1366986. |
| [42] | Sparkes I A, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants[J]. Nature Protocols, 2006, 1(4): 2019-2025. |
| [43] | Aslanidis C, Jong P J, Schmitz G. Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products[J]. Genome Research, 1994, 4(3): 172-177. |
| [44] | Raychaudhuri P, Stringer E A, Valenzuela D M, Maitra U. Ribosomal subunit antiassociation activity in rabbit reticulocyte lysates. Evidence for a low molecular weight ribosomal subunit antiassociation protein factor (Mr= 25,000)[J]. Journal of Biological Chemistry, 1984, 259(19): 11930-11935. |
| [45] | Xing Y, Zhang Q. Genetic and molecular bases of rice yield[J]. Annual Review of Plant Biology, 2010, 61:421-442. |
| [46] | 何秀英, 伍时照, 宋美芳, 林贤琛. 水稻籽粒发育和胚乳淀粉粒形成的研究[J]. 广东农业科学, 2000(2): 8-10. |
| He X Y, Wu S Z, Song M F, Lin X C. Research on rice grain development and endosperm starch grain formation[J]. Guangdong Agricultural Science, 2000, (2): 8-10. (in Chinese with English abstract) | |
| [47] | 黄海祥, 钱前. 水稻粒形遗传与长粒型优质粳稻育种进展[J]. 中国水稻科学, 2017, 31(6): 665-672. |
| Huang H X, Qian Q. Progress in Rice Grain Shape Genetics and Breeding of Long Grain High Quality Japonica Rice[J]. Chinese Journal of Rice Science, 2017, 31(6): 665-672. (in Chinese with English abstract) | |
| [48] | 况慧云, 张慧, 许寿增, 黄英金. 水稻产量基因设计育种及其发展前景[J]. 安徽农学通报, 2014, 20(8):42-45. |
| Kuang H Y, Zhang H, Xu S Z, Huang Y J. Design and breeding of rice yield genes and their development prospects[J]. Anhui Agricultural Bulletin, 2014, 20(8): 42-45. (in Chinese with English abstract) | |
| [49] | 李孝琼, 韦宇, 高国庆, 邓国富, 郭嗣斌. 水稻遗传图谱构建及粒形相关性状的QTL定位[J]. 南方农业学报, 2014, 45(7): 1154-1159. |
| Li X Q, Wei Y, Gao G Q, Deng G F, Guo S B. Construction of rice genetic map and QTL mapping of grain shape related traits[J]. Journal of Southern Agriculture, 2014, 45(7): 1154-1159. (in Chinese with English abstract) | |
| [50] | Wood L C, Ashby M N, Grunfeld C, Feingold K R. Cloning of murine translation initiation factor 6 and functional analysis of the homologous sequence YPR016c in Saccharomyces cerevisiae[J]. Journal of Biological Chemistry, 1999, 274(17): 11653-11659. |
| [51] | Guo H M, Cui Y C, Huang L J, Ge L, Xu X R, Xue D Y, Tang M, Zheng J S, Yi Y, Chen L. The RNA binding protein OsLa influences grain and anther development in rice[J]. The Plant Journal, 2022, 110(5): 1397-1414. |
| [1] | 朱鹏, 凌溪铁, 王金彦, 张保龙, 杨郁文, 许轲, 裘实. 机直播条件下不同控草方式对抗除草剂水稻产量和品质差异性研究 [J]. 中国水稻科学, 2025, 39(4): 501-515. |
| [2] | 董立强, 张义凯, 杨铁鑫, 冯莹莹, 马亮, 梁潇, 张玉屏, 李跃东. 北方粳稻密苗机插育秧对秧苗素质及取秧特性的影响 [J]. 中国水稻科学, 2025, 39(4): 516-528. |
| [3] | 周洋, 叶凡, 刘立军. 典型促生微生物提高盐胁迫水稻抗性的研究进展 [J]. 中国水稻科学, 2025, 39(4): 529-542. |
| [4] | 朱建平, 李霞, 李文奇, 许扬, 王芳权, 陶亚军, 蒋彦婕, 陈智慧, 范方军, 杨杰. 水稻粉质胚乳突变体we1的表型分析与基因定位 [J]. 中国水稻科学, 2025, 39(4): 543-551. |
| [5] | 陈嘉乐, 于清涛, 郑琛凡, 汪庆, 谭瑗瑗, 陈百翠, 李承欣, 蒋萌, 舒庆尧. 水稻OsNF-YC10自然变异及其与谷粒宽度的相关性 [J]. 中国水稻科学, 2025, 39(4): 552-562. |
| [6] | 黄福灯, 吴春艳, 郝媛媛, 韩一飞, 张小斌, 孙会锋, 潘刚. 不同氮肥水平下水稻倒二叶叶鞘的转录组分析[J]. 中国水稻科学, 2025, 39(4): 563-571. |
| [7] | 卢椰子, 邱结华, 蒋楠, 寇艳君, 时焕斌. 稻瘟病菌效应子研究进展[J]. 中国水稻科学, 2025, 39(3): 287-294. |
| [8] | 王超瑞, 周宇琨, 温雅, 张瑛, 法晓彤, 肖治林, 张耗. 秸秆还田方式对稻田土壤特性和温室气体排放的影响及其水肥互作调控[J]. 中国水稻科学, 2025, 39(3): 295-305. |
| [9] | 王雅宣, 王新峰, 杨后红, 刘芳, 肖晶, 蔡玉彪, 魏琪, 傅强, 万品俊. 稻飞虱适应水稻抗性机制的研究进展[J]. 中国水稻科学, 2025, 39(3): 306-321. |
| [10] | 黄涛, 魏兆根, 陈玘, 程泽, 刘欣, 王广达, 胡珂鸣, 谢文亚, 陈宗祥, 冯志明, 左示敏. 水稻类病斑突变体lm52的基因克隆及其广谱抗病性分析[J]. 中国水稻科学, 2025, 39(3): 322-330. |
| [11] | 张彬涛, 刘聪聪, 郭明亮, 杨绍华, 吴世强, 郭龙彪, 朱义旺. 水稻OsDR8基因的稻瘟病抗性评价及优异单倍型鉴定[J]. 中国水稻科学, 2025, 39(3): 343-351. |
| [12] | 韦新宇, 曾跃辉, 肖长春, 黄建鸿, 阮宏椿, 杨旺兴, 邹文广, 许旭明. 水稻康丰B抗稻瘟病基因Pi-kf2(t)的克隆与功能验证[J]. 中国水稻科学, 2025, 39(3): 352-364. |
| [13] | 李文奇, 许扬, 王芳权, 朱建平, 陶亚军, 李霞, 范方军, 蒋彦婕, 陈智慧, 杨杰. 广谱抗稻瘟病基因PigmR的KASP标记开发及应用[J]. 中国水稻科学, 2025, 39(3): 365-372. |
| [14] | 韦还和, 汪璐璐, 马唯一, 张翔, 左博源, 耿孝宇, 朱旺, 朱济邹, 孟天瑶, 陈英龙, 高平磊, 许轲, 戴其根. 盐−旱复合胁迫下粳稻品种南粳9108籽粒灌浆特性及其与产量形成的关系[J]. 中国水稻科学, 2025, 39(3): 373-386. |
| [15] | 沈智达, 余秋华, 张斌, 曹玉东, 王少华, 王红飞, 伍永清, 戴志刚, 李小坤. 磷肥施用量对湖北省直播水稻产量、磷素积累及利用率的影响[J]. 中国水稻科学, 2025, 39(3): 399-411. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||