
中国水稻科学 ›› 2026, Vol. 40 ›› Issue (1): 51-60.DOI: 10.16819/j.1001-7216.2026.250206
王轶欣1,2, 林参3, 马刘洋2, 陈龙2, 奉保华2, 倪深2, 魏祥进2, 贺记外1,*( ), 陈天晓2,*(
), 陈天晓2,*( )
)
收稿日期:2025-02-21
修回日期:2025-03-20
出版日期:2026-01-10
发布日期:2026-01-21
通讯作者:
*email: hejiwai@hunau.edu.cn;基金资助:
WANG Yixin1,2, LIN Shen3, MA Liuyang2, CHEN Long2, FENG Baohua2, NI Shen2, WEI Xiangjin2, HE Jiwai1,*( ), CHEN Tianxiao2,*(
), CHEN Tianxiao2,*( )
)
Received:2025-02-21
Revised:2025-03-20
Online:2026-01-10
Published:2026-01-21
摘要:
【目的】研究OsAlaAT4在调控水稻氮素利用中的功能,有助于完善水稻氮调控网络,同时为水稻高氮利用率提供理论依据。【方法】根据OsAlaAT1的氨基酸序列在NCBI数据库找同源基因,构建敲除载体,通过农杆菌介导的遗传转化方法转化日本晴愈伤组织,从而获得水稻转基因植株并筛选纯合突变体,将野生型日本晴与突变系通过不同氮浓度的水培试验和田间试验来验证OsAlaAT4与氮吸收利用相关。【结果】查找到同源基因OsAlaAT4,成功构建敲除载体并转入日本晴(NIP),获得2个纯合突变系。不同氮浓度水培结果显示,突变系株高、干质量和鲜质量低于相同处理的野生型,根长长于相同处理的野生型,在1/2倍标准氮(N1/2)、4倍标准氮(N4)、8倍标准氮(N8)处理下突变系alaat4-1和alaat4-2的整株氮含量分别提高了27.7%、6.6%、7.7%和26.0%、7.8%、4.5%。正常施氮条件下突变系籽粒的蛋白质含量分别降低了3.0%和3.3%。在低氮田条件下,突变系的株高显著低于野生型。与野生型相比,两个突变系的叶和茎氮含量分别提高了12.1%、13.5%和14.4%、6.9%,穗部氮含量则分别降低了2.6%和4.1%,产量分别降低了7.2%和7.6%;高氮田条件下,突变系的分蘖数显著高于野生型,与野生型相比,突变系的叶和茎氮含量分别提高了7.9%、6.7%和16.6%、16.1%,穗部氮含量则降低了4.9%、4.5%,产量下降了6.5%和5.4%。无论低氮还是高氮处理下,突变系的氮素利用率(NUE)均显著低于野生型。【结论】敲除OsAlaAT4导致水稻叶、茎中的氮含量增加,但穗部氮含量、蛋白质含量和产量下降,最终导致氮素利用率(NUE)下降。
王轶欣, 林参, 马刘洋, 陈龙, 奉保华, 倪深, 魏祥进, 贺记外, 陈天晓. 谷丙转氨酶基因OsAlaAT4调控水稻氮素吸收和产量[J]. 中国水稻科学, 2026, 40(1): 51-60.
WANG Yixin, LIN Shen, MA Liuyang, CHEN Long, FENG Baohua, NI Shen, WEI Xiangjin, HE Jiwai, CHEN Tianxiao. Regulation of Nitrogen Uptake and Yield in Rice by the Alanine Aminotransferase Gene OsAlaAT4[J]. Chinese Journal OF Rice Science, 2026, 40(1): 51-60.
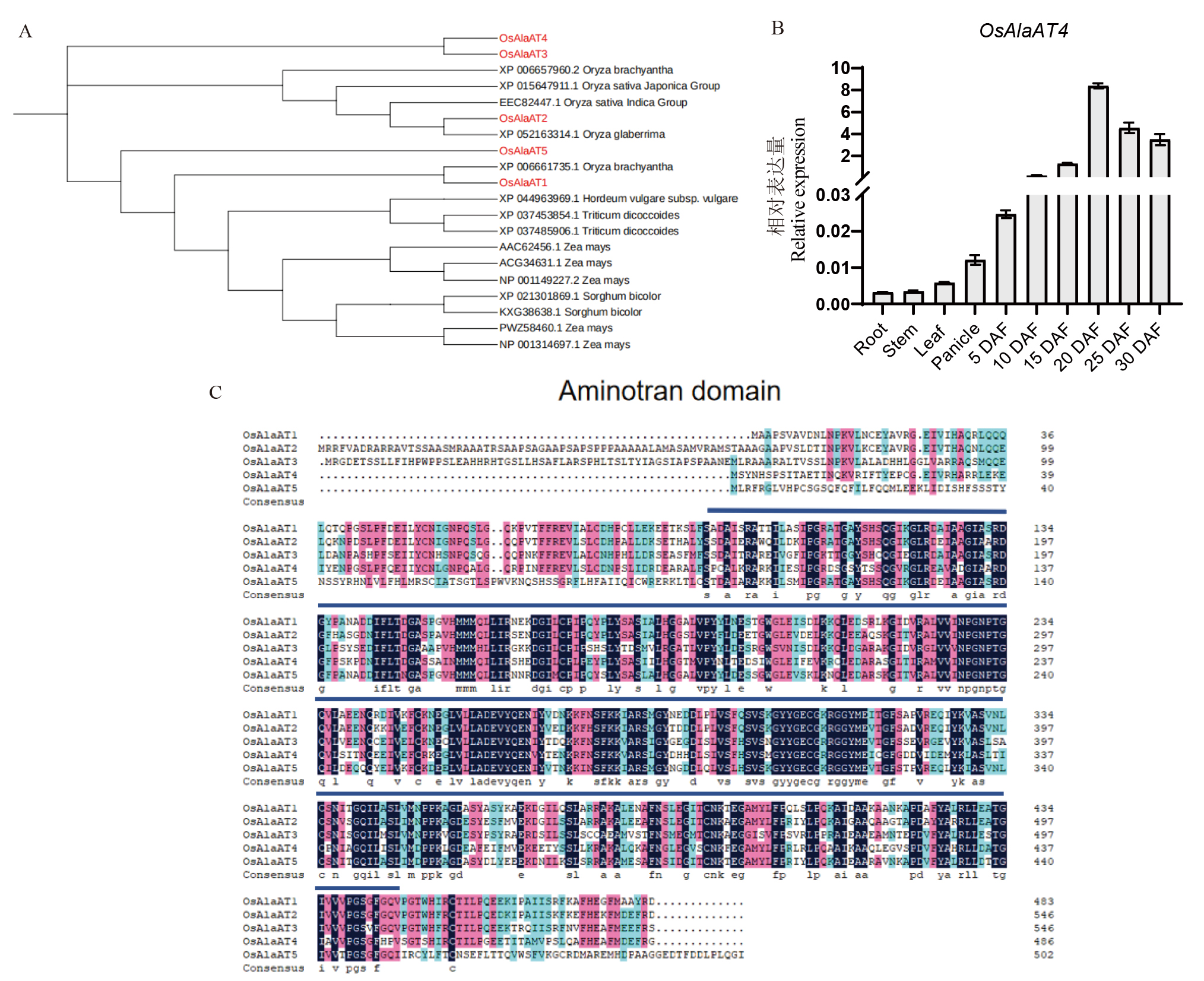
图1 进化树、表达量分析及保守结构域分析 A: 进化树分析,红色字体表示在水稻中的AlaAT家族基因;B: OsAlaAT4不同组织部位的相对表达量;DAF: 受精后天数;C: 保守结构域分析,蓝色线条下为保守氨基酸序列。
Fig. 1. Phylogenetic tree, expression analysis, and conserved domain analysis A, Phylogenetic tree analysis, with red font indicating the AlaAT family genes in rice; B, Relative expression levels of OsAlaAT4 in different tissues; DAF, Days after fertilization; C, Conserved domain analysis, with conserved amino acid sequences under the blue lines.
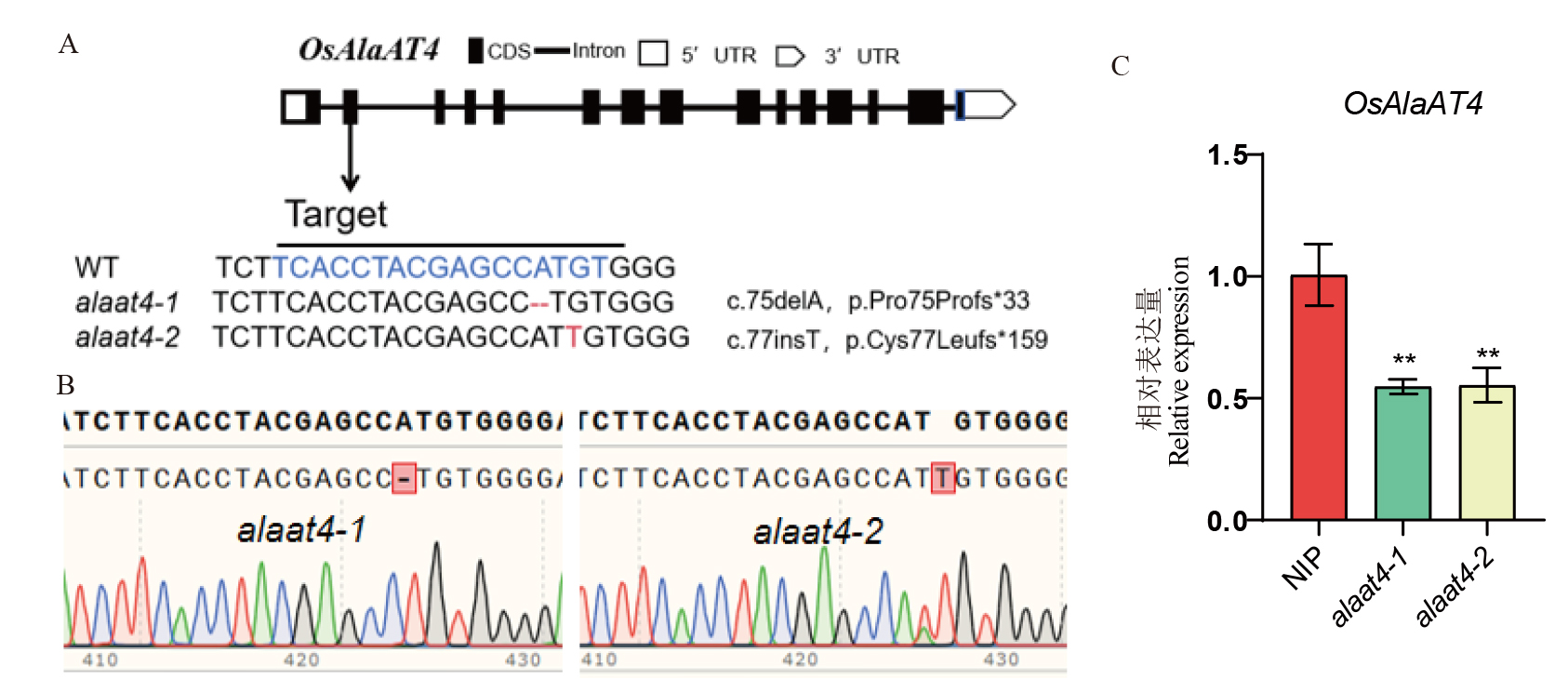
图2 OsAlaAT4的突变系鉴定 A: 2个突变系的碱基类型,---表示缺失的碱基序列,红色字体表示插入的碱基;B: 2个突变系的测序结果;C: 野生型与突变系OsAlaAT4表达量。
Fig. 2. Identification of OsAlaAT4 mutant lines A, Base types of the two mutant lines, --- indicates the deleted base sequence, and red font indicates the inserted bases; B, Sequencing results of the two mutant lines; C, Expression levels of OsAlaAT4 in the wild-type and mutant lines.
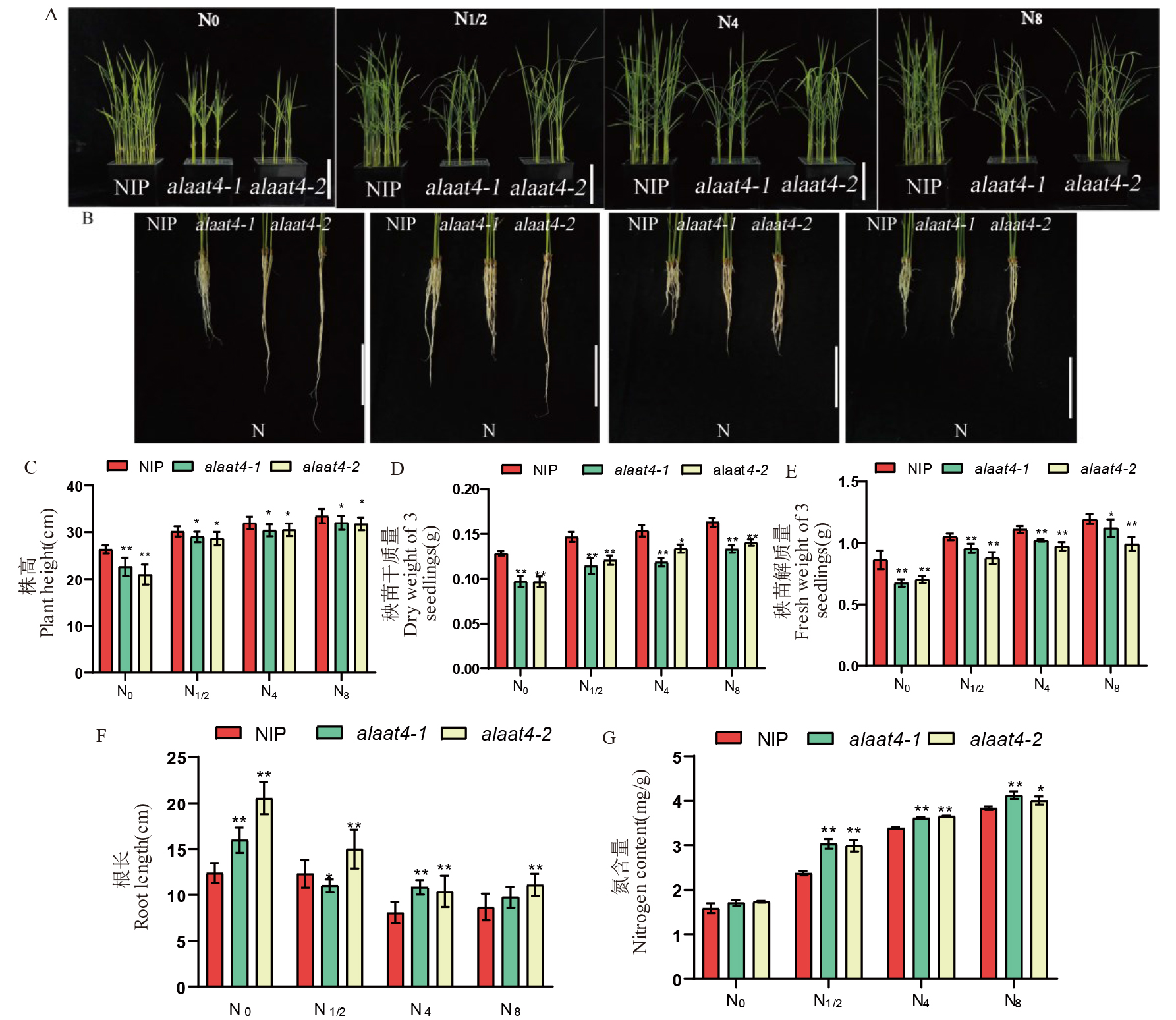
图3 突变系幼苗表型及氮含量 A:野生型和OsAlaAT4突变系水培14 d的地上部,标尺=10 cm;B:野生型和OsAlaAT4突变系水培14 d的根长,标尺=10 cm;C:野生型和OsAlaAT4突变系的苗高;D:野生型和OsAlaAT4突变系的干质量;E:野生型和OsAlaAT4突变系的鲜质量;F:野生型和OsAlaAT4突变系的根长;G:野生型和OsAlaAT4突变系氮含量。N0: 无氮; N1/2: 1/2倍标准氮; N4: 4倍标准氮; N8: 8倍标准氮。
Fig. 3. Phenotype and nitrogen content of mutant seedlings A, Above-ground part of wild-type and OsAlaAT4 mutant lines after 14 days of hydroponic cultivation, bar=10 cm; B, Root length of wild-type and OsAlaAT4 mutant lines after 14 days of hydroponic cultivation, bar=10 cm; C, Seedling height of wild-type and OsAlaAT4 mutant lines; D, Dry weight of wild-type and OsAlaAT4 mutant lines; E, Fresh weight of wild-type and OsAlaAT4 mutant lines; F, Root length of wild-type and OsAlaAT4 mutant lines; G, Nitrogen content of wild-type and OsAlaAT4 mutant lines. N0, No nitrogen; N1/2, 1/2 standard nitrogen; N4, 4 times standard nitrogen; N8, 8 times standard nitrogen.
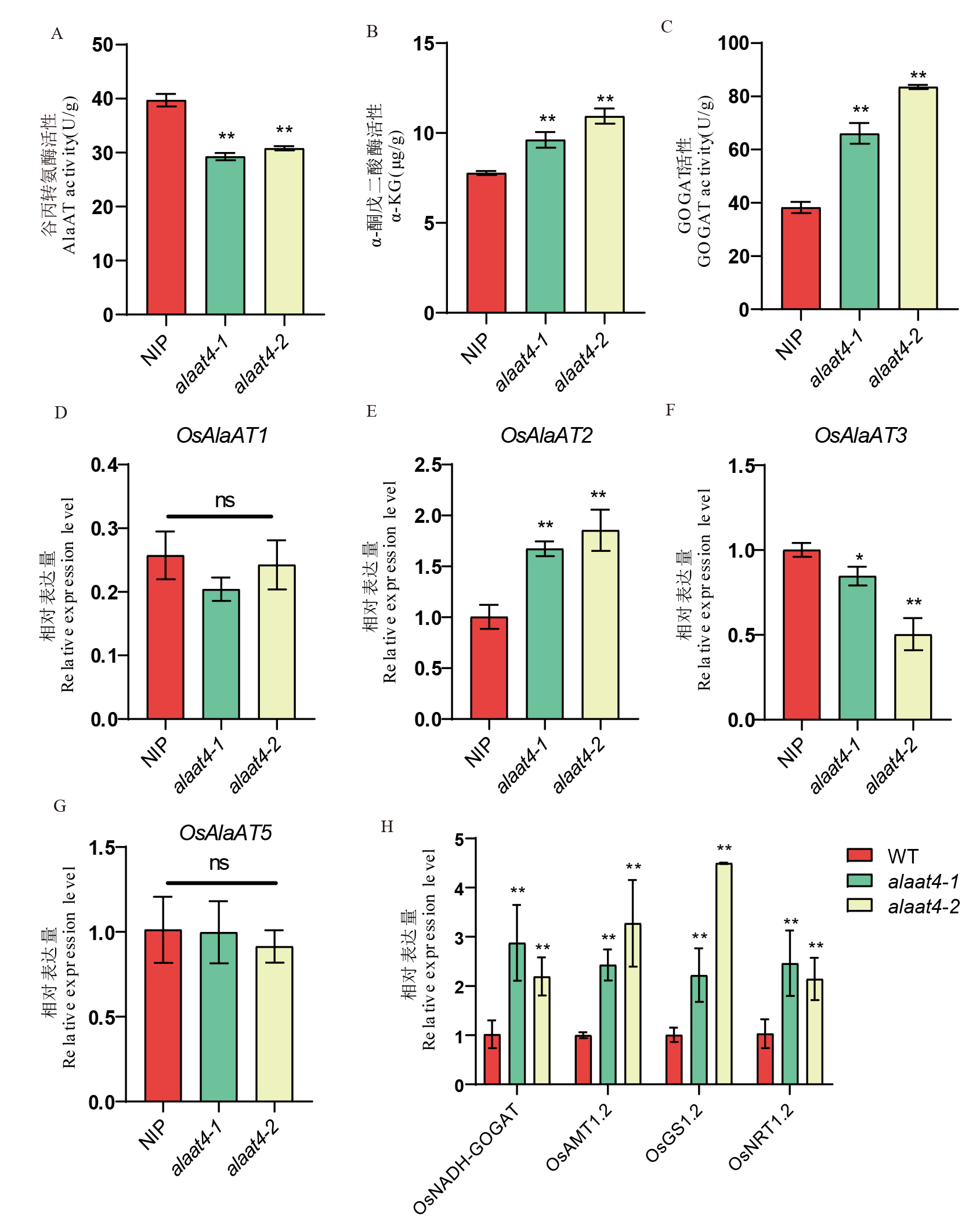
图4 关键酶活性及相关基因表达量测定 A:谷丙转氨酶活性;B:α-酮戊二酸酶活性;C:GOGAT酶活性;D-G:野生型与突变系中OsAlaAT1、OsAlaAT2、OsAlaAT3、OsAlaAT5的表达量;H为野生型与突变系中氮转运相关基因的表达量。
Fig. 4. Enzyme activity and related gene expression level measurement A, Alanine aminotransferase (ALT) enzyme activity; B, α-Ketoglutarate enzyme activity; C, Glutamate synthase (GOGAT) enzyme activity; D-G, Expression levels of OsAlaAT1, OsAlaAT2, OsAlaAT3, OsAlaAT5 in wild-type and mutant lines; H, Expression levels of nitrogen transport-related genes in wild-type and mutant lines.
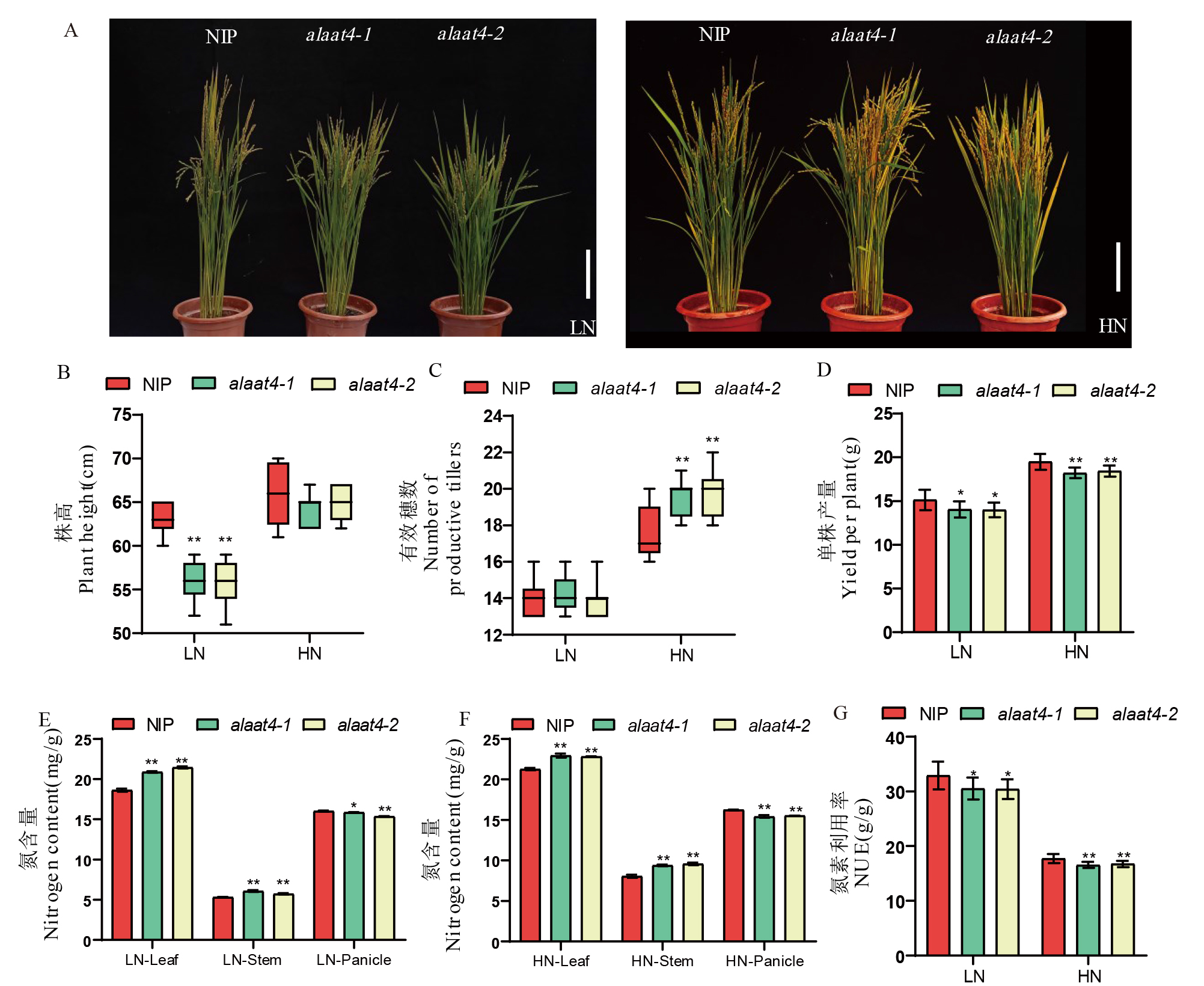
图5 突变系田间表型、氮含量及产量 A:成熟期野生型和OsAlaAT4突变系的表型;B:野生型和OsAlaAT4突变系的株高;C:野生型和OsAlaAT4突变系的分蘖数,n=12;D:成熟期野生型和OsAlaAT4突变系的单株产量;E:低氮条件下野生型和OsAlaAT4突变系的叶、茎和穗的氮含量;F:高氮条件下野生型和OsAlaAT4突变系的叶、茎和穗的氮含量;G:野生型和OsAlaAT4突变系的氮素利用率。LN:低氮; HN:高氮。
Fig. 5. Field phenotype, nitrogen content, and yield of mutant lines A, Phenotype of wild-type and OsAlaAT4 mutant lines at maturity; B, Plant height of wild-type and OsAlaAT4 mutant lines; C, Number of tillers in wild-type and OsAlaAT4 mutant lines, n=12; D, Single-plant yield of wild-type and OsAlaAT4 mutant lines at maturity; E, Nitrogen content in leaves, stems, and panicles of wild-type and OsAlaAT4 mutant lines under low nitrogen conditions; F, Nitrogen content in leaves, stems, and panicles of wild-type and OsAlaAT4 mutant lines under high nitrogen conditions; G, Nitrogen use efficiency (NUE) of wild-type and OsAlaAT4 mutant lines. LN, Low nitrogen; HN, High nitrogen.
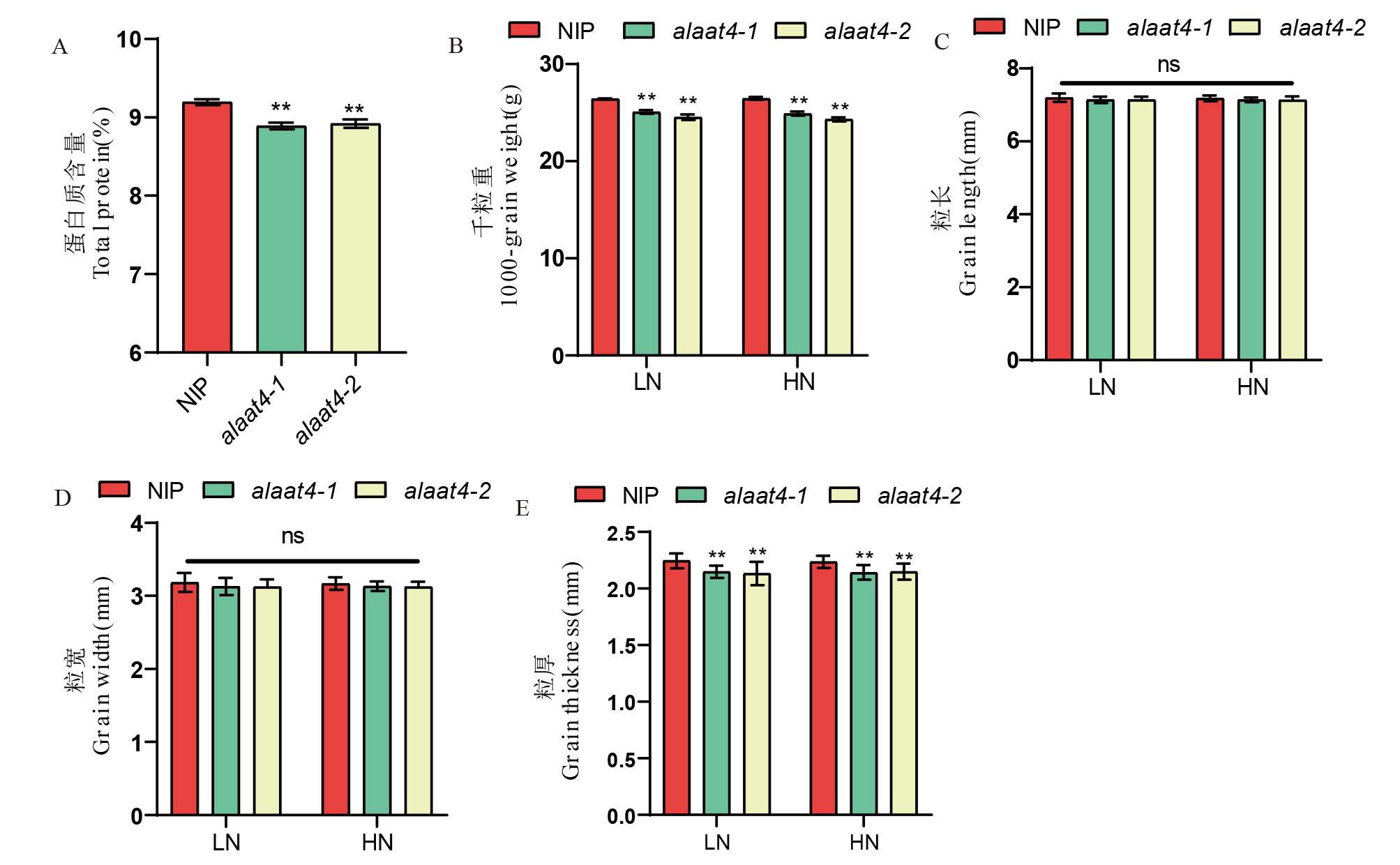
图6 OsAlaAT4突变对蛋白质和粒型的影响 A:蛋白质含量;B:千粒重;C-E:粒长、粒宽、粒厚。
Fig. 6. Impact of OsAlaAT4 mutation on protein and grain shape A, Protein content; B, Thousand-grain weight; C-E, Grain length, grain width, grain thickness.
| [1] | Tilman D, Cassman K G, Matson P A, Naylor R, Polasky S. Agricultural sustainability and intensive production practices[J]. Nature, 2002, 418(6898): 671-677. |
| [2] | Fukushima A, Kusano M. A network perspective on nitrogen metabolism from model to crop plants using integrated ‘omics’ approaches[J]. Journal of Experimental Botany, 2014, 65(19): 5619-5630. |
| [3] | Wang Q, Nian J, Xie X, Yu H, Zhang J, Bai J, Dong G, Hu J, Bai B, Chen L, Xie Q, Feng J, Yang X, Peng J, Chen F, Qian Q, Li J, Zuo J. Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice[J]. Nature Communications, 2018, 9: 735. |
| [4] | Chen X, Cui Z, Fan M, Vitousek P, Zhao M, Ma W, Wang Z, Zhang W, Yan X, Yang J, Deng X, Gao Q, Zhang Q, Guo S, Ren J, Li S, Ye Y, Wang Z, Huang J, Tang Q, Sun Y, Peng X, Zhang J, He M, Zhu Y, Xue J, Wang G, Wu L, An N, Wu L, Ma L, Zhang W, Zhang F. Producing more grain with lower environmental costs[J]. Nature, 2014, 514(7523): 486-489. |
| [5] | Cai S, Zhao X, Pittelkow C M, Fan M, Zhang X, Yan X. Optimal nitrogen rate strategy for sustainable rice production in China[J]. Nature, 2023, 615(7950): 73-79. |
| [6] | Raun W R, Solie J B, Johnson G V, Stone M L, Mullen R W, Freeman K W, Thomason W E, Lukina E V. Improving nitrogen use efficiency in cereal grain production with optical sensing and variable rate application[J]. Agronomy Journal, 2002, 94(4): 815-820. |
| [7] | Guo J H, Liu X J, Zhang Y, Shen J L, Han W X, Zhang W F, Christie P, Goulding K W T, Vitousek P M, Zhang F S. Significant acidification in major Chinese croplands[J]. Science, 2010, 327(5968): 1008-1010. |
| [8] | Kusano M, Fukushima A, Redestig H, Saito K. Metabolomic approaches toward understanding nitrogen metabolism in plants[J]. Journal of Experimental Botany, 2011, 62(4): 1439-1453. |
| [9] | Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture[J]. Annals of Botany, 2010, 105(7): 1141-1157. |
| [10] | Wang W, Hu B, Yuan D, Liu Y, Che R, Hu Y, Ou S, Liu Y, Zhang Z, Wang H, Li H, Jiang Z, Zhang Z, Gao X, Qiu Y, Meng X, Liu Y, Bai Y, Liang Y, Wang Y, Zhang L, Li L, Jing H, Li J, Chu C. Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice[J]. The Plant Cell, 2018, 30(3): 638-651. |
| [11] | Hu B, Wang W, Ou S, Tang J, Li H, Che R, Zhang Z, Chai X, Wang H, Wang Y, Liang C, Liu L, Piao Z, Deng Q, Deng K, Xu C, Liang Y, Zhang L, Li L, Chu C. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies[J]. Nature Genetics, 2015, 47(7): 834-838. |
| [12] | Wu W, Dong X, Chen G, Lin Z, Chi W, Tang W, Yu J, Wang S, Jiang X, Liu X, Wu Y, Wang C, Cheng X, Zhang W, Xuan W, Terzaghi W, Ronald P C, Wang H, Wang C, Wan J. The elite haplotype OsGATA8-H coordinates nitrogen uptake and productive tiller formation in rice[J]. Nature Genetics, 2024, 56(7): 1516-1526. |
| [13] | Kumar A, Silim S N, Okamoto M, Siddiqi M Y, Glass A D M. Differential expression of three members of the AMT1 gene family encoding putative high-affinity NH4+ transporters in roots of Oryza sativa subspecies indica[J]. Plant, Cell & Environment, 2003, 26(6): 907-914. |
| [14] | Yu J, Xuan W, Tian Y, Fan L, Sun J, Tang W, Chen G, Wang B, Liu Y, Wu W, Liu X, Jiang X, Zhou C, Dai Z, Xu D, Wang C, Wan J. Enhanced OsNLP4-OsNiR cascade confers nitrogen use efficiency by promoting tiller number in rice[J]. Plant Biotechnology Journal, 2021, 19(1): 167-176. |
| [15] | Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y, Harberd N P, Fu X. Modulating plant growth-metabolism coordination for sustainable agriculture[J]. Nature, 2018, 560(7720): 595-600. |
| [16] | Wu K, Wang S, Song W, Zhang J, Wang Y, Liu Q, Yu J, Ye Y, Li S, Chen J, Zhao Y, Wang J, Wu X, Wang M, Zhang Y, Liu B, Wu Y, Harberd N P, Fu X. Enhanced sustainable green revolution yield via nitrogen- responsive chromatin modulation in rice[J]. Science, 2020, 367(6478): eaaz2046. |
| [17] | Liu Y, Wang H, Jiang Z, Wang W, Xu R, Wang Q, Zhang Z, Li A, Liang Y, Ou S, Liu X, Cao S, Tong H, Wang Y, Zhou F, Liao H, Hu B, Chu C. Genomic basis of geographical adaptation to soil nitrogen in rice[J]. Nature, 2021, 590(7847): 600-605. |
| [18] | Igarashi D, Miwa T, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Ohsumi C. Identification of photorespiratory glutamate: Glyoxylate aminotransferase (GGAT) gene in Arabidopsis[J]. The Plant Journal, 2003, 33(6): 975-987. |
| [19] | Rech J, Crouzet J. Partial purification and initial studies of the tomato l-alanine: 2-oxoglutarate aminotransferase[J]. Biochimica et Biophysica Acta, 1974, 350(2): 392-399. |
| [20] | Kikuchi H, Hirose S, Toki S, Akama K, Takaiwa F. Molecular characterization of a gene for alanine aminotransferase from rice (Oryza sativa)[J]. Plant Molecular Biology, 1999, 39(1): 149-159. |
| [21] | Ricoult C, Echeverria L O, Cliquet J B, Limami A M. Characterization of alanine aminotransferase (AlaAT) multigene family and hypoxic response in young seedlings of the model legume Medicago truncatula[J]. Journal of Experimental Botany, 2006, 57(12): 3079-3089. |
| [22] | Liepman A H, Olsen L J. Alanine aminotransferase homologs catalyze the glutamate: Glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis[J]. Plant Physiology, 2003, 131(1): 215-227. |
| [23] | Good A G, Johnson S J, De Pauw M, Carroll R T, Savidov N, Vidmar J, Lu Z, Taylor G, Stroeher V. Engineering nitrogen use efficiency with alanine aminotransferase[J]. Canadian Journal of Botany, 2007, 85(3): 252-262. |
| [24] | 段迎辉, 郭军, 王淑娟. 小麦丙氨酸氨基转移酶基因TaAlaAT1的克隆及表达特征分析[J]. 植物病理学报, 2009, 39(2): 139-146. |
| Duan Y H, Guo J, Wang S J. Cloning and expression characteristics analysis of the wheat alanine aminotransferase gene TaAlaAT1[J]. Journal of Plant Pathology, 2009, 39(2): 139-146. (in Chinese with English abstract) | |
| [25] | 谢月兰, 崔欢, 王长龙. 水稻种子总RNA提取质量问题的探究[J]. 中国农学通报, 2020, 36(29): 37-46. |
| Xie Y L, Cui H, Wang C L. Investigation on the quality issues of total RNA extraction from rice seeds[J]. Chinese Agricultural Science Bulletin, 2020, 36(29): 37-46. (in Chinese with English abstract) | |
| [26] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method[J]. Methods, 2001, 25(4): 402-408. |
| [27] | Yang J, Kim S R, Lee S K, Choi H, Jeon J S, An G. Alanine aminotransferase 1 (OsAlaAT1) plays an essential role in the regulation of starch storage in rice endosperm[J]. Plant Science, 2015, 240: 79-89. |
| [28] | McAllister C H, Good A G. Alanine aminotransferase variants conferring diverse NUE phenotypes in Arabidopsis thaliana[J]. PLoS One, 2015, 10(4): e0121830. |
| [29] | Zhong M, Liu X, Liu F, Ren Y, Wang Y, Zhu J, Teng X, Duan E, Wang F, Zhang H, Wu M, Hao Y, Zhu X, Jing R, Guo X, Jiang L, Wang Y, Wan J. FLOURY ENDOSPERM12 encoding alanine aminotransferase 1 regulates carbon and nitrogen metabolism in rice[J]. Journal of Plant Biology, 2019, 62(1): 61-73. |
| [30] | Fang L, Ma L, Zhao S, Cao R, Jiao G, Hu P, Wei X. Alanine aminotransferase (OsAlaAT1) modulates nitrogen utilization, grain yield, and quality in rice[J]. Journal of Genetics and Genomics, 2022, 49(5): 510-513. |
| [31] | 鲁丝琼. OsNLP基因家族在水稻不同生育期表达分析研究[D]. 长沙: 湖南农业大学, 2022. |
| Lu S Q. Expression analysis of the OsNLP gene family in rice at different growth stages[D]. Changsha: Hunan Agricultural University, 2022. (in Chinese with English abstract) | |
| [32] | Tian Q, Chen F, Zhang F, Mi G. Possible involvement of cytokinin in nitrate-mediated root growth in maize[J]. Plant and Soil, 2005, 277(1): 185-196. |
| [33] | 姜苏育. 低氮营养对小麦幼苗根系生长与氮素吸收利用的影响及其生理机制[D]. 南京: 南京农业大学, 2018. |
| Jiang S Y. Effects of low nitrogen nutrition on root growth and nitrogen uptake and utilization in wheat seedlings and their physiological mechanisms[D]. Nanjing: Nanjing Agricultural University, 2018. (in Chinese with English abstract) | |
| [34] | 谢桂先, 荣湘民, 刘强. 肥料不同配比对水稻产量与蛋白质含量的影响[J]. 湖南农业大学学报(自然科学版), 2004(5): 405-410. |
| Xie G X, Rong X M, Liu Q. Effects of different fertilizer ratios on rice yield and protein content[J]. Journal of Hunan Agricultural University (Natural Science Edition), 2004(5): 405-410. (in Chinese with English abstract) | |
| [35] | 余文. 水稻OsHXK6克隆表达和催化特性研究[D]. 武汉: 华中师范大学, 2015. |
| Yu W. Research on cloning, expression, and catalytic properties of rice OsHXK6[D]. Wuhan: Central China Normal University, 2015. (in Chinese with English abstract) |
| [1] | 李兴沂, 陈玲, 邵建韬, 肖素勤, 李金璐, 付惠仙, 殷富有, 张建红, 程在全, 刘丽. 水稻产量与淀粉品质协同调控的分子遗传研究进展[J]. 中国水稻科学, 2026, 40(1): 1-17. |
| [2] | 岳轩宇, 谢文亚, 冯志明, 陈宗祥, 胡珂鸣, 左示敏. OsERF93参与调控水稻纹枯病抗性的研究[J]. 中国水稻科学, 2026, 40(1): 37-50. |
| [3] | 黄奇娜, 姜鸿瑞, 杨婕, 于坤宇, 杨长登, 梁燕. 种子休眠基因Sdr4的生物信息学分析与分子标记开发和应用[J]. 中国水稻科学, 2026, 40(1): 61-71. |
| [4] | 程朝平, 何旎清, 白康呈, 林少俊, 黄凤凰, 刘军化, 程祖锌, 黄成志, 杨德卫. 聚合稻瘟病抗性基因Pigm-1和Pid2的水稻三系不育系福梦A的选育与利用[J]. 中国水稻科学, 2026, 40(1): 72-84. |
| [5] | 刘亚萍, 董译词, 郑君妍, 邱绚, 刘鹏程, 叶亚峰, 刘斌美, 陈析丰, 马伯军. 水稻类病变早衰突变体lmes7的鉴定与基因精细定位[J]. 中国水稻科学, 2026, 40(1): 85-94. |
| [6] | 肖无为, 张雨晴, 朱辰光, 田贵生, 蔡岳宏, 王飞, 熊栋梁, 黄见良, 彭少兵, 崔克辉. 促芽肥施用时期和施用量对再生稻产量和头季稻米品质的影响[J]. 中国水稻科学, 2026, 40(1): 118-130. |
| [7] | 谢世民, 周誉株, 薛晓迪, 朱广飞, 孙良, 陈建能. 水稻钵苗取栽协同作业式移栽机构设计与试验[J]. 中国水稻科学, 2026, 40(1): 131-144. |
| [8] | 陈玲, 林文英, 梁丽梅, 欧阳由男, 叶胜海, 季芝娟. 水稻开花习性及其在粳型三系不育系选育中的应用[J]. 中国水稻科学, 2025, 39(6): 731-743. |
| [9] | 王娟, 吴丽娟, 洪海波, 姚志文, 王磊, 鄂志国. 水稻泛素结合酶E2的生物学功能研究进展[J]. 中国水稻科学, 2025, 39(6): 744-750. |
| [10] | 陶士博, 许娜, 徐正进, 刘畅, 徐铨. 水稻发芽期耐冷基因Cold6的克隆[J]. 中国水稻科学, 2025, 39(6): 751-759. |
| [11] | 陈伟, 叶元妹, 赵剑华, 冯志明, 陈宗祥, 胡珂鸣, 左示敏. 利用CRISPR/Cas9技术改良南粳46抽穗期[J]. 中国水稻科学, 2025, 39(6): 760-770. |
| [12] | 侯桂花, 周立国, 雷建国, 陈虹, 聂元元. 水稻OsRDR5基因功能及作用机制初步解析[J]. 中国水稻科学, 2025, 39(6): 779-788. |
| [13] | 陆帅, 陶涛, 刘冉, 周文玉, 曹蕾, 杨青青, 张明秋, 任鑫哲, 杨芝笛, 徐福祥, 环海东, 龚远航, 张皓程, 金素奎, 蔡秀玲, 高继平, 冷语佳. 水稻长护颖小粒突变体lsg8的表型鉴定与基因克隆[J]. 中国水稻科学, 2025, 39(6): 813-824. |
| [14] | 邓欢, 刘亚培, 王春连, 郭威, 陈析丰, 纪志远. 水稻抗白叶枯病新基因Xa49(t)的定位分析[J]. 中国水稻科学, 2025, 39(6): 825-831. |
| [15] | 卞金龙, 任高磊, 裘实, 许方甫, 胡忠磊, 张洪程, 魏海燕. 不同机插方式下控混肥施用方式对淮北地区优质食味粳稻产量及氮素利用的影响[J]. 中国水稻科学, 2025, 39(6): 847-862. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||