
Chinese Journal OF Rice Science ›› 2020, Vol. 34 ›› Issue (5): 406-412.DOI: 10.16819/j.1001-7216.2020.0104
• Research Papers • Previous Articles Next Articles
Shanbin XU1, Hongliang ZHENG1, Lifeng LIU3, Qingyun BU2, Xiufeng Li2,*( ), Detang ZOU1,*(
), Detang ZOU1,*( )
)
Received:2020-01-10
Revised:2020-04-16
Online:2020-09-10
Published:2020-09-10
Contact:
Xiufeng Li, Detang ZOU
徐善斌1, 郑洪亮1, 刘利锋3, 卜庆云2, 李秀峰2,*( ), 邹德堂1,*(
), 邹德堂1,*( )
)
通讯作者:
李秀峰,邹德堂
基金资助:CLC Number:
Shanbin XU, Hongliang ZHENG, Lifeng LIU, Qingyun BU, Xiufeng Li, Detang ZOU. Improvement of Grain Shape and Fragrance by Using CRISPR/Cas9 System[J]. Chinese Journal OF Rice Science, 2020, 34(5): 406-412.
徐善斌, 郑洪亮, 刘利锋, 卜庆云, 李秀峰, 邹德堂. 利用CRISPR/Cas9技术高效创制长粒香型水稻[J]. 中国水稻科学, 2020, 34(5): 406-412.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2020.0104
| 引物名称 Primer name | 引物序列(5'-3') Primer sequence(5'-3') |
|---|---|
| B1-F | GCCGAGTGACATGGCAATGGCGG |
| B1-R | AAACCCGCCATTGCCATGTCACT |
| B2-F | GCCGCGATTGCTTCCTGCTCGGTT |
| B2-R | AAACAACCGAGCAGGAAGCAATCG |
| B3-F | GCCGCAAGTACCTCCGCGCAATCG |
| B3-R | AAACCGATTGCGCGGAGGTACTTG |
| U-F | CTCCGTTTTACCTGTGGAATCG |
| gRNA-R | CGGAGGAAAATTCCATCCAC |
| B1' | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| B2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| B2' | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| B3 | AGCGTGGGTCTCGTCTTGGTCCATCCACTCCAAGCTC |
| B3' | TTCAGAGGTCTCTAAGACACTGGAATCGGCAGCAAAGG |
| BL | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| Seq1-F | TTCAAAGCAAAGCACCAAGC |
| Seq1-R | GCTGGGGAAAACTTACAATG |
| Seq2-F | TCCACTCGACTCCTCACTCA |
| Seq2-R | GATTAGCGTAGGCCCTCACG |
| Seq3-F | ATCCATCTCCGTATCTCT |
| Seq3-R | GGTAGTCACCACCCTA |
| Hyg-F | ACGGTGTCGTCCATCACAGTTTGCC |
| Hyg-R | TTCCGGAAGTGCTTGACATTGGGGA |
| NOFS | GCGGTGTCATCTATGTTACTAG |
| M13F-47 | CGCCAGGGTTTTCCCAGTCACGAC |
Table 1 Primers used in this research.
| 引物名称 Primer name | 引物序列(5'-3') Primer sequence(5'-3') |
|---|---|
| B1-F | GCCGAGTGACATGGCAATGGCGG |
| B1-R | AAACCCGCCATTGCCATGTCACT |
| B2-F | GCCGCGATTGCTTCCTGCTCGGTT |
| B2-R | AAACAACCGAGCAGGAAGCAATCG |
| B3-F | GCCGCAAGTACCTCCGCGCAATCG |
| B3-R | AAACCGATTGCGCGGAGGTACTTG |
| U-F | CTCCGTTTTACCTGTGGAATCG |
| gRNA-R | CGGAGGAAAATTCCATCCAC |
| B1' | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| B2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| B2' | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| B3 | AGCGTGGGTCTCGTCTTGGTCCATCCACTCCAAGCTC |
| B3' | TTCAGAGGTCTCTAAGACACTGGAATCGGCAGCAAAGG |
| BL | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| Seq1-F | TTCAAAGCAAAGCACCAAGC |
| Seq1-R | GCTGGGGAAAACTTACAATG |
| Seq2-F | TCCACTCGACTCCTCACTCA |
| Seq2-R | GATTAGCGTAGGCCCTCACG |
| Seq3-F | ATCCATCTCCGTATCTCT |
| Seq3-R | GGTAGTCACCACCCTA |
| Hyg-F | ACGGTGTCGTCCATCACAGTTTGCC |
| Hyg-R | TTCCGGAAGTGCTTGACATTGGGGA |
| NOFS | GCGGTGTCATCTATGTTACTAG |
| M13F-47 | CGCCAGGGTTTTCCCAGTCACGAC |
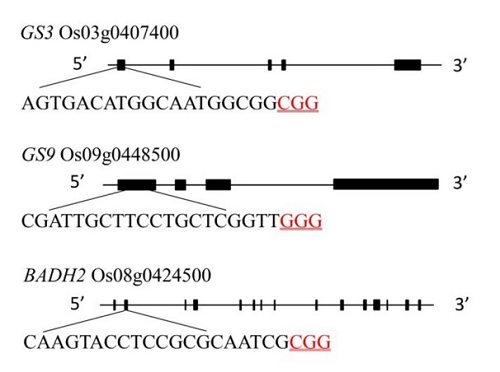
Fig. 1. Gene structure and target site of GS3, GS9 and Badh2. The black sequence is the target sequence and the red underlined sequence is the PAM sequence.
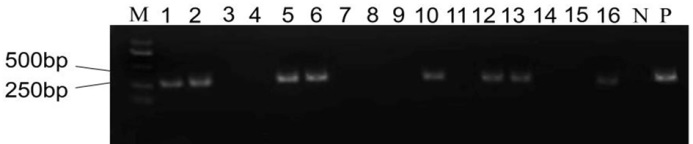
Fig. 3. Transgenic detection of T0 generation plants. M, DM2000 DNA marker; N, Negative control; P, Positive control; Lanes 1-16, T0 generation plants.
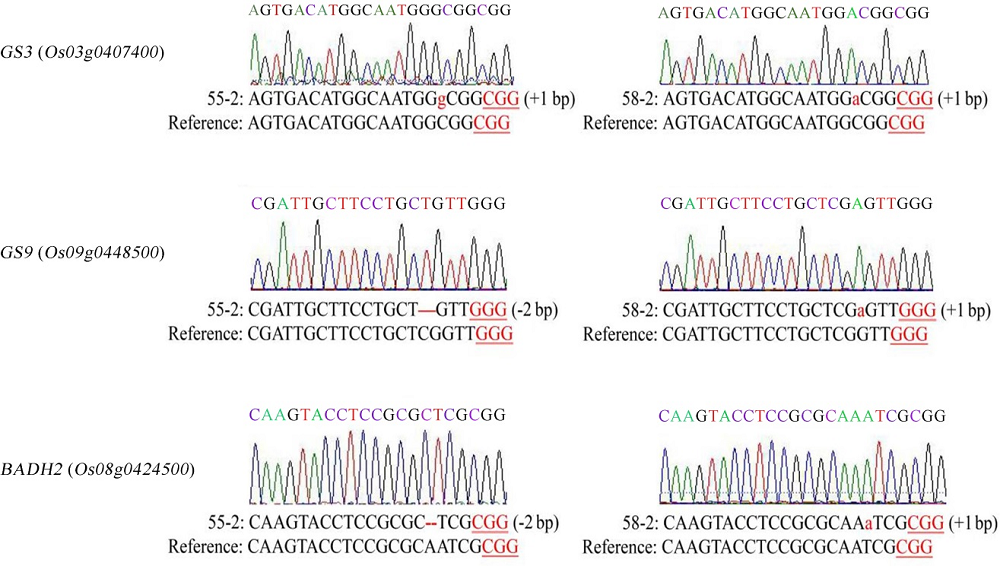
Fig. 4. Sequencing result of homozygous plants with triple gene mutation. Red lowercase letters represent 1 bp insertions and the deleted sequences are shown by red hyphens.
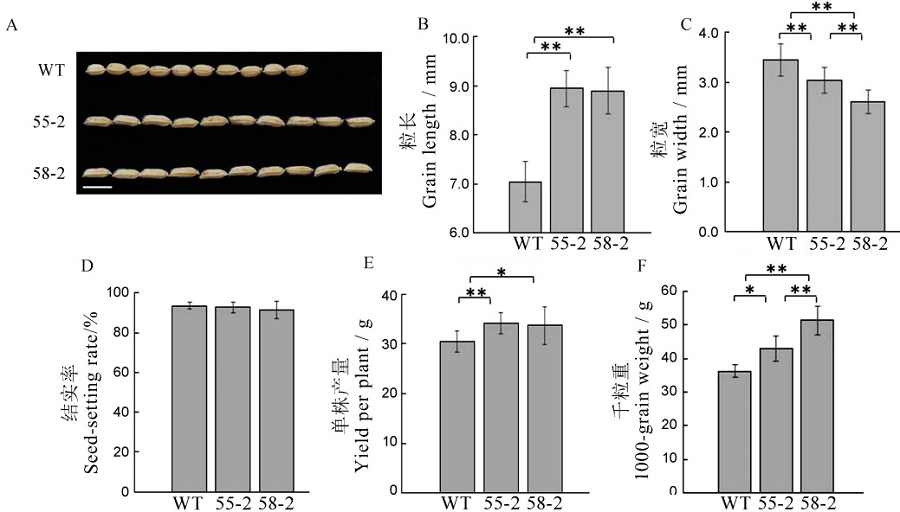
Fig. 5. Agronomic traits of T2 generation plants. Values are shown as mean ± SD.*,**Significantly different at 0.05 and 0.01 levels by t-test, respectively.
| [1] | 郭韬, 余泓, 邱杰, 李家洋, 韩斌, 林鸿宣. 中国水稻遗传学研究进展与分子设计育种[J]. 中国科学: 生命科学, 2019, 49(10): 1185-1212. |
| Guo T, Yu H, Qiu J, Li J Y, Han B, Lin H X.Advances in rice genetics and breeding by molecular design in China[J]. Science in China: Life Sciences, 2019, 49(10): 1185-1212. (in Chinese with English abstract) | |
| [2] | Wang M G, Mao Y F, Lu Y M, Wang Z D, Tao X P, Zhu J K.Multiplex gene editing in rice with simplified CRISPR-Cpf1 and CRISPR-Cas9 systems[J]. Journal of Integrative Plant Biology, 2018, 60(8): 626-631. |
| [3] | Cai Y P, Chen L, Liu X J, Guo C, Sun S, Wu C X, Jiang B J, Han T F, Hou W S.CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean[J]. Plant Biotechnology Journal, 2018, 16(1): 176-185. |
| [4] | Qi W W, Zhu T, Tian Z R, Li C B, Zhang W, Song R T.High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize[J/OL].BMC Biotechnology, 2016, 16(1): 58. |
| [5] | Zhang S J, Zhang R Z, Song G Q, Gao J, Li W, Han X D, Chen M L, Li Y L, Li G Y.Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat[J/OL].BMC Plant Biology, 2018, 18(1): 302. |
| [6] | Wang P C, Zhang J, Sun L, Ma Y Z, Xu J, Liang S J, Deng J W, Tan J F, Zhang Q H, Tu L L, Daniell H, Jin S X, Zhang X L.High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system[J]. Plant Biotechnology Journal, 2018, 16(1): 137-150. |
| [7] | Liu G Q, Li J Q, Godwin I D.Genome editing by CRISPR/Cas9 in sorghum through biolistic bombardment[J]. Methods in Molecular Biology, 2019, 1931: 169-183. |
| [8] | Samanta M K, Dey A, Gayen S.CRISPR/Cas9: An advanced tool for editing plant genomes[J]. Transgenic Research, 2016, 25(5): 561-573. |
| [9] | Ji X, Zhang H W, Zhang Y, Wang Y P, Gao C X.Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants[J/OL].Nature Plants, 2015, 1: 15144. |
| [10] | Hsu P D, Lander E S, Zhang F.Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6): 1262-1278. |
| [11] | Smih F, Rouet P, Romanienko P J, Jasin M.Double strand breaks at the target locus stimulate gene targeting in embryonic stem cells[J]. Nucleic Acids Research, 1995, 23(24): 5012-5019. |
| [12] | Song X J, Huang W, Shi M, Zhu M Z, Lin H X.A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase[J]. Nature Genetics, 2007, 39(5): 623-630. |
| [13] | Fan C C, Xing Y Z, Mao H L, Lu T T, Han B, Xu C G, Li X H, Zhang Q F.GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein[J]. Theoretical and Applied Genetics, 2006, 112(6): 1164-1171. |
| [14] | Zhao D S, Li Q F, Zhang C Q, Zhang C, Yang Q Q, Pan L X, Ren X Y, Lu J, Gu M H, Liu Q Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality[J/OL]. Nature Communications, 2018, 9(1): 1240. |
| [15] | Liu J F, Chen J, Zheng X M, Wu F Q, Lin Q B, Heng Y Q, Tian P, Cheng Z J, Yu X W, Zhou K N, Zhang X, Guo X P, Wang J L, Wang H Y, Wan J M.GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice[J/OL].Nature Plants, 2017, 3: 17043. |
| [16] | Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B I, Onishi A, Miyagawa H, Katoh E.Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield[J]. Nature Genetics, 2013, 45(6): 707-711. |
| [17] | Che R H, Tong H N, Shi B H, Liu Y Q, Fang S R, Liu D P, Xiao Y H, Hu B, Liu L C, Wang H R, Zhao M F, Chu C C. Control of grain size and rice yield by GL2-mediated brassinosteroid responses[J/OL].Nature Plants, 2015, 2: 15195. |
| [18] | Qi P, Lin Y S, Song X J, Shen J B, Huang W, Shan J X, Zhu M Z, Jiang L, Gao J P, Lin H X.The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3[J]. Cell Research, 2012, 22(12): 1666-1680. |
| [19] | Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu C J, Li X H, Xiao J H, He Y Q, Zhang Q F.Natural variation in GS5 plays an important role in regulating grain size and yield in rice[J]. Nature Genetics, 2011, 43(12): 1266-1269. |
| [20] | Wang Y X, Xiong G S, Hu J, Jiang L, Yu H, Xu J, Fang Y X, Zeng L J, Xu E B, Xu J, Ye W J, Meng X B, Liu R F, Chen H Q, Jing Y H, Wang Y H, Zhu X D, Li J Y, Qian Q.Copy number variation at the GL7 locus contributes to grain size diversity in rice[J]. Nature Genetics, 2015, 47(8): 944-948. |
| [21] | Si L Z, Chen J Y, Huang X H, Gong H, Luo J H, Hou Q Q, Zhou T Y, Lu T T, Zhu J J, Shang G Y, Chen E W, Gong C X, Zhao Q, Jing Y F, Zhao Y, Li Y, Cui L L, Fan D L, Lu Y Q, Weng Q J, Wang Y C, Zhan Q L, Liu K Y, Wei X H, An K, An G, Han B.OsSPL13 controls grain size in cultivated rice[J]. Nature Genetics, 2016, 48(4): 447-456. |
| [22] | Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D.Control of grain size, shape and quality by OsSPL16 in rice[J]. Nature Genetics, 2012, 44(8): 950-954. |
| [23] | Li M R, Li X X, Zhou Z J, Wu P Z, Fang M C, Pan X P, Lin Q P, Luo W B, Wu G J, Li H Q.Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system[J/OL].Frontiers in Plant Science, 2016, 7: 377. |
| [24] | Sakthivel K, Sundaram R M, Rani N S, Balachandran S M, Neeraja C N.Genetic and molecular basis of fragrance in rice[J]. Biotechnology Advances, 2009, 27(4): 468-473. |
| [25] | Chen S H, Yang Y, Shi W W, Ji Q, He F, Zhang Z D, Cheng Z K, Liu X N, Xu M L.Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance[J]. Plant Cell, 2008, 20(7): 1850-1861. |
| [26] | Shan Q W, Zhang Y, Chen K L, Zhang K, Gao C X.Creation of fragrant rice by targeted knockout of the OsBadh2 gene using TALEN technology[J]. Plant Biotechnology Journal, 2015, 13(6): 791-800. |
| [27] | 孙慧宇, 宋佳, 王敬国, 刘化龙, 孙健, 莫天宇, 徐善斌, 郑洪亮, 邹德堂. 利用CRISPR/Cas9技术编辑Badh2基因改良粳稻香味[J]. 华北农学报, 2019, 34(4): 1-8. |
| Sun H Y, Song J, Wang J G, Liu H L, Sun J, Mo T Y, Xu S B, Zheng H L, Zou D T.Editing Badh2 gene to improve the fragrance of japonica rice by CRISPR/Cas9 technology[J]. Acta Agricultuae Boreali-Sinica, 2019, 34(4): 1-8. (in Chinese with English abstract) | |
| [28] | 曾栋昌, 马兴亮, 谢先荣, 祝钦泷, 刘耀光. 植物CRISPR/Cas9多基因编辑载体构建和突变分析的操作方法[J]. 中国科学: 生命科学, 2018, 48(7): 783-794. |
| Zeng D C, Ma X L, Xie X R, Zhu Q L, Liu Y G.A protocol for CRISPR/Cas9-based multi-gene editing and sequence decoding of mutant sites in plants[J]. Science in China: Life Sciences, 2018, 48(7): 783-794. (in Chinese with English abstract) | |
| [29] | Xie X R, Ma X, Zhu Q L, Zeng D C, Li G S, Liu Y G, CRISPR-GE: A convenient software toolkit for CRISPR- based genome editing[J]. Molecular Plant, 2017, 10(9): 1246-1249. |
| [30] | Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G.A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants[J]. Molecular Plant, 2015, 8(8): 1274-1284. |
| [31] | Liu W Z, Xie X R, Ma X L, Li J, Chen J H, Liu Y G.DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations[J]. Molecular Plant, 2015, 8(9): 1431-1433. |
| [32] | Zhang Y C, Li S M, Xue S J, Yang S H, Huang J, Wang L.Phylogenetic and CRISPR/Cas9 studies in deciphering the evolutionary trajectory and phenotypic impacts of rice ERECTA genes[J/OL]. Frontiers in Plant Science, 2018, 9: 473. |
| [33] | Wang M, Mao Y F, Lu Y M, Wang Z D, Tao X P, Zhu J K.Multiplex gene editing in rice with simplified CRISPR-Cpf1 and CRISPR-Cas9 systems[J]. Journal of Integrative Plant Biology, 2018, 60(8): 626-631. |
| [34] | Zhang J S, Zhang H, Botella J R, Zhu J K.Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties[J]. Journal of Integrative Plant Biology, 2018, 60(5): 369-375. |
| [35] | Wang Y, Geng L Z, Yuan M L, Wei J, Jin C, Li M, Yu K, Zhang Y, Jin H B, Wang E, Chai Z J, Fu X D, Li X G.Deletion of a target gene in indica rice via CRISPR/Cas9[J]. Plant Cell Reports, 2017, 36(8): 1333-1343. |
| [36] | Nan J Z, Feng X M, Wang C, Zhang X H, Wang R S, Liu J X, Yuan Q B, Jiang G Q, Lin S Y.Improving rice grain length through updating the GS3 locus of an elite variety Kongyu 131[J]. Rice, 2018, 11(1): 21. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||