
Chinese Journal OF Rice Science ›› 2024, Vol. 38 ›› Issue (3): 256-265.DOI: 10.16819/j.1001-7216.2024.231012
• Research Papers • Previous Articles Next Articles
YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue*( ), WANG Wenming*(
), WANG Wenming*( )
)
Received:2023-10-30
Revised:2023-12-12
Online:2024-05-10
Published:2024-05-13
Contact:
*email: zhixuezhao@sicau.edu.cn;
j316wenmingwang@163.com
尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学*( ), 王文明*(
), 王文明*( )
)
通讯作者:
*email: zhixuezhao@sicau.edu.cn;
j316wenmingwang@163.com
基金资助:YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens[J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265.
尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2024.231012
| 引物名称 Primer name | 引物序列(5′-3′) Primer sequence(5′-3′) | 用途 Usage | |
|---|---|---|---|
| Psuc2-44-F | AATTCATGAAGGCCTCGACGCTGCTGGCTCTCCTGCCCTTTGCCGCGGCCC | 酵母系统鉴定信号肽功能 Functional validation of the signal peptides using yeast | |
| Psuc2-44-R | TCGAGGGCCGCGGCAAAGGGCAGGAGAGCCAGCAGCGTCGAGGCCTTCATG | ||
| Psuc2-1548-F | AATTCATGAACCCAATCCTGCTTCTCCTCGCCGCATCAGTCCCCGCCCGCAGCC | ||
| Psuc2-1548-R | TCGAGGCTGCGGGCGGGGACTGATGCGGCGAGGAGAAGCAGGATTGGGTTCATG | ||
| Psuc2-1567-F | AATTCATGCAGTTGTCCTACCTTCTCTCGCTCGTCTCCCTTGTGGCCGCTC | ||
| Psuc2-1567-R | TCGAGAGCGGCCACAAGGGAGACGAGCGAGAGAAGGTAGGACAACTGCATG | ||
| Psuc2-1990-F | AATTCATGCGCCCAGCAACCGTCATCTTCGCACTGGCCGTCTCCGTCAAGGCTC | ||
| Psuc2-1990-R | TCGAGAGCCTTGACGGAGACGGCCAGTGCGAAGATGACGGTTGCTGGGCGCATG | ||
| Psuc2-3667-F | AATTCATGAAGTACACTGCTGCTCTCGTCGCCCTGGCTGCTGCCGTCATGGCCC | ||
| Psuc2-3667-R | TCGAGGGCCATGACGGCAGCAGCCAGGGCGACGAGAGCAGCAGTGTACTTCATG | ||
| Psuc2-4213-F | AATTCATGATTCTGCTGCTCCTCTTGCTATGCACGACAACATGTTTAGGAC | ||
| Psuc2-4213-R | TCGAGTCCTAAACATGTTGTCGTGCATAGCAAGAGGAGCAGCAGAATCATG | ||
| Psuc2-4989-F | AATTCATGCACCAGCTCCTTGCACTACTCCTCGCCAGCCTCACCGCTGCCC | ||
| Psuc2-4989-R | TCGAGGGCAGCGGTGAGGCTGGCGAGGAGTAGTGCAAGGAGCTGGTGCATG | ||
| Psuc2-5215-F | AATTCATGAAGTTTATTTCCCTCGTAGCCGCGGCCGGTGTGGCGTATGCCC | ||
| Psuc2-5215-R | TCGAGGGCATACGCCACACCGGCCGCGGCTACGAGGGAAATAAACTTCATG | ||
| Psuc2-6754-F | AATTCATGGCTGCAGCATCAGCCCTTTTCTTCCTCACGGCGGCTGTCTCCTGGACAC | ||
| Psuc2-6754-R | TCGAGTGTCCAGGAGACAGCCGCCGTGAGGAAGAAAAGGGCTGATGCTGCAGCCATG | ||
| Psuc2-8184-F | AATTCATGCAAATCGTCCTCCCTCTCCTGGCCTGCGCAGCCGTGGCTGCCGCCC | ||
| Psuc2-8184-R | TCGAGGGCGGCAGCCACGGCTGCGCAGGCCAGGAGAGGGAGGACGATTTGCATG | ||
| P1300-44-F | CATGAAGGCCTCGACGCTGCTGGCTCTCCTGCCCTTTGCCGCGGCCGGTAC | 烟草瞬时表达鉴定信号肽功能 Functional validation of the signal peptides using N. benthamiana | |
| P1300-44-R | CGGCCGCGGCAAAGGGCAGGAGAGCCAGCAGCGTCGAGGCCTTCATGGTAC | ||
| P1300-1548-F | CATGAACCCAATCCTGCTTCTCCTCGCCGCATCAGTCCCCGCCCGCAGCGGTAC | ||
| P1300-1548-R | CGCTGCGGGCGGGGACTGATGCGGCGAGGAGAAGCAGGATTGGGTTCATGGTAC | ||
| P1300-1567-F | CATGCAGTTGTCCTACCTTCTCTCGCTCGTCTCCCTTGTGGCCGCTGGTAC | ||
| P1300-1567-R | CAGCGGCCACAAGGGAGACGAGCGAGAGAAGGTAGGACAACTGCATGGTAC | ||
| P1300-1990-F | CATGCGCCCAGCAACCGTCATCTTCGCACTGGCCGTCTCCGTCAAGGCTGGTAC | ||
| P1300-1990-R | CAGCCTTGACGGAGACGGCCAGTGCGAAGATGACGGTTGCTGGGCGCATGGTAC | ||
| P1300-3667-F | CATGAAGTACACTGCTGCTCTCGTCGCCCTGGCTGCTGCCGTCATGGCCGGTAC | ||
| P1300-3667-R | CGGCCATGACGGCAGCAGCCAGGGCGACGAGAGCAGCAGTGTACTTCATGGTAC | ||
| P1300-4213-F | CATGATTCTGCTGCTCCTCTTGCTATGCACGACAACATGTTTAGGAGGTAC | ||
| P1300-4213-R | CTCCTAAACATGTTGTCGTGCATAGCAAGAGGAGCAGCAGAATCATGGTAC | ||
| P1300-4989-F | CATGCACCAGCTCCTTGCACTACTCCTCGCCAGCCTCACCGCTGCCGGTAC | ||
| P1300-4989-R | CGGCAGCGGTGAGGCTGGCGAGGAGTAGTGCAAGGAGCTGGTGCATGGTAC | ||
| P1300-5215-F | CATGAAGTTTATTTCCCTCGTAGCCGCGGCCGGTGTGGCGTATGCCGGTAC | ||
| P1300-5215-R | CGGCATACGCCACACCGGCCGCGGCTACGAGGGAAATAAACTTCATGGTAC | ||
| P1300-6754-F | CATGGCTGCAGCATCAGCCCTTTTCTTCCTCACGGCGGCTGTCTCCTGGACAGGTAC | ||
| P1300-6754-R | CTGTCCAGGAGACAGCCGCCGTGAGGAAGAAAAGGGCTGATGCTGCAGCCATGGTAC | ||
| P1300-8184-F | CATGCAAATCGTCCTCCCTCTCCTGGCCTGCGCAGCCGTGGCTGCCGCCGGTAC | ||
| P1300-8184-R | CGGCGGCAGCCACGGCTGCGCAGGCCAGGAGAGGGAGGACGATTTGCATGGTAC | ||
| Uv_Tublin2α-2F/R | ACCAGCTCGTTGAGAACTCG / AATCAGAGTTGAGCTGGCCG | 实时荧光定量PCR RT-qPCR | |
| UV_44-RT-F/R | GCGTTGCCAAGAAGACCAAG / ATTCCAGCAATCACGGCAGA | ||
| UV_1548-RT- F/R | GACAACGGGCAGACAAACTG / CTAGCAATCCCAGCAGTCGT | ||
| UV_1567-RT-F/R | TGCTAGCGCTATCGTTCCTG / GTCGGCATGGTTTTGCAACT | ||
| UV_1990-RT-F/R | ACTCACTTCCAAACACGGCA / GCCTTGTCTTCTCAACACGC | ||
| UV_3667-RT-F/R | GCCTCCTCTGCAATGTGGAA / CTTGAGGCAGTTGAGTCCGA | ||
| UV_4213-RT-F/R | ATGCGAGAACTTGGTCGTGT / AACCAACGCCTCCAACTTGA | ||
| UV_4989-RT-F/R | CCTGCCTCCTTCGTCCAAAT / GGTCGATCTGGCAAAGTGGA | ||
| UV_5215-RT-F/R | TAGCGTACTGCGACGGTAAC / GAAACAGCGCCCAATTCCTG | ||
| UV_6754-RT-F/R | AAATGCGAGGACTAGGGCAC / CTTTTTGCCGGAGGAGGTCT | ||
| UV_8184-RT-F/R | CATCGCTGCGAAGACAAAGG / GCGCCTGTGAACTTTTCCTG | ||
Table 1. Primers used in this study
| 引物名称 Primer name | 引物序列(5′-3′) Primer sequence(5′-3′) | 用途 Usage | |
|---|---|---|---|
| Psuc2-44-F | AATTCATGAAGGCCTCGACGCTGCTGGCTCTCCTGCCCTTTGCCGCGGCCC | 酵母系统鉴定信号肽功能 Functional validation of the signal peptides using yeast | |
| Psuc2-44-R | TCGAGGGCCGCGGCAAAGGGCAGGAGAGCCAGCAGCGTCGAGGCCTTCATG | ||
| Psuc2-1548-F | AATTCATGAACCCAATCCTGCTTCTCCTCGCCGCATCAGTCCCCGCCCGCAGCC | ||
| Psuc2-1548-R | TCGAGGCTGCGGGCGGGGACTGATGCGGCGAGGAGAAGCAGGATTGGGTTCATG | ||
| Psuc2-1567-F | AATTCATGCAGTTGTCCTACCTTCTCTCGCTCGTCTCCCTTGTGGCCGCTC | ||
| Psuc2-1567-R | TCGAGAGCGGCCACAAGGGAGACGAGCGAGAGAAGGTAGGACAACTGCATG | ||
| Psuc2-1990-F | AATTCATGCGCCCAGCAACCGTCATCTTCGCACTGGCCGTCTCCGTCAAGGCTC | ||
| Psuc2-1990-R | TCGAGAGCCTTGACGGAGACGGCCAGTGCGAAGATGACGGTTGCTGGGCGCATG | ||
| Psuc2-3667-F | AATTCATGAAGTACACTGCTGCTCTCGTCGCCCTGGCTGCTGCCGTCATGGCCC | ||
| Psuc2-3667-R | TCGAGGGCCATGACGGCAGCAGCCAGGGCGACGAGAGCAGCAGTGTACTTCATG | ||
| Psuc2-4213-F | AATTCATGATTCTGCTGCTCCTCTTGCTATGCACGACAACATGTTTAGGAC | ||
| Psuc2-4213-R | TCGAGTCCTAAACATGTTGTCGTGCATAGCAAGAGGAGCAGCAGAATCATG | ||
| Psuc2-4989-F | AATTCATGCACCAGCTCCTTGCACTACTCCTCGCCAGCCTCACCGCTGCCC | ||
| Psuc2-4989-R | TCGAGGGCAGCGGTGAGGCTGGCGAGGAGTAGTGCAAGGAGCTGGTGCATG | ||
| Psuc2-5215-F | AATTCATGAAGTTTATTTCCCTCGTAGCCGCGGCCGGTGTGGCGTATGCCC | ||
| Psuc2-5215-R | TCGAGGGCATACGCCACACCGGCCGCGGCTACGAGGGAAATAAACTTCATG | ||
| Psuc2-6754-F | AATTCATGGCTGCAGCATCAGCCCTTTTCTTCCTCACGGCGGCTGTCTCCTGGACAC | ||
| Psuc2-6754-R | TCGAGTGTCCAGGAGACAGCCGCCGTGAGGAAGAAAAGGGCTGATGCTGCAGCCATG | ||
| Psuc2-8184-F | AATTCATGCAAATCGTCCTCCCTCTCCTGGCCTGCGCAGCCGTGGCTGCCGCCC | ||
| Psuc2-8184-R | TCGAGGGCGGCAGCCACGGCTGCGCAGGCCAGGAGAGGGAGGACGATTTGCATG | ||
| P1300-44-F | CATGAAGGCCTCGACGCTGCTGGCTCTCCTGCCCTTTGCCGCGGCCGGTAC | 烟草瞬时表达鉴定信号肽功能 Functional validation of the signal peptides using N. benthamiana | |
| P1300-44-R | CGGCCGCGGCAAAGGGCAGGAGAGCCAGCAGCGTCGAGGCCTTCATGGTAC | ||
| P1300-1548-F | CATGAACCCAATCCTGCTTCTCCTCGCCGCATCAGTCCCCGCCCGCAGCGGTAC | ||
| P1300-1548-R | CGCTGCGGGCGGGGACTGATGCGGCGAGGAGAAGCAGGATTGGGTTCATGGTAC | ||
| P1300-1567-F | CATGCAGTTGTCCTACCTTCTCTCGCTCGTCTCCCTTGTGGCCGCTGGTAC | ||
| P1300-1567-R | CAGCGGCCACAAGGGAGACGAGCGAGAGAAGGTAGGACAACTGCATGGTAC | ||
| P1300-1990-F | CATGCGCCCAGCAACCGTCATCTTCGCACTGGCCGTCTCCGTCAAGGCTGGTAC | ||
| P1300-1990-R | CAGCCTTGACGGAGACGGCCAGTGCGAAGATGACGGTTGCTGGGCGCATGGTAC | ||
| P1300-3667-F | CATGAAGTACACTGCTGCTCTCGTCGCCCTGGCTGCTGCCGTCATGGCCGGTAC | ||
| P1300-3667-R | CGGCCATGACGGCAGCAGCCAGGGCGACGAGAGCAGCAGTGTACTTCATGGTAC | ||
| P1300-4213-F | CATGATTCTGCTGCTCCTCTTGCTATGCACGACAACATGTTTAGGAGGTAC | ||
| P1300-4213-R | CTCCTAAACATGTTGTCGTGCATAGCAAGAGGAGCAGCAGAATCATGGTAC | ||
| P1300-4989-F | CATGCACCAGCTCCTTGCACTACTCCTCGCCAGCCTCACCGCTGCCGGTAC | ||
| P1300-4989-R | CGGCAGCGGTGAGGCTGGCGAGGAGTAGTGCAAGGAGCTGGTGCATGGTAC | ||
| P1300-5215-F | CATGAAGTTTATTTCCCTCGTAGCCGCGGCCGGTGTGGCGTATGCCGGTAC | ||
| P1300-5215-R | CGGCATACGCCACACCGGCCGCGGCTACGAGGGAAATAAACTTCATGGTAC | ||
| P1300-6754-F | CATGGCTGCAGCATCAGCCCTTTTCTTCCTCACGGCGGCTGTCTCCTGGACAGGTAC | ||
| P1300-6754-R | CTGTCCAGGAGACAGCCGCCGTGAGGAAGAAAAGGGCTGATGCTGCAGCCATGGTAC | ||
| P1300-8184-F | CATGCAAATCGTCCTCCCTCTCCTGGCCTGCGCAGCCGTGGCTGCCGCCGGTAC | ||
| P1300-8184-R | CGGCGGCAGCCACGGCTGCGCAGGCCAGGAGAGGGAGGACGATTTGCATGGTAC | ||
| Uv_Tublin2α-2F/R | ACCAGCTCGTTGAGAACTCG / AATCAGAGTTGAGCTGGCCG | 实时荧光定量PCR RT-qPCR | |
| UV_44-RT-F/R | GCGTTGCCAAGAAGACCAAG / ATTCCAGCAATCACGGCAGA | ||
| UV_1548-RT- F/R | GACAACGGGCAGACAAACTG / CTAGCAATCCCAGCAGTCGT | ||
| UV_1567-RT-F/R | TGCTAGCGCTATCGTTCCTG / GTCGGCATGGTTTTGCAACT | ||
| UV_1990-RT-F/R | ACTCACTTCCAAACACGGCA / GCCTTGTCTTCTCAACACGC | ||
| UV_3667-RT-F/R | GCCTCCTCTGCAATGTGGAA / CTTGAGGCAGTTGAGTCCGA | ||
| UV_4213-RT-F/R | ATGCGAGAACTTGGTCGTGT / AACCAACGCCTCCAACTTGA | ||
| UV_4989-RT-F/R | CCTGCCTCCTTCGTCCAAAT / GGTCGATCTGGCAAAGTGGA | ||
| UV_5215-RT-F/R | TAGCGTACTGCGACGGTAAC / GAAACAGCGCCCAATTCCTG | ||
| UV_6754-RT-F/R | AAATGCGAGGACTAGGGCAC / CTTTTTGCCGGAGGAGGTCT | ||
| UV_8184-RT-F/R | CATCGCTGCGAAGACAAAGG / GCGCCTGTGAACTTTTCCTG | ||
| 基因名称 Gene name | 蛋白大小(氨基酸) Length of protein (aa) | 信号肽长度(氨基酸) Length of signal peptide (aa) | 信号肽评分 Signal peptide scoring(Sec/SPI) |
|---|---|---|---|
| UV_44 | 386 | 15 | 0.8501 |
| UV_1548 | 104 | 16 | 0.9929 |
| UV_1567 | 388 | 15 | 0.9722 |
| UV_1990 | 279 | 16 | 0.9845 |
| UV_3667 | 89 | 16 | 0.9995 |
| UV_4213 | 216 | 15 | 0.9642 |
| UV_4989 | 231 | 15 | 0.9924 |
| UV_5215 | 88 | 15 | 0.9981 |
| UV_6754 | 211 | 17 | 0.4707 |
| UV_8184 | 106 | 16 | 0.9982 |
Table 2. Signal peptide prediction of ten effectors from U. virens
| 基因名称 Gene name | 蛋白大小(氨基酸) Length of protein (aa) | 信号肽长度(氨基酸) Length of signal peptide (aa) | 信号肽评分 Signal peptide scoring(Sec/SPI) |
|---|---|---|---|
| UV_44 | 386 | 15 | 0.8501 |
| UV_1548 | 104 | 16 | 0.9929 |
| UV_1567 | 388 | 15 | 0.9722 |
| UV_1990 | 279 | 16 | 0.9845 |
| UV_3667 | 89 | 16 | 0.9995 |
| UV_4213 | 216 | 15 | 0.9642 |
| UV_4989 | 231 | 15 | 0.9924 |
| UV_5215 | 88 | 15 | 0.9981 |
| UV_6754 | 211 | 17 | 0.4707 |
| UV_8184 | 106 | 16 | 0.9982 |
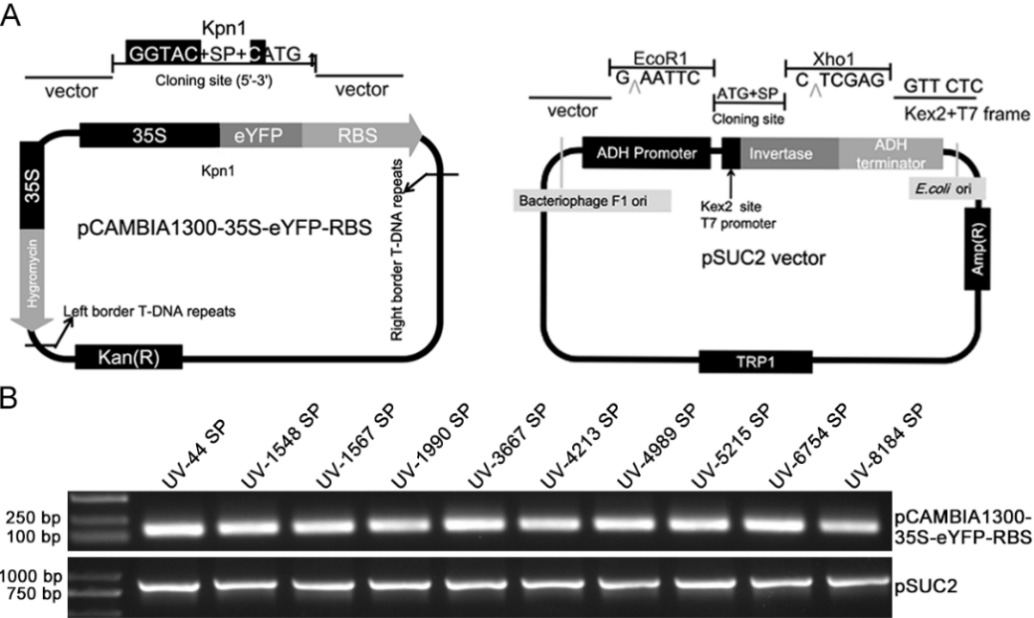
Fig. 1. Construction of vectors and identification of the positive clones A, Diagrams of the pCAMBIA1300-35S-eYFP-RBS and pSUC2 plasmids; B, Positive clones were identified by PCR and agarose gel electrophoresis.
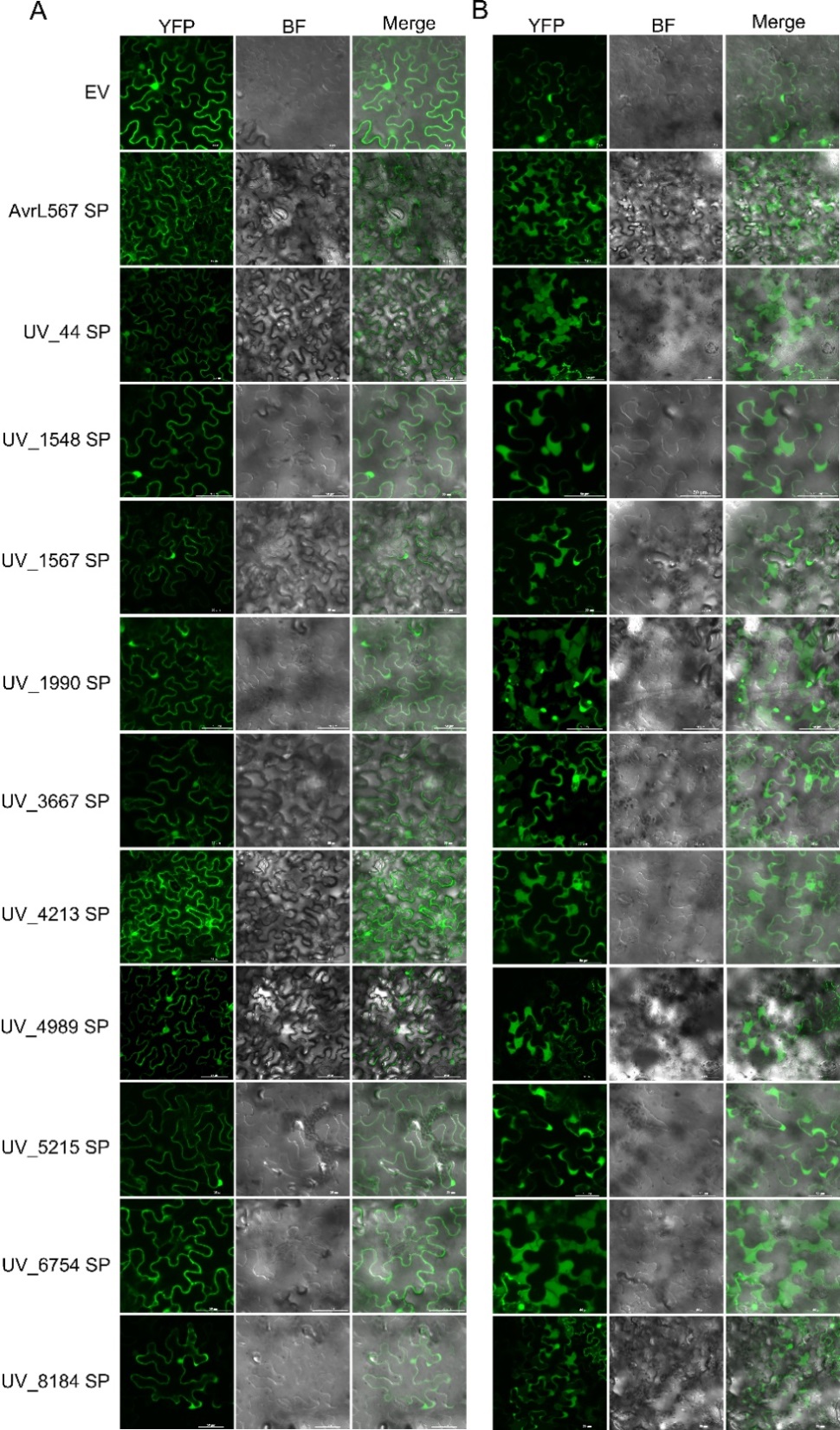
Fig. 2. Subcellular localization of the transiently expressed signal peptides of ten effectors from U. virens in N. benthamiana Subcellular localization of the fusion proteins containing the predicted signal peptide before (A) and after (B) plasmolysis induced by 0.8 mol/L mannitol. The images were captured at 36 h after infiltration. EV was used as a negative control, and AvrL567SP was used as a positive control.

Fig. 3. Functional evaluation of the signal peptides using the yeast invertase assay A, Diagrams showing the sample location on Petri plates; B, C and D, Detection of the secretion of signal peptides by the yeast signal trap assay; E, TTC assay of the signal peptides. pSUC2-Avr1b was used as a positive control. YTK12 and pSUC2-Mg87 were used as the negative controls.
| [1] | Brooks S A, Anders M M, Yeater K M. Effect of cultural management practices on the severity of false smut and kernel smut of rice[J]. Plant Disease, 2009, 93(11): 1202-1208. |
| [2] | Tang Y X, Jin J, Hu D W, Yong M L, Xu Y, He L P. Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets[J]. Plant Pathology, 2013, 62(1): 1-8. |
| [3] | Cooke M C. Some extra-european fungi[J]. Grevillea, 1878, 7: 13-15. |
| [4] | Sun W, Fan J, Fang A, Li Y, Tariqjaveed M, Li D, Hu D, Wang W M. Ustilaginoidea virens: Insights into an emerging rice pathogen[J]. Annual Review of Phytopathology, 2020, 58(1): 363-385. |
| [5] | Fan J, Liu J, Gong Z Y, Xu P Z, Hu X H, Wu J L, Li G B, Yang J, Wang Y Q, Zhou Y F, Li S C, Wang L, Chen X Q. The false smut pathogen Ustilaginoidea virens requires rice stamens for false smut ball formation[J]. Environmental Microbiology, 2020, 22(2): 646-659. |
| [6] | Fan J, Guo X Y, Li L, Huang F, Sun W X, Li Y, Huang Y Y, Xu Y J, Shi J, Zheng A P, Wang W M. Infection of Ustilaginoidea virens intercepts rice seed formation but activates grain-filling-related genes[J]. Journal of Integrative Plant Biology, 2015, 57(6): 577-590. |
| [7] | Fan J, Yang J, Wang Y Q, Li G B, Li Y, Wang W M. Current understanding on Villosiclava virens, a unique flower-infecting fungus causing rice false smut disease[J]. Molecular Plant Pathology, 2016, 17(9): 1321-1330. |
| [8] | Wang B, Liu L, Li Y, Zou J, Li D, Zhao D, Li W, Sun W. Ustilaginoidin D induces hepatotoxicity and behaviour aberrations in zebrafish larvae[J]. Toxicology, 2021, 456: 152786. |
| [9] | Lin X, Bian Y, Mou R, Cao Z, Cao Z, Zhu Z, Chen M. Isolation, identification, and characterization of Ustilaginoidea virens from rice false smut balls with high ustilotoxin production potential[J]. Journal of Basic Microbiology, 2018, 58(8): 670-678. |
| [10] | Rafiqi M, Ellis J G, Ludowici V A, Hardham A R, Dodds P N. Challenges and progress towards understanding the role of effectors in plant-fungal interactions[J]. Current Opinion in Plant Biology, 2012, 15(4): 477-482. |
| [11] | Zhang K, Zhao Z, Zhang Z, Li Y, Li S, Yao N, Hsiang T. Insights into genomic evolution from chromosomal and mitochondrial genomes of Ustilaginoidea virens[J]. Phytopathology Research, 2021, 3(1): 9. |
| [12] | Zhang N, Yang J, Fang A, Wang J, Li D, Li Y, Wang S, Cui F, Yu J, Liu Y, Peng Y L, Sun W. The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region[J]. Molecular Plant Pathology, 2020, 21(4): 445-459. |
| [13] | Zheng X, Fang A, Qiu S, Zhao G, Wang J, Wang S, Wei J, Gao H, Yang J, Mou B, Cui F, Zhang J, Liu J, Sun W. Ustilaginoidea virens secretes a family of phosphatases that stabilize the negative immune regulator OsMPK6 and suppress plant immunity[J]. The Plant Cell, 2022, 34(8): 3088-3109. |
| [14] | Chen X, Duan Y, Qiao F, Liu H, Huang J, Luo C, Chen X, Li G, Xie K, Hsiang T, Zheng L. A secreted fungal effector suppresses rice immunity through host histone hypoacetylation[J]. New Phytologist, 2022, 235(5): 1977. |
| [15] | Li G B, He J X, Wu J L, Wang H, Zhang X, Liu J, Hu X H, Zhu Y, Shen S, Bai Y F, Wang W M, Fan J. Overproduction of OsRACK1A, an effector-targeted scaffold protein promoting OsRBOHB-mediated ROS production, confers rice floral resistance to false smut disease without yield penalty[J]. Molecular Plant, 2022, 15(11): 1790-1806. |
| [16] | Chen X, Li X, Duan Y, Pei Z, Liu H, Yin W, Huang J, Luo C, Chen X, Li G, Xie K, Hsiang T, Zheng L. A secreted fungal subtilase interferes with rice immunity via degradation of SUPPRESSOR OF G2 ALLELE OF skp1[J]. Plant Physiology, 2022, 190(2): 1474-1489. |
| [17] | Owji H, Nezafat N, Negahdaripour M, Hajiebrahimi A, Ghasemi Y. A comprehensive review of signal peptides: Structure, roles, and applications[J]. European Journal of Cell Biology, 2018, 97(6): 422-441. |
| [18] | von Heijne G. Signal sequences: Limits of variation[J]. Journal of Molecular Biology, 1985, 184(1): 99-105. |
| [19] | Rafiqi M, Gan P H P, Ravensdale M, Lawrence G J, Ellis J G, Jones D A, Hardham A R, Dodds P N. Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen[J]. The Plant Cell, 2010, 22(6): 2017-2032. |
| [20] | Yin W, Wang Y, Chen T, Lin Y, Luo C. Functional evaluation of the signal peptides of secreted proteins[J]. Bio-protocol, 2018, 8(9): e2839. |
| [21] | Gu B, Kale S D, Wang Q, Wang D, Pan Q, Cao H, Meng Y, Kang Z, Tyler B M, Shan W. Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells[J]. PLOS ONE, 2011, 6(11): e27217. |
| [22] | Bao J, Wang R, Gao S, Wang Z, Fang Y, Wu L, Wang M. High-Quality genome sequence resource of a rice false smut fungus Ustilaginoidea virens isolate, UV-FJ-1[J]. Phytopathology, 2021, 111(10): 1889-1892. |
| [23] | Zhang Y, Zhang K, Fang A, Han Y, Yang J, Xue M, Bao J, Hu D, Zhou B, Sun X, Li S, Wen M, Yao N, Ma L J, Liu Y, Zhang M, Huang F, Luo C, Zhou L, Li J, Chen Z. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics[J]. Nature Communications, 2014, 5(1): 3849. |
| [24] | Wang H, Yang X, Wei S, Wang Y. Proteomic analysis of mycelial exudates of Ustilaginoidea virens[J]. Pathogens, 2021, 10(3): 364. |
| [25] | Fang A, Han Y, Zhang N, Zhang M, Liu L, Li S, Lu F, Sun W. Identification and characterization of plant cell death-inducing secreted proteins from Ustilaginoidea virens[J]. Molecular Plant-Microbe Interactions, 2016, 29(5): 405-416. |
| [26] | Wei S, Wang Y, Zhou J, Xiang S, Sun W, Peng X, Li J, Hai Y, Wang Y, Li S. The conserved effector UvHrip1 interacts with OsHGW and infection of Ustilaginoidea virens regulates defense- and heading date-related signaling pathway[J]. International Journal of Molecular Sciences, 2020, 21(9): 3376. |
| [27] | Qiu S, Fang A, Zheng X, Wang S, Wang J, Fan J, Sun Z, Gao H, Yang J, Zeng Q, Cui F, Wang W M, Chen J, Sun W. Ustilaginoidea virens nuclear effector SCRE4 suppresses rice immunity via inhibiting expression of a positive immune regulator OsARF17[J]. International Journal of Molecular Sciences, 2022, 23(18): 10527. |
| [28] | Fang A, Gao H, Zhang N, Zheng X, Qiu S, Li Y, Zhou S, Cui F, Sun W. A novel effector gene SCRE2 contributes to full virulence of Ustilaginoidea virens to rice[J]. Frontiers in Microbiology, 2019, 10. |
| [29] | Presti L L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R. Fungal Effectors and plant susceptibility[J]. Annual Review of Plant Biology, 2015, 66(1): 513-545. |
| [30] | Fan J, Du N, Li L, Li G B, Wang Y Q, Zhou Y F, Hu X H, Liu J, Zhao J Q, Li Y, Huang F, Wang W M. A core effector UV_1261 promotes Ustilaginoidea virens infection via spatiotemporally suppressing plant defense[J]. Phytopathology Research, 2019, 1(1): 11. |
| [1] |
WANG Yichen, ZHU Benshun, ZHOU Lei, ZHU Jun, YANG Zhongnan.
Sterility Mechanism of Photoperiod/Thermo-sensitive Genic Male Sterile Lines and Development and Prospects of Two-line Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(5): 463-474. |
| [2] |
XU Yongqiang XU Jun, FENG Baohua, XIAO Jingjing, WANG Danying, ZENG Yuxiang, FU Guanfu.
Research Progress of Pollen Tube Growth in Pistil of Rice and Its Response to Abiotic stress [J]. Chinese Journal OF Rice Science, 2024, 38(5): 495-506. |
| [3] |
HE Yong, LIU Yaowei, XIONG Xiang, ZHU Danchen, WANG Aiqun, MA Lana, WANG Tingbao, ZHANG Jian, LI Jianxiong, TIAN Zhihong.
Creation of Rice Grain Size Mutants by Editing OsOFP30 via CRISPR/Cas9 System [J]. Chinese Journal OF Rice Science, 2024, 38(5): 507-515. |
| [4] |
LÜ Yang, LIU Congcong, YANG Longbo, CAO Xinglan, WANG Yueying, TONG Yi, Mohamed Hazman, QIAN Qian, SHANG Lianguang, GUO Longbiao.
Identification of Candidate Genes for Rice Nitrogen Use Efficiency by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(5): 516-524. |
| [5] |
YANG Hao, HUANG Yanyan, WANG Jian, YI Chunlin, SHI Jun, TAN Chutian, REN Wenrui, WANG Wenming.
Development and Application of Specific Molecular Markers for Eight Rice Blast Resistance Genes in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(5): 525-534. |
| [6] |
JIANG Peng, ZHANG Lin, ZHOU Xingbing, GUO Xiaoyi, ZHU Yongchuan, LIU Mao, GUO Chanchun, XIONG Hong, XU Fuxian.
Yield Formation Characteristics of Ratooning Hybrid Rice Under Simplified Cultivation Practices in Winter Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(5): 544-554. |
| [7] |
YANG Mingyu, CHEN Zhicheng, PAN Meiqing, ZHANG Bianhong, PAN Ruixin, YOU Lindong, CHEN Xiaoyan, TANG Lina, HUANG Jinwen.
Effects of Nitrogen Reduction Combined with Biochar Application on Stem and Sheath Assimilate Translocation and Yield Formation in Rice Under Tobacco-rice Rotation [J]. Chinese Journal OF Rice Science, 2024, 38(5): 555-566. |
| [8] |
XIONG Jiahuan, ZHANG Yikai, XIANG Jing, CHEN Huizhe, XU Yicheng, WANG Yaliang, WANG Zhigang, YAO Jian, ZHANG Yuping .
Effect of Biochar-based Fertilizer Application on Rice Yield and Nitrogen Utilization in Film- mulched PaddyFields [J]. Chinese Journal OF Rice Science, 2024, 38(5): 567-576. |
| [9] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [10] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [11] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [12] | FU Rongtao, CHEN Cheng, WANG Jian, ZHAO Liyu, CHEN Xuejuan, LU Daihua. Combined Transcriptome and Metabolome Analyses Reveals the Pathogenic Factors of Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(4): 375-385. |
| [13] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [14] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [15] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||