
Chinese Journal OF Rice Science ›› 2018, Vol. 32 ›› Issue (5): 427-436.DOI: 10.16819/j.1001-7216.2018.7037
• 研究论文 • Previous Articles Next Articles
Zhantian ZHANG, Yafei SUN, Hao AI, Wenzhen LUO, Bing FENG, Wenxian SUN, Guohua XU, Shubin SUN*( )
)
Received:2017-03-30
Revised:2017-08-18
Online:2018-09-10
Published:2018-09-10
Contact:
Shubin SUN
张占田, 孙雅菲, 艾昊, 罗闻真, 冯冰, 孙文献, 徐国华, 孙淑斌*( )
)
通讯作者:
孙淑斌
基金资助:CLC Number:
Zhantian ZHANG, Yafei SUN, Hao AI, Wenzhen LUO, Bing FENG, Wenxian SUN, Guohua XU, Shubin SUN. Expression Patterns and Regulation of Transcription Factor Gene OsSHR2 in Vegetative Growth in Rice[J]. Chinese Journal OF Rice Science, 2018, 32(5): 427-436.
张占田, 孙雅菲, 艾昊, 罗闻真, 冯冰, 孙文献, 徐国华, 孙淑斌. 水稻转录因子基因OsSHR2的表达特征及其在营养生长中的调控作用[J]. 中国水稻科学, 2018, 32(5): 427-436.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2018.7037
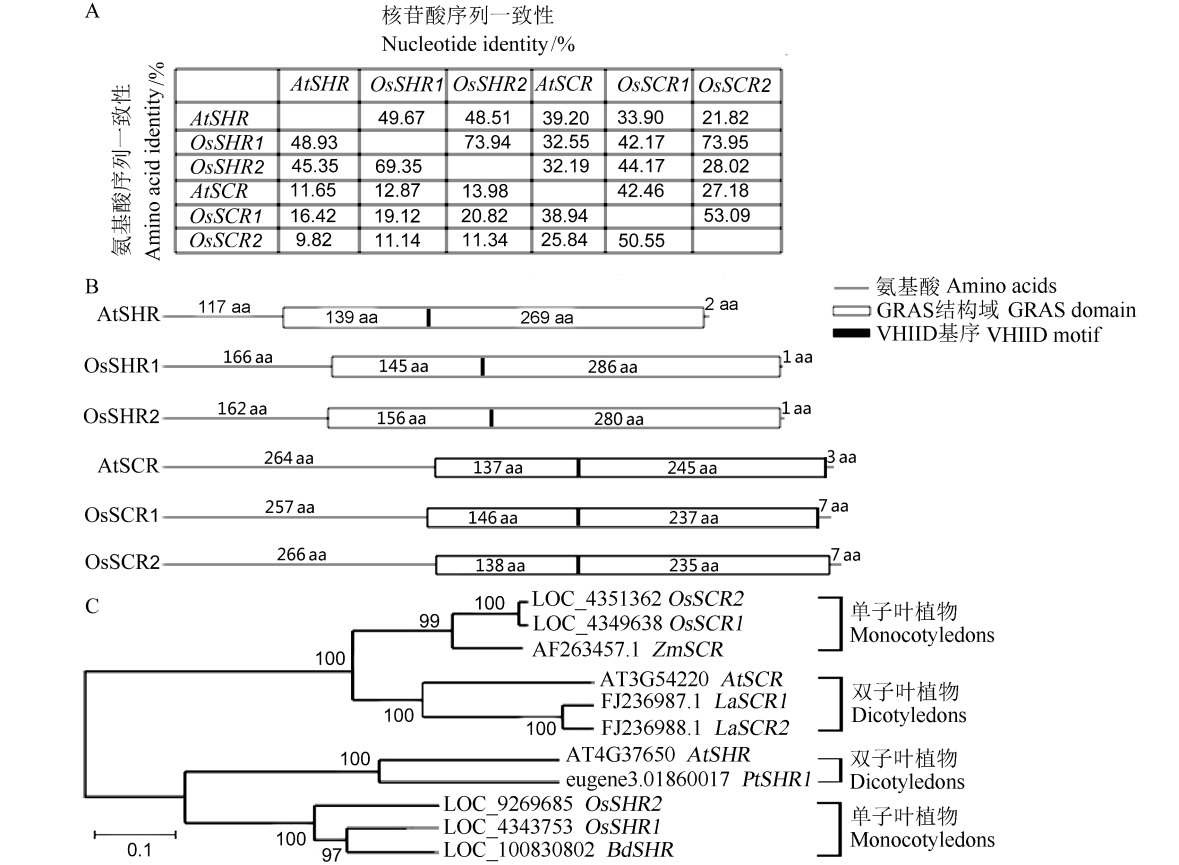
Fig. 1. Bioinformatics analysis of SHR and SCR homologous genes of different species. A, Sequence homology analysis of amino acids and nucleic acid of SHR and SCR homologous genes in Arabidopsis thaliana and rice; B, Prediction of the positions of SHR and SCR homologous GRAS conserved domains and conserved motifs in Arabidopsis thaliana and rice by InterPro (http://www.ebi.ac.uk/interpro/); C, Phylogenetic tree analysis of SHR and SCR homologous genes of different species.
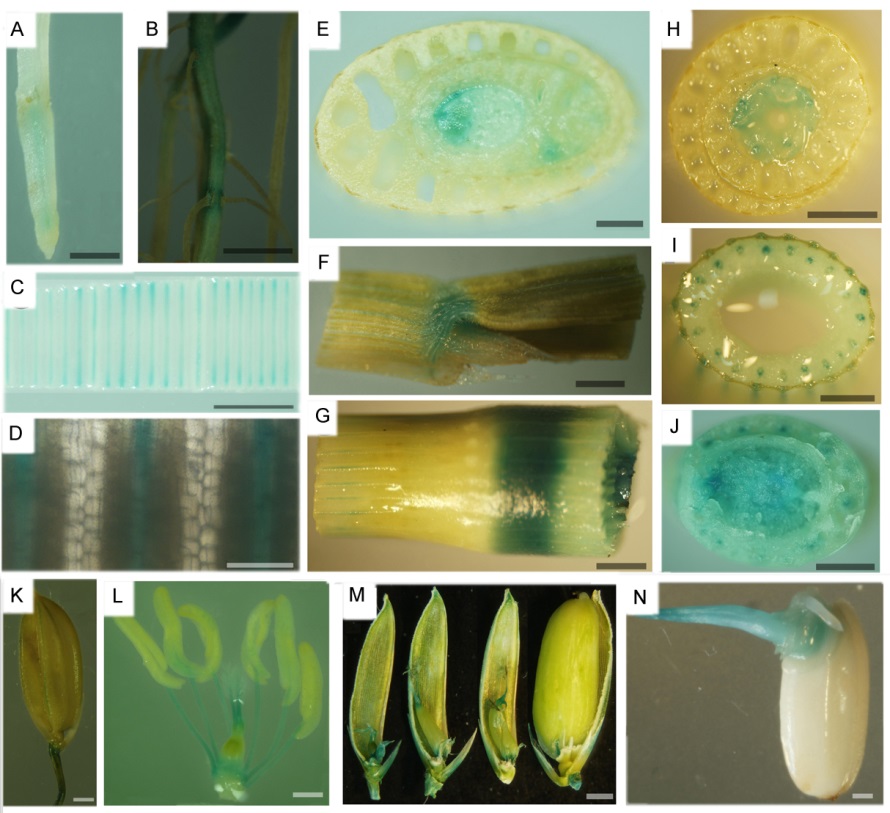
Fig. 3. Identification of OsSHR2 promoter-driven tissue-specific GUS staining. A, Seed root; B, Seed root and lateral root; C, Young leaf; D, The enlarged view of the young leaf; E, Leaf primordium; F, Ligule; G, Node; H, Stem and leaf sheath; I, Stem; J, Basal stem; K, Husk and rachilla; L, Spikelet; M, Ovary and stigma; N, Embryo(three days after germination). A-E, J: Seedling stage; F, G, I, K-N: Grain filling stage; H, Tilling stage. A-J, Bar=2 mm; K-N, Bar=0.5 mm.
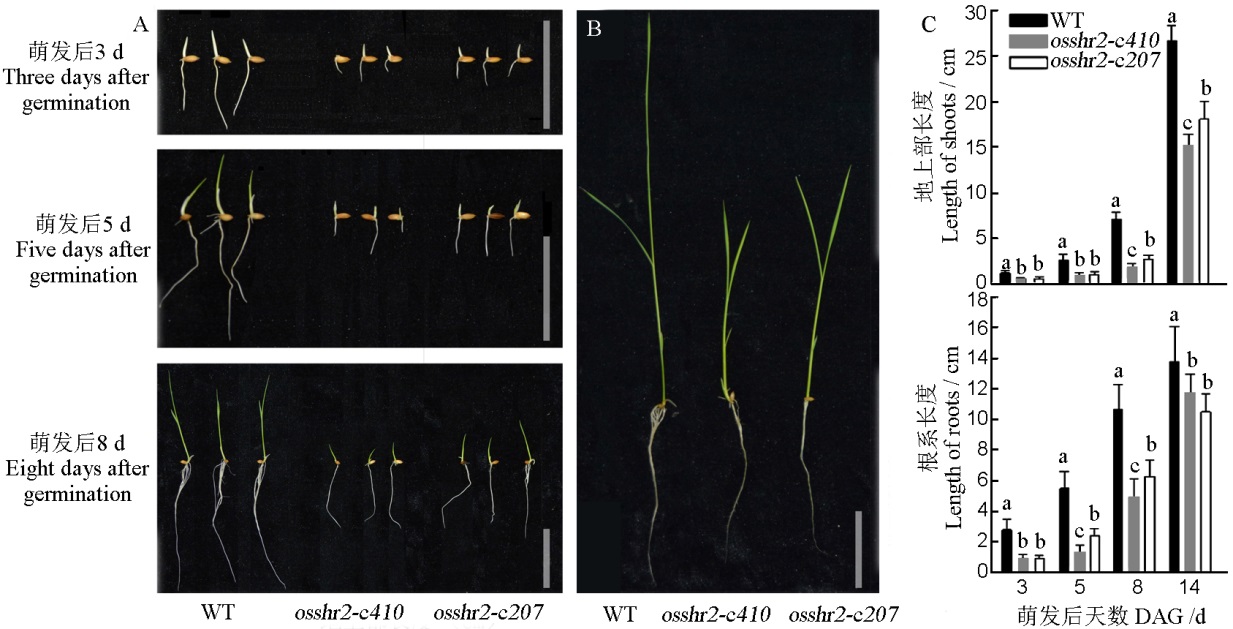
Fig. 4. Phenotype and statistics of seed germination and hydroponics experiments of WT and osshr2. A, Germination phenotype(3, 5 and 8 days after germination), bar=5 cm; B, Hydroponics phenotype(14 days after germination), bar=5 cm; C, Shoot and root length (3, 5, 8 and 14 days after germination); DAG, Day after germination.
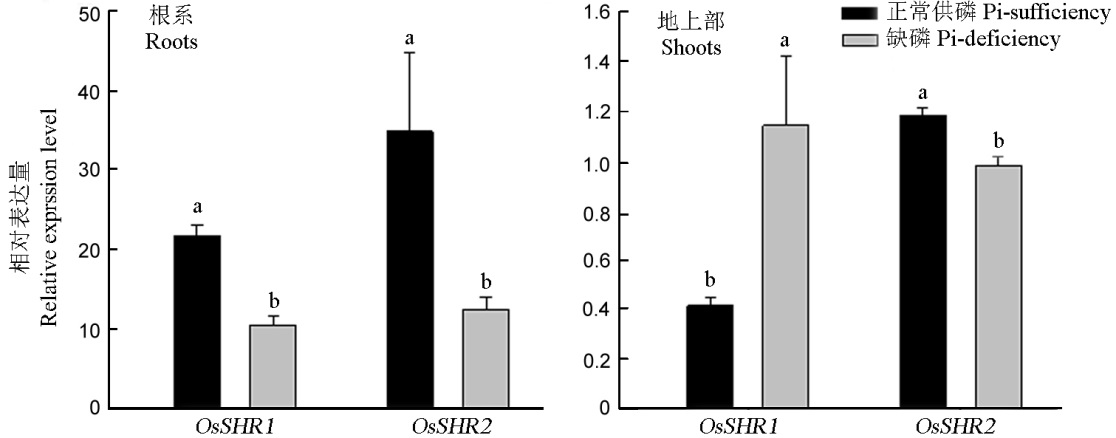
Fig. 5. Relative expression level of OsSHR1 and OsSHR2 under Pi-sufficient and Pi-deficient conditions in rice. A, The relative expression of OsSHR1 and OsSHR2 in the roots; B, The relative expression of OsSHR1 and OsSHR2 in the shoots.
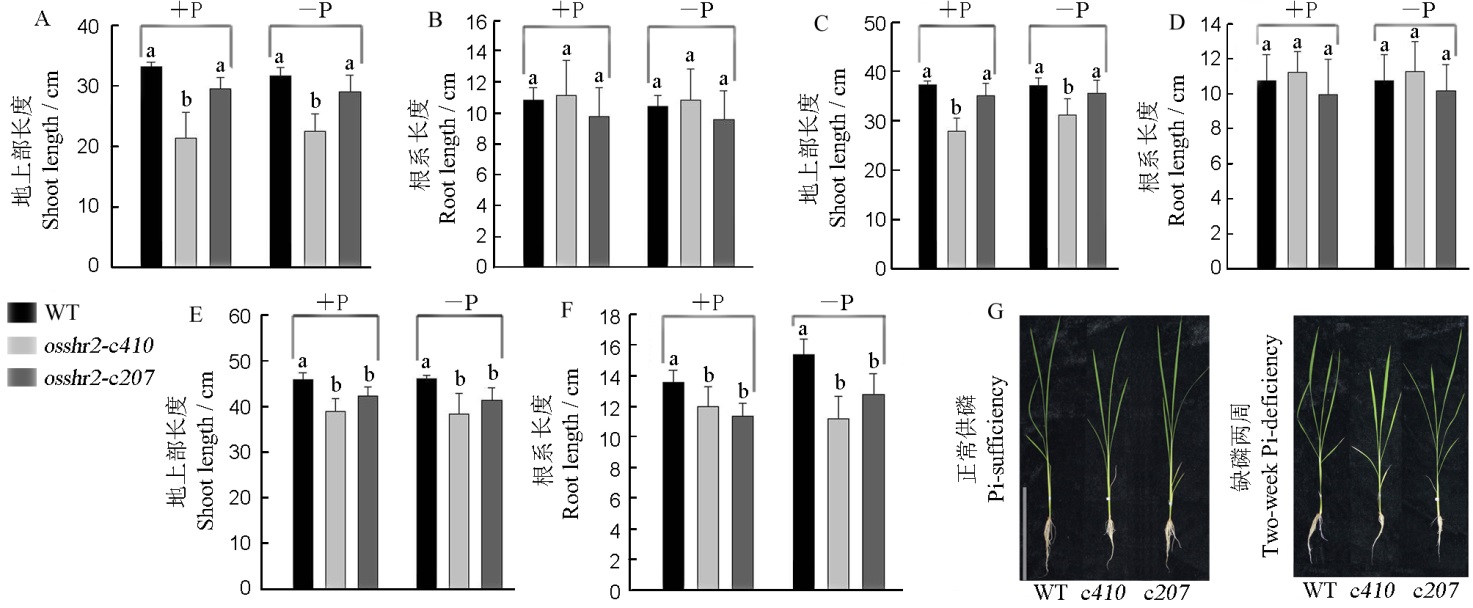
Fig. 6. Phenotype of the wild type(WT) and osshr2 under Pi-sufficient and Pi-deficient conditions. A and B, Pi-deficient lasting three days; C and D, Pi-deficient lasting one week; E and F, Pi-deficient lasting two weeks; G, The phenotype of material under Pi-sufficient and Pi-deficient lasting two weeks, bar=20 cm. c410, osshr2-c410; c207, osshr2-c207.
| [1] | 朱义旺, 林雅容, 陈亮. 我国水稻分子育种研究进展. 厦门大学学报, 2016, 55(5): 661-671. |
| Zhu Y W, Lin Y R, Chen L.Research progress of rice molecular breeding in China.J Xiamen Univ, 2016, 55(5): 661-671. (in Chinese with English abstract) | |
| [2] | Riechmann J L, Heard J, Martin G, Reuber L, Jiang C Z, Keddie J, Adam L, Pineda O, Ratcliffe O J, Samaha R R, Creelman R, Pilgrim M, Broun P, Zhang J Z, Ghandehari D, Sherman B K, Yu G L.Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science, 2000, 290(5499): 2105-2110. |
| [3] | Pysh L D, Wysocka-Diller J W, Christine C, David B, Benfey P N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J Cell & Mol Biol, 1999, 18(1): 111. |
| [4] | Cui H, Levesque M P, Vernoux T, Jung J W, Paquette A J, Gallagher K L, Wang J Y, Blilou I, Scheres B, Benfey P N.An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants.Science, 2007, 316(5823): 421-425. |
| [5] | Dolan L.SCARECROWs at the Border.Science, 2007, 316(5823): 377-378. |
| [6] | Wu S, Lee C M, Hayashi T, Pricea S, Divolb F, Henryb S, Pauluzzib G, Perinb C, Gallaghera K L.A plausible mechanism, based upon short-root movement, for regulating the number of cortex cell layers in roots.Proc Natl Acad Sci USA, 2014, 111(45): 16184-16189. |
| [7] | Benfey P N, Scheres B.Root development.Curr Biol, 2000, 10(22): 813-815. |
| [8] | Benfey P N, Linstead P J, Roberts K, Schiefelbein J W, Hauser M T, Aeschbacher R A.Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis.Development, 1993, 119(1): 57-70. |
| [9] | Laurenzio L D, Wysockadiller J, Malamy J E, Pysh L, Helariutta Y, Freshour G, Hahn M G, Feldmann K A, Benfey P N.The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell, 1996, 86(3): 423-433. |
| [10] | 倪君. OsIAA23介导的生长素信号胚后维持水稻根静止中心. 杭州: 浙江大学, 2011. |
| Ni J.OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in primary in rice. Hangzhou: Zhejiang University, 2011. (in Chinese with English abstract) | |
| [11] | Lim J, Benfey P N.Molecular analysis of the SCARECROW gene in maize reveals a common basis for radial patterning in diverse meristems.Discuss Pap, 2000, 12(8): 1307-1318. |
| [12] | Sbabou L, Bucciarelli B, Miller S, Liu J, Berhada F, Filali-Maltouf A, Allan D, Vance C.Molecular analysis of SCARECROW genes expressed in white lupin cluster roots. J Exp Bot, 2010, 61(5): 1351-1363. |
| [13] | Wang J, Anderssongunneras S, Gaboreanu I, Hertzberg M, Tucker M R, Zheng B, Lesniewska J, Mellerowicz E J, Laux T, Sandberg G, Jones B.Reduced expression of the SHORT-ROOT gene increases the rates of growth and development in hybrid poplar and Arabidopsis. PloS ONE, 2011, 6(12): e28878. |
| [14] | Wysockadiller J W, Helariutta Y, Fukaki H, Malamy J E, Benfey P N.Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development, 2000, 127(3): 595-603. |
| [15] | 霍胜楠. 水稻胚胎发生相关基因的表达及其功能鉴定. 济南: 山东农业大学, 2008. |
| Huo S N.Isolation and characterization of rice genes involved in embryo development. Jinan: Shandong Agricultural University, 2008. (in Chinese with English abstract) | |
| [16] | Cui H, Kong D, Liu X, Hao Y.SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J Cell & Mol Biol, 2014, 78(2): 319-327. |
| [17] | Gao X R, Wang C L, Cui H C.Identification of bundle sheath cell fate factors provides new tools for C3-to-C4 engineering.Plant Signal & Behav, 2014, 9(6): e29163. |
| [18] | Morikami A.The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J, 2003, 36(1): 45-54. |
| [19] | Lucas M, Swarup R, Paponov I A, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, Wang J H, Ljung K, Marchant A, Sandberg G, Holdsworth M J, Palme K, Pridmore T, Mooney S, Bennett M J.Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol, 2011, 155(1): 384-398. |
| [20] | Tian H, Jia Y, Niu T, Yu Q, Ding Z.The key players of the primary root growth and development also function in lateral roots in Arabidopsis. Plant Cell Rep, 2014, 33(5): 745-753. |
| [21] | Goh T, Toyokura K, Wells D M, Swarup K, Yamamoto M, Mimura T, Weijers D, Fukaki H, Laplaze L, Bennett M J, Guyomarc’h S.Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development, 1991, 143(18): 3363. |
| [22] | Lavenus J, Goh T, Guyomarc’h S, Hill K, Lucas M, Voß U, Kenobi K, Wilson M H, Farcot E, Hagen G, Guilfoyle T J, Fukaki H, Laplaze L, Bennettb M J.Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. Plant Cell, 2015, 27(5): 1368-1388. |
| [23] | Bieleski R.Phosphate pools, phosphate transport, and phosphate availability.Ann Rev Plant Physiol, 1973, 24(1): 225-252. |
| [24] | Muchhal U S, Pardo J M, Raghothama K G.Phosphate transporters from the higher plant Arabidopsis thaliana.Proc Natl Acad Sci USA, 1996, 93(19): 10519-105123. |
| [25] | Wang L, Shan L, Ye Z, Li Z, Du X, Liu D. Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12,AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol, 2014, 56(3): 299-314. |
| [26] | Rausch C, Bucher M.Molecular mechanisms of phosphate transport in plants.Planta, 2002, 216(1): 23-37. |
| [27] | Paszkowski U, Kroken S, Roux C, Briggs S P.Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis.Proc Natl Acad Sci USA, 2002, 99(20): 13324-13329. |
| [28] | Liu F, Chang X J, Ye Y, Xie W B, Wu P, Lian X M.Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice.Mol Plant, 2011, 4(6): 1105-1122. |
| [29] | Zhang F, Sun Y, Pei W, Jain A, Sun R, Cao Y, Wu X N, Jiang T T, Zhang L, Fan X R, Chen A Q, Shen Q R, Xu G H, Sun S B.Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J Cell & Mol Biol, 2015, 82(4): 556. |
| [30] | Rubio V, Linhares F, Solano R, Martín A C, Iglesias J, Leyva A, Paz-Ares J.A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae.Genes & Dev, 2001, 15(16): 2122-2133. |
| [31] | Bustos R, Castrillo G, Linhares F, Puga M I, Rubio V, Perez-Perez J, Solano R, Leyva A, Paz-Ares J.A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PloS Genet, 2010, 6(9): e1001102. |
| [32] | Wu P, Wang X.Role of OsPHR2 on phosphorus homeostasis and root hairs development in rice(Oryza sativa L.). Plant Signal & Behav, 2008, 3(9): 674-675. |
| [33] | Zhou J, Jiao F, Wu Z C, Li Y Y, Wang X M, He X W, Zhong W Q, Wu P.OsPHR2 is involved in phosphate- starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol, 2008, 146(4): 1673-1686. |
| [34] | Raghothama K G, Maggio A, Narasimhan M L, Kononowicz A K, Wang G, D’Urzo M P, Hasegawa P M, Bressanl R A. Tissue-specific activation of the OsMotin gene by ABA, C2H4 and NaCl involves the same promoter region. Plant Mol Biol, 1997, 34(3): 393-402. |
| [35] | Liao H, Rubio G, Yan X, Cao A, Brown K M, Lynch J P.Effect of phosphorus availability on basal root shallowness in common bean.Plant & Soil, 2001, 232(1): 69-79. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||