
中国水稻科学 ›› 2025, Vol. 39 ›› Issue (3): 343-351.DOI: 10.16819/j.1001-7216.2025.240509
张彬涛1,2,3, 刘聪聪1,3, 郭明亮3, 杨绍华2, 吴世强5, 郭龙彪4,*( ), 朱义旺2,3,*(
), 朱义旺2,3,*( )
)
收稿日期:2024-05-16
修回日期:2024-07-19
出版日期:2025-05-10
发布日期:2025-05-21
通讯作者:
*email: guolongbiao@caas.cn;zhuyiwang@caas.cn基金资助:
ZHANG Bintao1,2,3, LIU Congcong1,3, GUO Mingliang3, YANG Shaohua2, WU Shiqiang5, GUO Longbiao4,*( ), ZHU Yiwang2,3,*(
), ZHU Yiwang2,3,*( )
)
Received:2024-05-16
Revised:2024-07-19
Online:2025-05-10
Published:2025-05-21
Contact:
*email: guolongbiao@caas.cn;zhuyiwang@caas.cn摘要:
【目的】水稻(Oryza sativa L.)是全球重要的主粮作物,其生产安全受到稻瘟病的严重威胁。抗稻瘟病材料的鉴定及相关抗稻瘟病基因的解析,有助于揭示稻瘟病抗性机制。本研究旨在系统创制和鉴定水稻抗稻瘟病基因OsDR8的遗传材料,并对其进行单倍型和进化分析,以期筛选优异单倍型,探究其亚群分化,为OsDR8基因的抗病育种应用提供理论依据。【方法】创制了OsDR8基因的过表达株系和敲除株系,并用RO1-1和RB22稻瘟菌对过表达株系、纯合敲除株系和对应野生型材料进行稻瘟病抗性鉴定。同时,基于水稻超级泛基因组变异图谱,进行OsDR8基因的单倍型分析和进化分析。【结果】对这些材料进行症状观察和稻瘟病抗性鉴定,发现相比野生型株系,敲除突变体株系病斑显著增大,发病叶面积占比也显著上升;而过表达株系病斑显著减小,发病叶面积占比也显著降低。结合单倍型及进化分析,说明携带优异单倍型Hap2的材料抗病性较强,籼稻亚群中95.52%材料为Hap2单倍型。【结论】OsDR8基因正调控水稻稻瘟病抗性,优异单倍型Hap2可能与籼稻稻瘟病抗性较强有关。
张彬涛, 刘聪聪, 郭明亮, 杨绍华, 吴世强, 郭龙彪, 朱义旺. 水稻OsDR8基因的稻瘟病抗性评价及优异单倍型鉴定[J]. 中国水稻科学, 2025, 39(3): 343-351.
ZHANG Bintao, LIU Congcong, GUO Mingliang, YANG Shaohua, WU Shiqiang, GUO Longbiao, ZHU Yiwang. Evaluation of Blast Resistance and Identification of Superior Haplotype of OsDR8 in Rice[J]. Chinese Journal OF Rice Science, 2025, 39(3): 343-351.
| 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|
| pEGFP-N3 | GTCGCCGTCCAGCTCGACCAG |
| Ubi-F | TTAGCCCTGCCTTCATACGC |
| OsDR8-g1-F | CAGCTTGATGGGGCTGAACCTGA |
| OsDR8-g1-R | AACTCAGGTTCAGCCCCATCAAG |
| OsDR8-g2-F | CAGCTTGACGATGAGGTCCTCGA |
| OsDR8-g2-R | AACTCGAGGACCTCATCGTCAAG |
| OsDR8-F1 | CACTCTCCTCATCCCAGAGCA |
| OsDR8-R1 | CGATCATGCCGATGTCCTGCA |
| OsDR8-F2 | TGTTACTTCTGCAGGGATCCATGGCAGCCATGGCCACC |
| OsDR8-R2 | CTCACCATAGGCCTCACGTGGGCGTCCACGATCTCGCC |
| VKOO5-F | GATGAAGTGGACGGAAGGAAGGAG |
表1 本研究所用引物
Table 1. Primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|
| pEGFP-N3 | GTCGCCGTCCAGCTCGACCAG |
| Ubi-F | TTAGCCCTGCCTTCATACGC |
| OsDR8-g1-F | CAGCTTGATGGGGCTGAACCTGA |
| OsDR8-g1-R | AACTCAGGTTCAGCCCCATCAAG |
| OsDR8-g2-F | CAGCTTGACGATGAGGTCCTCGA |
| OsDR8-g2-R | AACTCGAGGACCTCATCGTCAAG |
| OsDR8-F1 | CACTCTCCTCATCCCAGAGCA |
| OsDR8-R1 | CGATCATGCCGATGTCCTGCA |
| OsDR8-F2 | TGTTACTTCTGCAGGGATCCATGGCAGCCATGGCCACC |
| OsDR8-R2 | CTCACCATAGGCCTCACGTGGGCGTCCACGATCTCGCC |
| VKOO5-F | GATGAAGTGGACGGAAGGAAGGAG |
| 类型 Type | 元件名称 Element | 数目 Number | 功能注释 Functional annotation |
|---|---|---|---|
| 光响应元件 Light-responsive elements | G-Box | 5 | 光响应元件Light-responsive elements |
| GATA-motif | 2 | 光响应元件Light-responsive elements | |
| 激素响应元件 Hormone-responsive elements | ABRE | 4 | ABA响应元件 ABA-responsive element |
| CGTCA-motif | 3 | MeJA响应的顺式作用元件 MeJA-responsive cis-acting element | |
| TGACG-motif | 3 | MeJA响应的顺式作用元件 MeJA-responsive cis-acting element | |
| as-1 | 3 | 水杨酸响应元件 Salicylic acid-responsive element | |
| TCA | 2 | 水杨酸响应元件 Salicylic acid-responsive element | |
| MRE | 3 | 雌激素反应元件 Estrogen response element | |
| ERE | 2 | 雌激素反应元件 Estrogen response element | |
| 结合元件 Binding elements | MYB | 3 | MYB结合元件 MYB binding elements |
| MYC | 4 | MYC结合元件 MYC binding elements | |
| 胁迫相关元件 Stress-responsive elements | LTR | 2 | 低温响应元件 Low-temperature-responsive element |
| GC-motif | 2 | 防御与应激反应响应元件 Defense- and stress-responsive element | |
| TC-rich repeats | 2 | 抗病和胁迫响应元件 Disease- and stress-responsive element | |
| STRE | 5 | 应激响应元件 Stress-responsive element |
表2 OsDR8基因启动子区域顺式作用元件
Table 2. Cis-acting elements in the promoter region of OsDR8
| 类型 Type | 元件名称 Element | 数目 Number | 功能注释 Functional annotation |
|---|---|---|---|
| 光响应元件 Light-responsive elements | G-Box | 5 | 光响应元件Light-responsive elements |
| GATA-motif | 2 | 光响应元件Light-responsive elements | |
| 激素响应元件 Hormone-responsive elements | ABRE | 4 | ABA响应元件 ABA-responsive element |
| CGTCA-motif | 3 | MeJA响应的顺式作用元件 MeJA-responsive cis-acting element | |
| TGACG-motif | 3 | MeJA响应的顺式作用元件 MeJA-responsive cis-acting element | |
| as-1 | 3 | 水杨酸响应元件 Salicylic acid-responsive element | |
| TCA | 2 | 水杨酸响应元件 Salicylic acid-responsive element | |
| MRE | 3 | 雌激素反应元件 Estrogen response element | |
| ERE | 2 | 雌激素反应元件 Estrogen response element | |
| 结合元件 Binding elements | MYB | 3 | MYB结合元件 MYB binding elements |
| MYC | 4 | MYC结合元件 MYC binding elements | |
| 胁迫相关元件 Stress-responsive elements | LTR | 2 | 低温响应元件 Low-temperature-responsive element |
| GC-motif | 2 | 防御与应激反应响应元件 Defense- and stress-responsive element | |
| TC-rich repeats | 2 | 抗病和胁迫响应元件 Disease- and stress-responsive element | |
| STRE | 5 | 应激响应元件 Stress-responsive element |
| 阳性株系 Transgenic-positive lines | 编辑率 Mutation rate(%) | 纯合型 Homozygous | 杂合型 Heterozygous | 野生型 Wild-type |
|---|---|---|---|---|
| 36 | 77.78 | 5 | 23 | 8 |
表3 农杆菌介导的OsDR8基因编辑效率及基因型公布
Table 3. Editing efficiency and genotypic distribution of OsDR8 through Agrobacterium-mediated genetic transformation
| 阳性株系 Transgenic-positive lines | 编辑率 Mutation rate(%) | 纯合型 Homozygous | 杂合型 Heterozygous | 野生型 Wild-type |
|---|---|---|---|---|
| 36 | 77.78 | 5 | 23 | 8 |
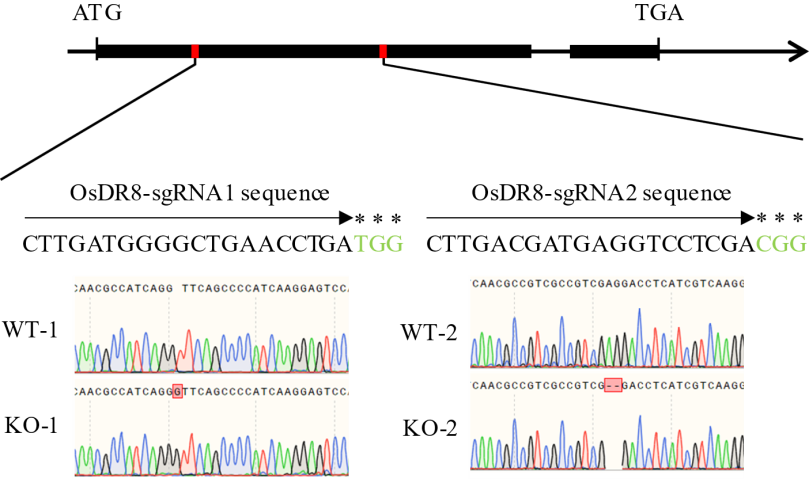
图3 OsDR8敲除突变体鉴定结果 图中的箭头代表gRNA识别序列,星号代表PAM区域。WT-1, WT-2: 野生型; KO-1, KO-2: 敲除株系。
Fig. 3. Identification results of OsDR8 knockout mutants The arrows represent gRNA recognition sequences, and the asterisks denote PAM regions. WT-1 and WT-2, Wild type; KO-1 and KO-2, Knockout mutant lines.
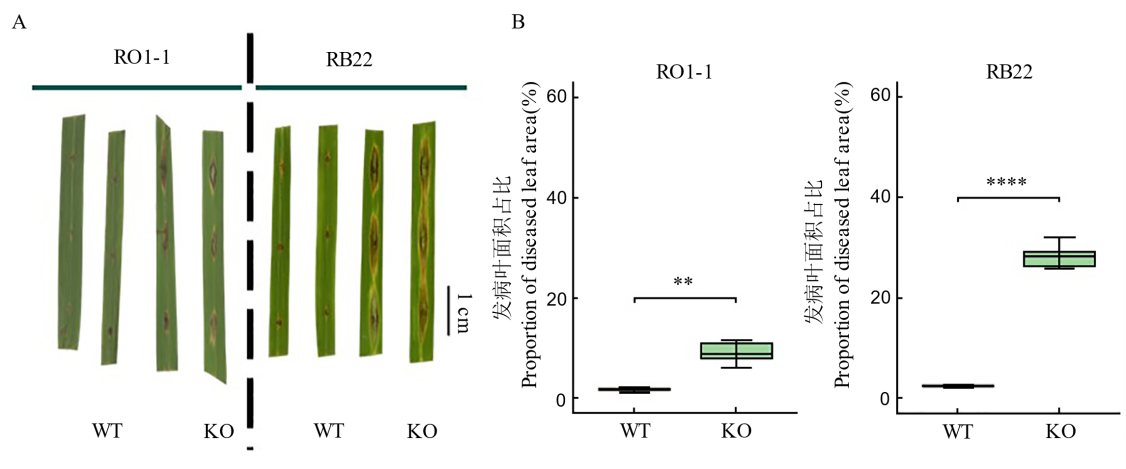
图4 稻瘟病菌侵染后野生型和基因编辑材料发病症状(A)和病叶面积占比(B) **和****分别代表处理间在P<0.01和P<0.0001水平上差异达显著水平。WT: 野生型; KO: OsDR8敲除株系; RO1-1, RB22: 稻瘟病菌株。
Fig. 4. Disease symptoms (A) and proportion of diseased leaf area (B) in wild-type (WT) and gene-edited materials after infection with Magnaporthe oryzae ** and **** represent significant differences between treatments at the levels of P<0.01 and P<0.0001, respectively. WT, Wild type; KO, OsDR8 knockout mutant lines. RO1-1 and RB22, Magnaporthe isolates.
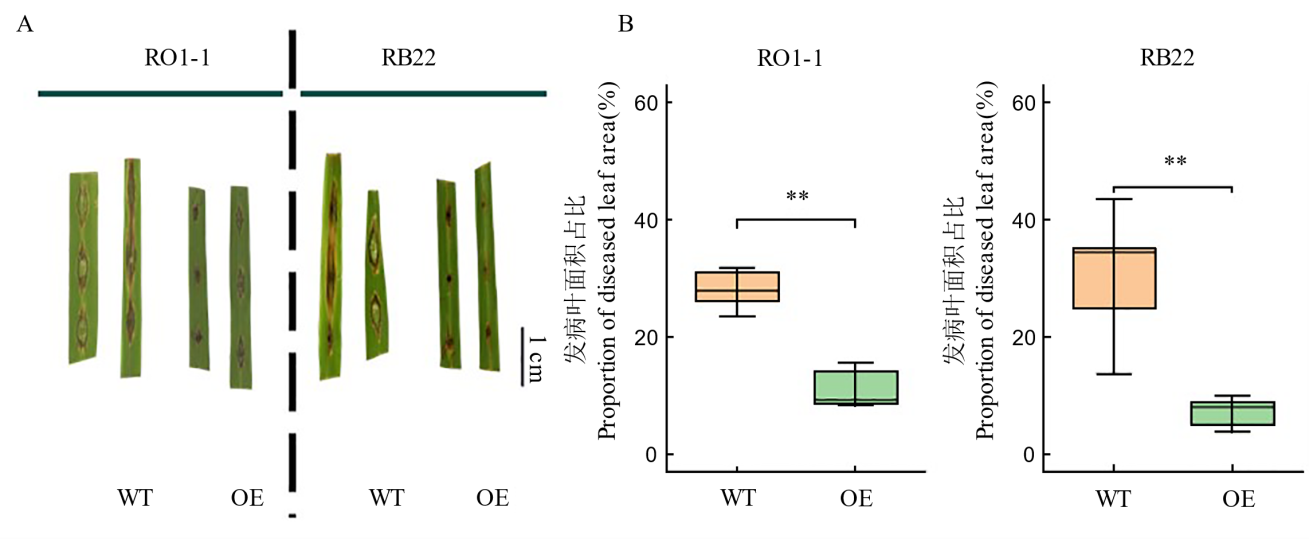
图5 稻瘟病菌侵染后野生型和过表达材料发病症状(A)和发病叶面积占比(B) **代表处理间在P<0.01水平上差异达显著水平。WT: 野生型; OE: OsDR8过表达株系; RO1-1, RB22: 稻瘟病菌株。
Fig. 5. Disease symptoms (A) and proportion of diseased leaf area (B) in wild-type (WT) and overexpression rice materials after infection with Magnaporthe oryzae ** represents significant differences between treatments at the level of P<0.01. WT, Wild type; OE, OsDR8 overexpression lines; RO1-1, RB22, Magnaporthe isolates.
| 单倍型Haplotype | 物理位置Physical position(bp) | 群体数量Number | ||
|---|---|---|---|---|
| 20,724,692 | 20,724,742 | 20,725,760 | ||
| Hap1 | CC | AA | AA | 58 |
| Hap2 | TT | AA | AA | 155 |
| Hap3 | CC | AA | GG | 5 |
| Hap4 | CT | AA | AA | 8 |
| Hap5 | CC | GG | AA | 4 |
表4 OsDR8基因的单倍型分析
Table 4. Haplotype analysis of OsDR8
| 单倍型Haplotype | 物理位置Physical position(bp) | 群体数量Number | ||
|---|---|---|---|---|
| 20,724,692 | 20,724,742 | 20,725,760 | ||
| Hap1 | CC | AA | AA | 58 |
| Hap2 | TT | AA | AA | 155 |
| Hap3 | CC | AA | GG | 5 |
| Hap4 | CT | AA | AA | 8 |
| Hap5 | CC | GG | AA | 4 |
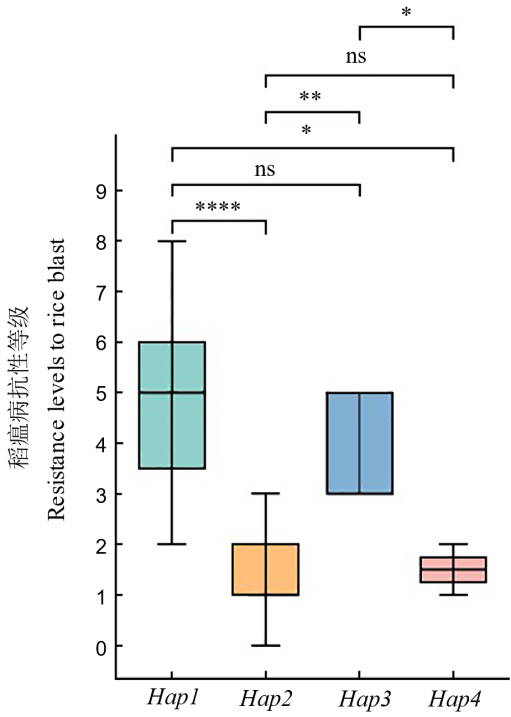
图6 OsDR8基因的不同单倍型对水稻稻瘟病抗性的影响 ns、*、**和****分别代表单倍型间差异不显著、在P<0.05、P<0.01和P<0.0001水平差异达显著水平。
Fig. 6. Effect of haplotypes of OsDR8 on rice blast resistance ns, *, **, and **** in the figure represent non-significant difference between haplotypes and significant differences at the levels of P<0.05, P<0.01, and P<0.0001, respectively.
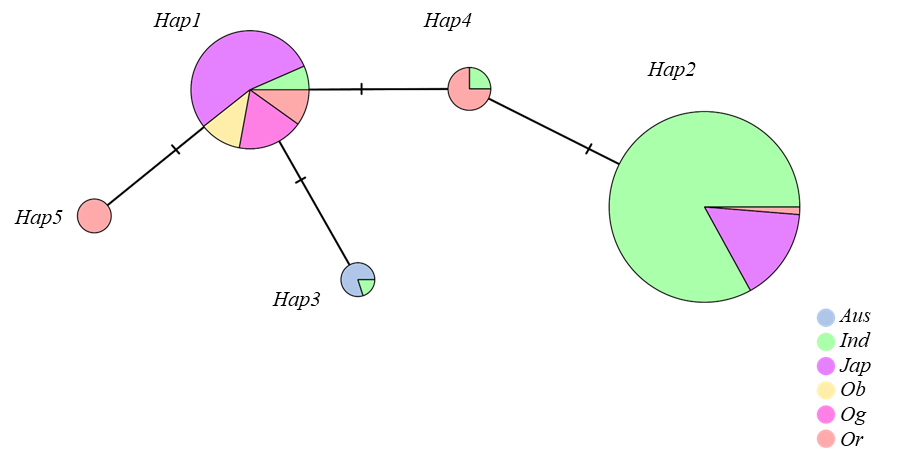
图7 OsDR8基因的单倍型进化分析 Aus、Ind、Jap、Ob、Og、Or分别指代Oryza sativa aus、O. sativa indica、O. sativa japonica、O. barthii、O. glaberrima、O. rufipogon。
Fig. 7. Haplotype evolutionary analysis of OsDR8 In the figure, Aus, Ind, Jap, Ob, Og, and Or refer to Oryza sativa aus, O. sativa indica, O. sativa japonica, O. barthii, O. glaberrima, and O. rufipogon, respectively.
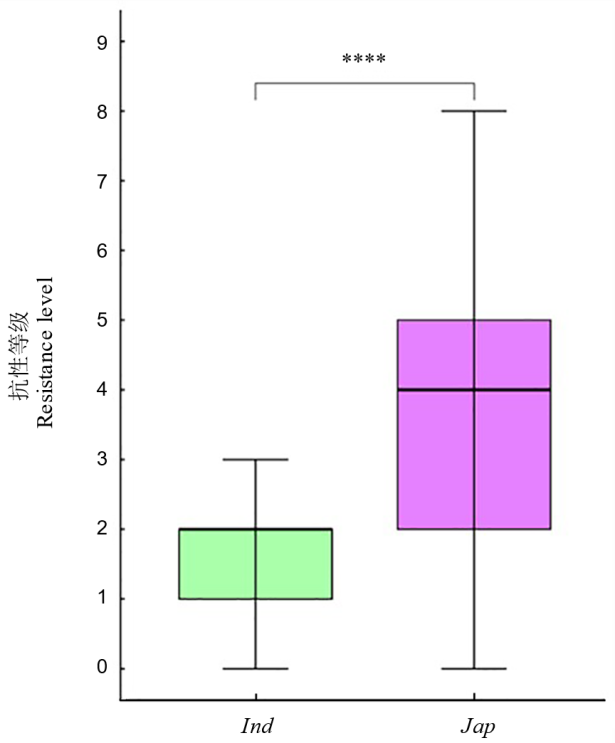
图9 亚洲栽培稻不同亚群的稻瘟病抗性等级差异 ****代表亚群间在P<0.0001水平差异达显著水平。
Fig. 9. Differences in resistance levels to rice blast among different subgroups of Asian cultivated rice **** in the figure represents significant differences between subgroups at the level of P<0.0001.
| [1] | Skamnioti P, Gurr S J. Against the grain: Safeguarding rice from rice blast disease[J]. Trends in Biotechnology, 2009, 27(3): 141-150. |
| [2] | Jones J D G, Dangl J L. The plant immune system[J]. Nature, 2006, 444(7117): 323-329. |
| [3] | Zhou J M, Zhang Y. Plant immunity: Danger perception and signaling[J]. Cell, 2020, 181(5): 978-989. |
| [4] | Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou J M, He S Y, Xin X F. Pattern-recognition receptors are required for NLR-mediated plant immunity[J]. Nature, 2021, 592(7852): 105-109. |
| [5] | Ngou B P M, Ahn H K, Ding P, Jones J D G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors[J]. Nature, 2021, 592(7852): 110-115. |
| [6] | Wang J, Song W, Chai J. Structure, biochemical function, and signaling mechanism of plant NLRs[J]. Molecular Plant, 2023, 16(1): 75-95. |
| [7] | Ding L N, Li Y T, Wu Y Z, Li T, Geng R, Cao J, Zhang W, Tan X L. Plant disease resistance-related signaling pathways: Recent progress and future prospects[J]. International Journal of Molecular Sciences, 2022, 23(24): 16200. |
| [8] | Dong W, Stockwell V O, Goyer A. Enhancement of thiamin content in Arabidopsis thaliana by metabolic engineering[J]. Plant and Cell Physiology, 2015, 56(12): 2285-2296. |
| [9] | Boubakri H, Gargouri M, Mliki A, Brini F, Chong J, Jbara M J P. Vitamins for enhancing plant resistance[J]. Planta, 2016, 244(3): 529-543. |
| [10] | Rapala-Kozik M, Wolak N, Kujda M, Banas A K. The upregulation of thiamine (vitamin B1) biosynthesis in Arabidopsis thaliana seedlings under salt and osmotic stress conditions is mediated by abscisic acid at the early stages of this stress response[J]. BMC Plant Biology, 2012, 12(1): 2. |
| [11] | Ahn I P, Kim S, Lee Y H. Vitamin B1 functions as an activator of plant disease resistance[J]. Plant Physiology, 2005, 138(3): 1505-1515. |
| [12] | Ahn I P, Kim S, Lee Y H, Suh S C. Vitamin B1-induced priming is dependent on hydrogen peroxide and the NPR1 gene in Arabidopsis[J]. Plant Physiology, 2007, 143(2): 838-848. |
| [13] | Bahuguna R N, Joshi R, Shukla A, Pandey M, Kumar J. Thiamine primed defense provides reliable alternative to systemic fungicide carbendazim against sheath blight disease in rice (Oryza sativa L.)[J]. Plant Physiology and Biochemistry, 2012, 57: 159-167. |
| [14] | Wang G, Ding X, Yuan M, Qiu D, Li X, Xu C, Wang S. Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation[J]. Plant Molecular Biology, 2006, 60(3): 437-449. |
| [15] | Xie X, Ma X, Zhu Q, Zeng D, Li G, Liu Y G. CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing[J]. Molecular Plant, 2017, 10(9): 1246-1249. |
| [16] | Shang L, He W, Wang T, Yang Y, Xu Q, Zhao X, Yang L, Zhang H, Li X, Lü Y, Chen W, Cao S, Wang X, Zhang B, Liu X, Yu X, He H, Wei H, Leng Y, Shi C, Guo M, Zhang Z, Zhang B, Yuan Q, Qian H, Cao X, Cui Y, Zhang Q, Dai X, Liu C, Guo L, Zhou Y, Zheng X, Ruan J, Cheng Z, Pan W, Qian Q. A complete assembly of the rice Nipponbare reference genome[J]. Molecular Plant, 2023, 16(8): 1232-1236. |
| [17] | Shang L, Li X, He H, Yuan Q, Song Y, Wei Z, Lin H, Hu M, Zhao F, Zhang C, Li Y, Gao H, Wang T, Liu X, Zhang H, Zhang Y, Cao S, Yu X, Zhang B, Zhang Y, Tan Y, Qin M, Ai C, Yang Y, Zhang B, Hu Z, Wang H, Lü Y, Wang Y, Ma J, Wang Q, Lu H, Wu Z, Liu S, Sun Z, Zhang H, Guo L, Li Z, Zhou Y, Li J, Zhu Z, Xiong G, Ruan J, Qian Q. A super pan-genomic landscape of rice[J]. Cell Research, 2022, 32(10): 878-896. |
| [18] | Cingolani P, Platts A, Wang L L, Coon M, Nguyen T, Wang L, Land S J, Lu X, Ruden D M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila Melanogaster strain w1118; iso-2; iso-3[J]. Fly, 2012, 6(2): 80-92. |
| [19] | Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models[J]. Biometrical Journal, 2008, 50(3): 346-363. |
| [20] | Paradis E. PEGAS: An R package for population genetics with an integrated-modular approach[J]. Bioinformatics, 2010, 26(3): 419-420. |
| [21] | Leigh J W, Bryant D. PopART: Full-feature software for haplotype network construction[J]. Methods in Ecology and Evolution, 2015, 6(9): 1110-1116. |
| [22] | Katoh K, Standley D M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability[J]. Molecular Biology and Evolution, 2013, 30(4): 772-780. |
| [23] | Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11[J]. Molecular Biology and Evolution, 2021, 38(7): 3022-3027. |
| [24] | Ogata Y, Kimura N, Sano R. Gcorn plant: A database for retrieving functional and evolutionary traits of plant genes[J]. Plant Physiology, 2019, 180(2): 732-742. |
| [25] | Chow C N, Lee T Y, Hung Y C, Li G Z, Tseng K C, Liu Y H, Kuo P L, Zheng H Q, Chang W C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants[J]. Nucleic Acids Research, 2019, 47(D1): D1115-D1163. |
| [26] | Liu M H, Kang H, Xu Y, Peng Y, Wang D, Gao L, Wang X, Ning Y, Wu J, Liu W, Li C, Liu B, Wang G L. Genome-wide association study identifies an NLR gene that confers partial resistance to Magnaporthe oryzae in rice[J]. Plant Biotechnology Journal, 2020, 18(6): 1376-1383. |
| [1] | 朱鹏, 凌溪铁, 王金彦, 张保龙, 杨郁文, 许轲, 裘实. 机直播条件下不同控草方式对抗除草剂水稻产量和品质差异性研究 [J]. 中国水稻科学, 2025, 39(4): 501-515. |
| [2] | 董立强, 张义凯, 杨铁鑫, 冯莹莹, 马亮, 梁潇, 张玉屏, 李跃东. 北方粳稻密苗机插育秧对秧苗素质及取秧特性的影响 [J]. 中国水稻科学, 2025, 39(4): 516-528. |
| [3] | 周洋, 叶凡, 刘立军. 典型促生微生物提高盐胁迫水稻抗性的研究进展 [J]. 中国水稻科学, 2025, 39(4): 529-542. |
| [4] | 朱建平, 李霞, 李文奇, 许扬, 王芳权, 陶亚军, 蒋彦婕, 陈智慧, 范方军, 杨杰. 水稻粉质胚乳突变体we1的表型分析与基因定位 [J]. 中国水稻科学, 2025, 39(4): 543-551. |
| [5] | 黄福灯, 吴春艳, 郝媛媛, 韩一飞, 张小斌, 孙会锋, 潘刚. 不同氮肥水平下水稻倒二叶叶鞘的转录组分析[J]. 中国水稻科学, 2025, 39(4): 563-571. |
| [6] | 卢椰子, 邱结华, 蒋楠, 寇艳君, 时焕斌. 稻瘟病菌效应子研究进展[J]. 中国水稻科学, 2025, 39(3): 287-294. |
| [7] | 王超瑞, 周宇琨, 温雅, 张瑛, 法晓彤, 肖治林, 张耗. 秸秆还田方式对稻田土壤特性和温室气体排放的影响及其水肥互作调控[J]. 中国水稻科学, 2025, 39(3): 295-305. |
| [8] | 王雅宣, 王新峰, 杨后红, 刘芳, 肖晶, 蔡玉彪, 魏琪, 傅强, 万品俊. 稻飞虱适应水稻抗性机制的研究进展[J]. 中国水稻科学, 2025, 39(3): 306-321. |
| [9] | 黄涛, 魏兆根, 陈玘, 程泽, 刘欣, 王广达, 胡珂鸣, 谢文亚, 陈宗祥, 冯志明, 左示敏. 水稻类病斑突变体lm52的基因克隆及其广谱抗病性分析[J]. 中国水稻科学, 2025, 39(3): 322-330. |
| [10] | 马顺婷, 胡运高, 高方远, 刘利平, 牟昌铃, 吕建群, 苏相文, 刘松, 梁毓玉, 任光俊, 郭鸿鸣. 水稻真核翻译起始因子OseIF6.2调控粒型的功能研究[J]. 中国水稻科学, 2025, 39(3): 331-342. |
| [11] | 韦新宇, 曾跃辉, 肖长春, 黄建鸿, 阮宏椿, 杨旺兴, 邹文广, 许旭明. 水稻康丰B抗稻瘟病基因Pi-kf2(t)的克隆与功能验证[J]. 中国水稻科学, 2025, 39(3): 352-364. |
| [12] | 李文奇, 许扬, 王芳权, 朱建平, 陶亚军, 李霞, 范方军, 蒋彦婕, 陈智慧, 杨杰. 广谱抗稻瘟病基因PigmR的KASP标记开发及应用[J]. 中国水稻科学, 2025, 39(3): 365-372. |
| [13] | 韦还和, 汪璐璐, 马唯一, 张翔, 左博源, 耿孝宇, 朱旺, 朱济邹, 孟天瑶, 陈英龙, 高平磊, 许轲, 戴其根. 盐−旱复合胁迫下粳稻品种南粳9108籽粒灌浆特性及其与产量形成的关系[J]. 中国水稻科学, 2025, 39(3): 373-386. |
| [14] | 沈智达, 余秋华, 张斌, 曹玉东, 王少华, 王红飞, 伍永清, 戴志刚, 李小坤. 磷肥施用量对湖北省直播水稻产量、磷素积累及利用率的影响[J]. 中国水稻科学, 2025, 39(3): 399-411. |
| [15] | 何勇, 张诗骞, 王志成, 詹逍康, 丁一可, 刘晓瑞, 马素素, 田志宏. 印度梨形孢与复合肥组合施用对水稻机插秧秧苗素质的影响[J]. 中国水稻科学, 2025, 39(3): 412-422. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||