
Chinese Journal OF Rice Science ›› 2025, Vol. 39 ›› Issue (3): 352-364.DOI: 10.16819/j.1001-7216.2025.240502
• Research Papers • Previous Articles Next Articles
WEI Xinyu1,2,#, ZENG Yuehui1,2,#, XIAO Changchun1,2, HUANG Jianhong1,2, RUAN Hongchun4, YANG Wangxing2,3, ZOU Wenguang2,3, XU Xuming2,3,*( )
)
Received:2024-04-30
Revised:2024-07-23
Online:2025-05-10
Published:2025-05-21
Contact:
*email: fj63xxm@sina.com
About author:#These authors contributed equally to this work
韦新宇1,2,#, 曾跃辉1,2,#, 肖长春1,2, 黄建鸿1,2, 阮宏椿4, 杨旺兴2,3, 邹文广2,3, 许旭明2,3,*( )
)
通讯作者:
*email: fj63xxm@sina.com
作者简介:#共同第一作者
基金资助:WEI Xinyu, ZENG Yuehui, XIAO Changchun, HUANG Jianhong, RUAN Hongchun, YANG Wangxing, ZOU Wenguang, XU Xuming. Cloning and Functional Verification of Rice-Blast Resistance Gene Pi-kf2(t) in Kangfeng B[J]. Chinese Journal OF Rice Science, 2025, 39(3): 352-364.
韦新宇, 曾跃辉, 肖长春, 黄建鸿, 阮宏椿, 杨旺兴, 邹文广, 许旭明. 水稻康丰B抗稻瘟病基因Pi-kf2(t)的克隆与功能验证[J]. 中国水稻科学, 2025, 39(3): 352-364.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2025.240502
| 编号Number | 菌种名称Isolate | 来源 Origin | 编号Number | 菌种名称Isolate | 来源 Origin | 编号Number | 菌种名称Isolate | 来源 Origin | 编号Number | 菌种名称 Isolate | 来源 Origin |
|---|---|---|---|---|---|---|---|---|---|---|---|
| J01 | 2022075 | 福建福州 | J26 | 2021146 | 广东惠州 | J51 | 2021137 | 福建沙县 | J76 | 2022027 | 福建武夷山 |
| J02 | 2021024 | 福建武夷山 | J27 | 2021021 | 广东惠州 | J52 | 2022026 | 福建武夷山 | J77 | 2021033 | 江西婺源 |
| J03 | 2022048 | 湖北恩施 | J28 | 2021119 | 福建连城 | J53 | 2021048 | 福建将乐 | J78 | 2022037 | 福建浦城 |
| J04 | 2022036 | 湖北恩施 | J29 | 2021013 | 福建尤溪 | J54 | 2021018 | 江西婺源 | J79 | 2021036 | 福建宁化 |
| J05 | 2022024 | 福建建阳 | J30 | 2021121 | 福建连城 | J55 | 2021032 | 江西婺源 | J80 | 2023020 | 湖北恩施 |
| J06 | 2023090 | 福建柘荣 | J31 | 2020117 | 安徽肥西 | J56 | 2023112 | 福建宁化 | J81 | 2022006 | 福建上杭 |
| J07 | 2021017 | 江苏扬州 | J32 | 2022072 | 安徽肥西 | J57 | 2022005 | 福建上杭 | J82 | 2023019 | 江苏扬州 |
| J08 | 2020062 | 江苏扬州 | J33 | 2021001 | 福建福州 | J58 | 2020114 | 安徽肥西 | J83 | 2020156 | 江苏扬州 |
| J09 | 2021085 | 福建上杭 | J34 | 2023104 | 江西婺源 | J59 | 2020059 | 安徽肥西 | J84 | 2022044 | 湖北宜昌 |
| J11 | 2020010 | 福建宁化 | J36 | 2021078 | 湖北宜昌 | J61 | 2023131 | 福建将乐 | J86 | 2022063 | 福建莆田 |
| J12 | 2021068 | 福建莆田 | J37 | 2022065 | 湖北恩施 | J62 | 2020107 | 福建松溪 | J87 | 2021031 | 福建松溪 |
| J13 | 2021092 | 福建福安 | J38 | 2022021 | 福建建阳 | 163 | 2023034 | 江苏扬州 | J88 | 2020017 | 江西井冈山 |
| J14 | 2022078 | 福建漳州 | J39 | 2021037 | 福建宁化 | J64 | 2022032 | 福建浦城 | J89 | 2021016 | 江西井冈山 |
| J15 | 2021122 | 福建连城 | J40 | 2023018 | 江西婺源 | J65 | 2020061 | 福建将乐 | J90 | 2023035 | 江苏扬州 |
| J16 | 2022074 | 福建福州 | J41 | 2022049 | 福建福安 | J66 | 2020041 | 福建上杭 | J91 | 2021084 | 福建上杭 |
| J17 | 2021034 | 江西井冈山 | J42 | 2021138 | 福建沙县 | J67 | 2022045 | 湖北宜昌 | J92 | 2022085 | 福建南靖 |
| J18 | 2022008 | 江西井冈山 | J43 | 2021049 | 福建将乐 | J68 | 2022081 | 福建漳州 | J93 | 2021081 | 福建三明 |
| J19 | 2021080 | 福建三明 | J44 | 2021027 | 福建武夷山 | J69 | 2023029 | 江苏徐州 | J94 | 2022007 | 福建将乐 |
| J20 | 2021040 | 福建将乐 | J45 | 2021035 | 江西井冈山 | J70 | 2023004 | 福建福州 | J95 | 2023037 | 江西井冈山 |
| J21 | 2021009 | 福建沙县 | J46 | 2021020 | 江苏扬州 | J71 | 2021141 | 福建沙县 | J96 | 2020143 | 福建浦城 |
| J22 | 2022046 | 湖北恩施 | J47 | 2022015 | 江苏扬州 | J72 | 2023003 | 江苏徐州 | J97 | 2020105 | 福建松溪 |
| J23 | 2022011 | 湖北宜昌 | J48 | 2021014 | 福建尤溪 | J73 | 2023031 | 江苏徐州 | J98 | 2020039 | 福建上杭 |
| J24 | 2023014 | 福建三明 | J49 | 2023041 | 福建建阳 | J74 | 2021149 | 广东惠州 | J99 | 2022004 | 福建上杭 |
| J25 | 2022018 | 福建三明 | J50 | 2021072 | 福建莆田 | J75 | 2021005 | 广东惠州 | J100 | 2023042 | 江苏徐州 |
Table 1. 100 Magnaporthe oryzae isolates originated from different regions of China
| 编号Number | 菌种名称Isolate | 来源 Origin | 编号Number | 菌种名称Isolate | 来源 Origin | 编号Number | 菌种名称Isolate | 来源 Origin | 编号Number | 菌种名称 Isolate | 来源 Origin |
|---|---|---|---|---|---|---|---|---|---|---|---|
| J01 | 2022075 | 福建福州 | J26 | 2021146 | 广东惠州 | J51 | 2021137 | 福建沙县 | J76 | 2022027 | 福建武夷山 |
| J02 | 2021024 | 福建武夷山 | J27 | 2021021 | 广东惠州 | J52 | 2022026 | 福建武夷山 | J77 | 2021033 | 江西婺源 |
| J03 | 2022048 | 湖北恩施 | J28 | 2021119 | 福建连城 | J53 | 2021048 | 福建将乐 | J78 | 2022037 | 福建浦城 |
| J04 | 2022036 | 湖北恩施 | J29 | 2021013 | 福建尤溪 | J54 | 2021018 | 江西婺源 | J79 | 2021036 | 福建宁化 |
| J05 | 2022024 | 福建建阳 | J30 | 2021121 | 福建连城 | J55 | 2021032 | 江西婺源 | J80 | 2023020 | 湖北恩施 |
| J06 | 2023090 | 福建柘荣 | J31 | 2020117 | 安徽肥西 | J56 | 2023112 | 福建宁化 | J81 | 2022006 | 福建上杭 |
| J07 | 2021017 | 江苏扬州 | J32 | 2022072 | 安徽肥西 | J57 | 2022005 | 福建上杭 | J82 | 2023019 | 江苏扬州 |
| J08 | 2020062 | 江苏扬州 | J33 | 2021001 | 福建福州 | J58 | 2020114 | 安徽肥西 | J83 | 2020156 | 江苏扬州 |
| J09 | 2021085 | 福建上杭 | J34 | 2023104 | 江西婺源 | J59 | 2020059 | 安徽肥西 | J84 | 2022044 | 湖北宜昌 |
| J11 | 2020010 | 福建宁化 | J36 | 2021078 | 湖北宜昌 | J61 | 2023131 | 福建将乐 | J86 | 2022063 | 福建莆田 |
| J12 | 2021068 | 福建莆田 | J37 | 2022065 | 湖北恩施 | J62 | 2020107 | 福建松溪 | J87 | 2021031 | 福建松溪 |
| J13 | 2021092 | 福建福安 | J38 | 2022021 | 福建建阳 | 163 | 2023034 | 江苏扬州 | J88 | 2020017 | 江西井冈山 |
| J14 | 2022078 | 福建漳州 | J39 | 2021037 | 福建宁化 | J64 | 2022032 | 福建浦城 | J89 | 2021016 | 江西井冈山 |
| J15 | 2021122 | 福建连城 | J40 | 2023018 | 江西婺源 | J65 | 2020061 | 福建将乐 | J90 | 2023035 | 江苏扬州 |
| J16 | 2022074 | 福建福州 | J41 | 2022049 | 福建福安 | J66 | 2020041 | 福建上杭 | J91 | 2021084 | 福建上杭 |
| J17 | 2021034 | 江西井冈山 | J42 | 2021138 | 福建沙县 | J67 | 2022045 | 湖北宜昌 | J92 | 2022085 | 福建南靖 |
| J18 | 2022008 | 江西井冈山 | J43 | 2021049 | 福建将乐 | J68 | 2022081 | 福建漳州 | J93 | 2021081 | 福建三明 |
| J19 | 2021080 | 福建三明 | J44 | 2021027 | 福建武夷山 | J69 | 2023029 | 江苏徐州 | J94 | 2022007 | 福建将乐 |
| J20 | 2021040 | 福建将乐 | J45 | 2021035 | 江西井冈山 | J70 | 2023004 | 福建福州 | J95 | 2023037 | 江西井冈山 |
| J21 | 2021009 | 福建沙县 | J46 | 2021020 | 江苏扬州 | J71 | 2021141 | 福建沙县 | J96 | 2020143 | 福建浦城 |
| J22 | 2022046 | 湖北恩施 | J47 | 2022015 | 江苏扬州 | J72 | 2023003 | 江苏徐州 | J97 | 2020105 | 福建松溪 |
| J23 | 2022011 | 湖北宜昌 | J48 | 2021014 | 福建尤溪 | J73 | 2023031 | 江苏徐州 | J98 | 2020039 | 福建上杭 |
| J24 | 2023014 | 福建三明 | J49 | 2023041 | 福建建阳 | J74 | 2021149 | 广东惠州 | J99 | 2022004 | 福建上杭 |
| J25 | 2022018 | 福建三明 | J50 | 2021072 | 福建莆田 | J75 | 2021005 | 广东惠州 | J100 | 2023042 | 江苏徐州 |

Fig. 1. Resistance spectrum analysis of rice blast resistance gene Pi-kf2(t) in KFB A, Reaction of KFB, 75-1-127, C101A51, IRBLzt-T, EBZ, and GM-4 to 100 isolates of Magnaporthe oryzae. J01-J100, The 100 isolates were collected from different regions of China. R%, Resistance spectrum. R, MR, MS, S, and HS represent resistance, mid-resistance, mid-susceptibility, susceptibility, and high-susceptibility to rice blast, respectively; B, C, D, Reaction of KFB, 75-1-127, C101A51, IRBLzt-T, EBZ, and GM-4 to J71(B), J91(C), and J93(D), where 1, 2, 3, 4, 5, and 6 represent KFB, 75-1-127, C101A51, IRBLzt-T, EBZ, and GM-4, respectively.
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 用途 Usage | 预测PCR产物大小 Excepted PCR product size(bp) |
|---|---|---|---|
| NBS2-Q-F | CTGCTAATGTACAGTGACCGA | NBS-LRR2基因qPCR检测 | 111 |
| NBS2-Q-R | AGTGACTGATCCTTCGGCAT | qPCR detection for NBS-LRR2 | |
| NBS4-Q-F | TGATCACGACCTTGCTTGTG | NBS-LRR4基因qPCR检测 | 122 |
| NBS4-Q-R | CTGACCTGTTGTTCGACGTC | qPCR detection for NBS-LRR4 | |
| HPT-F | ATTTGTGTACGCCCGACAGT | 鉴定HPT | 577 |
| HPT-R | GTGCTTGACATTGGGGAGTT | PCR detection for HPT | |
| NBS2Seq-01F | CCCAAGTGAGGATGCAAAACA | NBS-LRR2基因测序 | 803 |
| NBS2Seq-01R | TGACATGCAAGAAACGGGTG | Sequencing for NBS-LRR2 | |
| NBS2Seq-02F | TGGCTAGGCTTACGTTTTGG | NBS-LRR2基因测序 | 2219 |
| NBS2Seq-02R | ACAACGAAGAGTATGCAGACA | Sequencing for NBS-LRR2 | |
| NBS2Seq-03F | AGGATGTTACGGGTCTTGGA | NBS-LRR2基因测序 | 2107 |
| NBS2Seq-03R | AACAACTGCAGGCCAAACAA | Sequencing for NBS-LRR2 | |
| OsActin-F | ACCTTCAACACCCCTGCTAT | 内参基因OsActin qPCR检测 | 105 |
| OsActin-R | CACCATCACCAGAGTCCAAC | qPCR detection for OsActin (as internal standard) |
Table 2. Sequences of primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 用途 Usage | 预测PCR产物大小 Excepted PCR product size(bp) |
|---|---|---|---|
| NBS2-Q-F | CTGCTAATGTACAGTGACCGA | NBS-LRR2基因qPCR检测 | 111 |
| NBS2-Q-R | AGTGACTGATCCTTCGGCAT | qPCR detection for NBS-LRR2 | |
| NBS4-Q-F | TGATCACGACCTTGCTTGTG | NBS-LRR4基因qPCR检测 | 122 |
| NBS4-Q-R | CTGACCTGTTGTTCGACGTC | qPCR detection for NBS-LRR4 | |
| HPT-F | ATTTGTGTACGCCCGACAGT | 鉴定HPT | 577 |
| HPT-R | GTGCTTGACATTGGGGAGTT | PCR detection for HPT | |
| NBS2Seq-01F | CCCAAGTGAGGATGCAAAACA | NBS-LRR2基因测序 | 803 |
| NBS2Seq-01R | TGACATGCAAGAAACGGGTG | Sequencing for NBS-LRR2 | |
| NBS2Seq-02F | TGGCTAGGCTTACGTTTTGG | NBS-LRR2基因测序 | 2219 |
| NBS2Seq-02R | ACAACGAAGAGTATGCAGACA | Sequencing for NBS-LRR2 | |
| NBS2Seq-03F | AGGATGTTACGGGTCTTGGA | NBS-LRR2基因测序 | 2107 |
| NBS2Seq-03R | AACAACTGCAGGCCAAACAA | Sequencing for NBS-LRR2 | |
| OsActin-F | ACCTTCAACACCCCTGCTAT | 内参基因OsActin qPCR检测 | 105 |
| OsActin-R | CACCATCACCAGAGTCCAAC | qPCR detection for OsActin (as internal standard) |
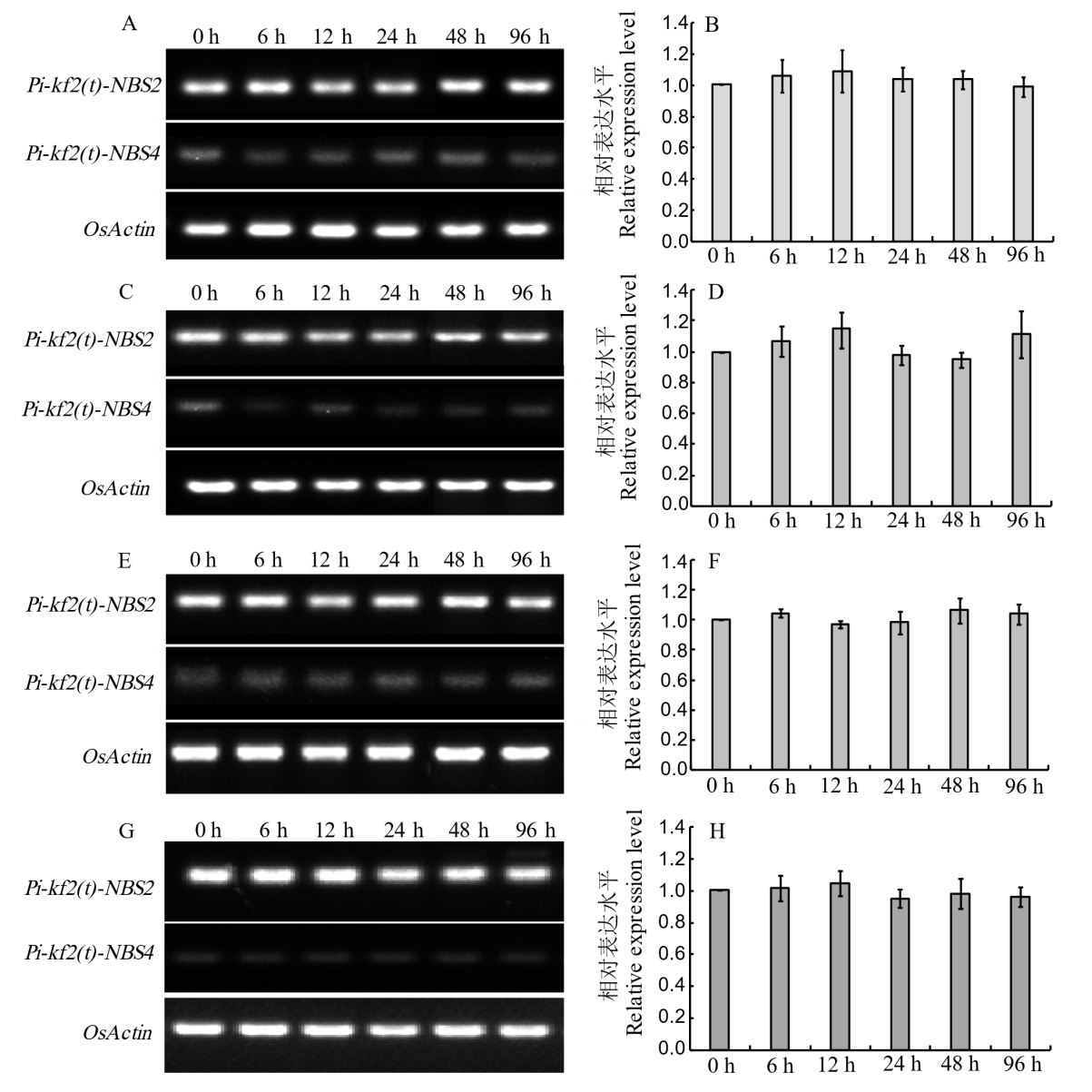
Fig. 2. Expression pattern analysis of Pi-kf2(t) candidate gene A, C, E, Expression patterns of candidate genes Pi-kf2(t)-NBS2 and Pi-kf2(t)-NBS4 in KFB seedling leaves at 0, 6, 12, 24, 48, and 96 h after inoculation with the isolates J71(A), J91(C), and J93(E), determined by RT-PCR; B, D, F, Expression patterns of candidate genes Pi-kf2(t)-NBS2 in KFB seedling leaves at 0, 6, 12, 24, 48, and 96 h after inoculation with the isolates J71(B), J91(D), and J93(F), determined by qRT-PCR;G, Expression patterns of candidate genes Pi-kf2(t)-NBS2 and Pi-kf2(t)-NBS4 in KFB seedling leaves at 0, 6, 12, 24, 48, and 96 h after mock inoculation, determined by RT-PCR; H, The expression patterns of candidate genes Pi-kf2(t)-NBS2 in KFB seedling leaves at 0, 6, 12, 24, 48, and 96 h after mock inoculation, determined by qRT-PCR.

Fig. 3. Amino-acid sequences alignment of Pi-kf2(t)-NBS2 with Pi9, Pi2, Piz-t/Pizh, Pi50, and Pigm Dark blue highlights identical residues, while similar residues are marked in pink or light blue, and red rectangles indicate the xxLxLxx motifs of LRR domain.
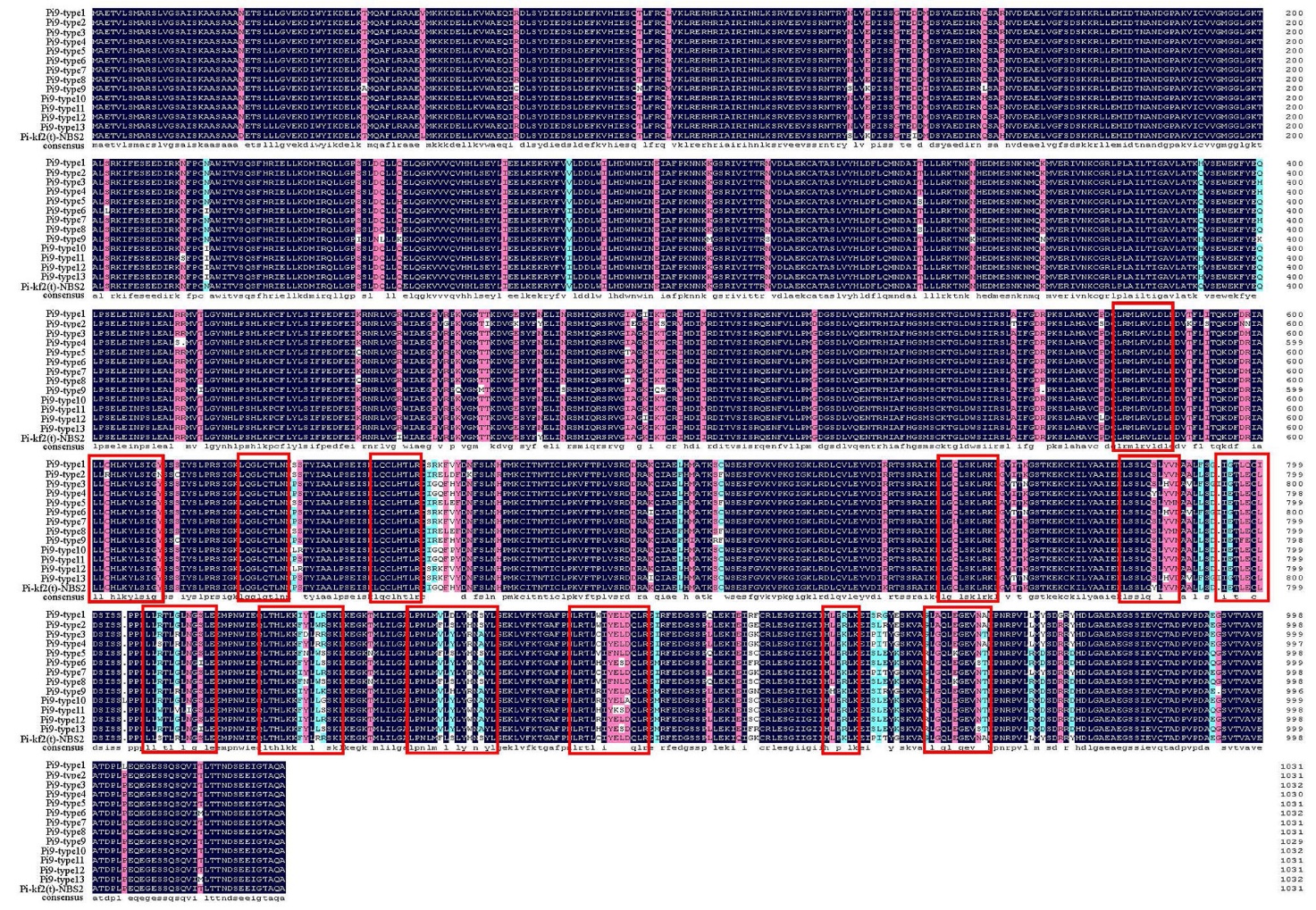
Fig. 4. Amino-acid sequences alignment of Pi-kf2(t)-NBS2 with Pi9-type1-Pi9-type13 Dark blue highlights identical residues, while similar residues are marked in pink or light blue, and red rectangles indicate the xxLxLxx motifs of LRR domain.
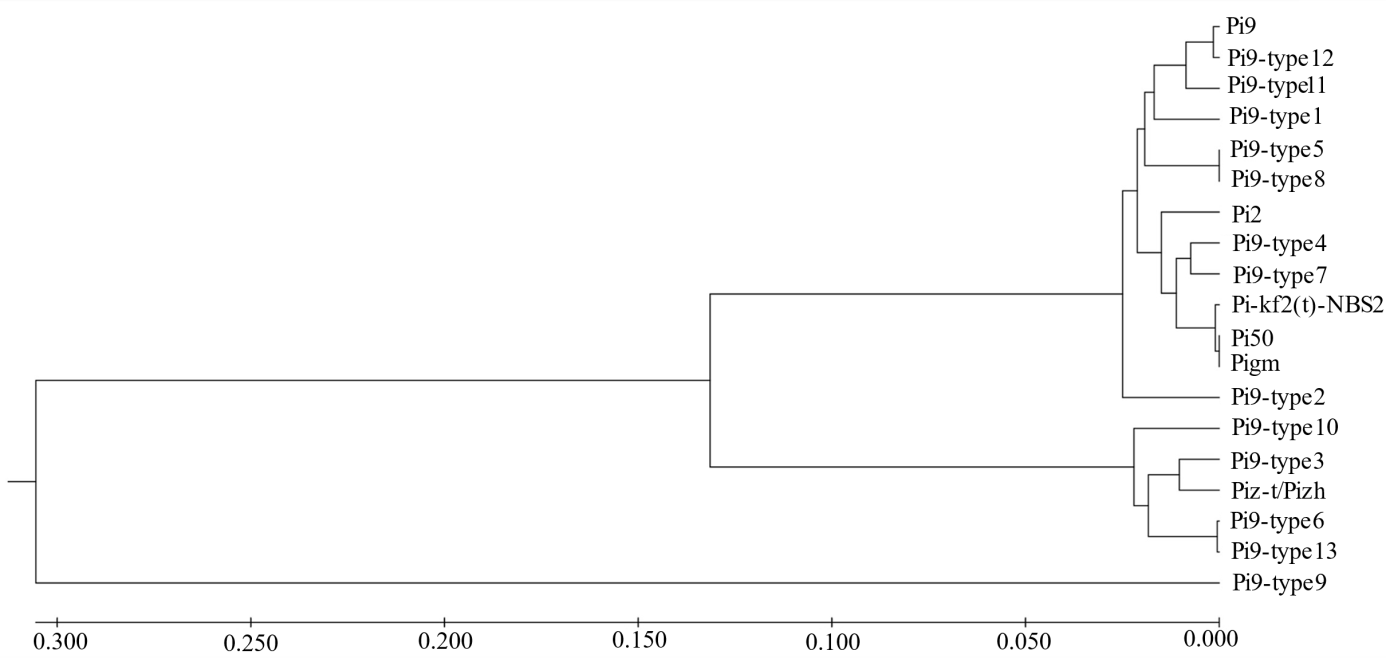
Fig. 5. A phylogenetic tree representing the alignment of Pi-kf2(t)-NBS2 and protein sequences of Pi9, Pi2, Piz-t/Pizh, Pi50, Pigm, and Pi9-type1-Pi9-type13
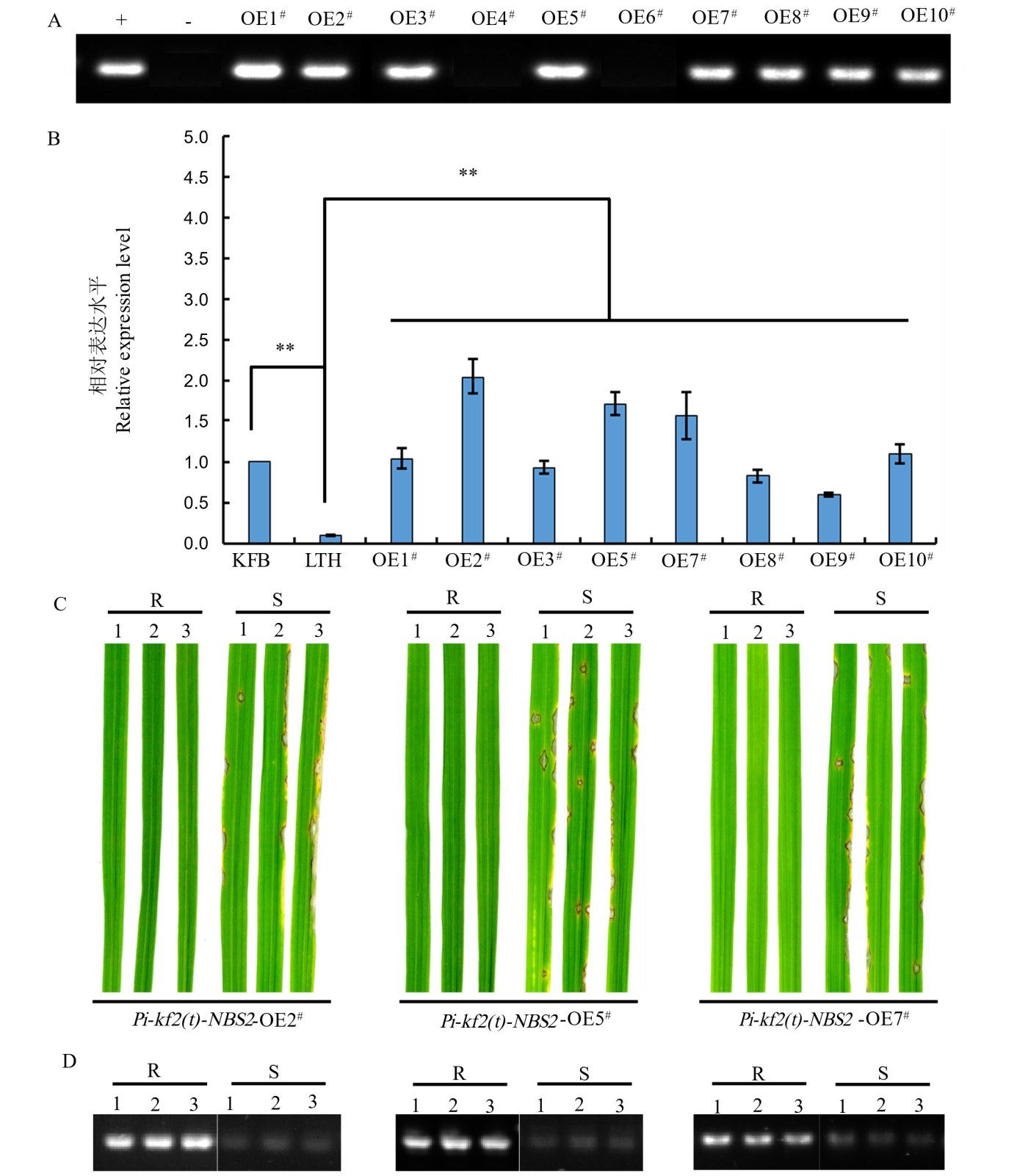
Fig. 6. Functional verification of candidate gene Pi-kf2(t)-NBS2 A, PCR identification of the positive transgenic plants in the overexpressed lines of T0 generation with HPT-F/R, where “+” and “−” indicate the positive control and negative control, respectively; B, Expression levels of Pi-kf2(t)-NBS2 candidate gene in the overexpressed transgenic lines, determined by qRT-PCR, where “**” indicates significant differences at 0.01 level and error bars represent the standard deviation (SD); C, Reaction of T1 transgenic lines inoculated with the isolates J71, J91, and J93, where “R” and “S” represent resistance and susceptibility to rice blast, respectively. Pi-kf2(t)-NBS2-OE2#, Pi-kf2(t)-NBS2-OE5#, and Pi-kf2(t)-NBS2-OE7# were inoculated with isolates J71, J91, and J81, respectively; D, Expression analysis of Pi-kf2(t)-NBS2 candidate gene in the individual plant of T1 transgenic lines, determined by RT-PCR.
| [1] | Valent B, Chumley F G. Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea[J]. Annual Review of Phytopathology, 1991, 29: 443-467. |
| [2] | Oliveira-Garcia E, Yan X, Oses-Ruiz M, de Paula S, Talbot N J. Effector-triggered susceptibility by the rice blast fungus Magnaporthe oryzae[J]. The New Phytologist, 2024, 241(3): 1007-1020. |
| [3] | 杨婕, 杨长登, 曾宇翔, 侯雨萱, 陈天晓, 梁燕. 水稻稻瘟病抗性基因挖掘与利用研究进展[J]. 中国水稻科学, 2024, 38(6): 5: 591-603. |
| Yang J, Yang C D, Zeng Y X, Hou Y X, Chen T X, Liang Y. Research progress on mining and utilization of rice blast resistance genes[J]. Chinese Journal of Rice Science, 2024, 38(6): 591-603. (in Chinese with English abstract) | |
| [4] | 殷得所, 夏明元, 李进波, 万丙良, 查中萍, 杜雪树, 戚华雄. 抗稻瘟病基因Pi9的STS连锁标记开发及在分子标记辅助育种中的应用[J]. 中国水稻科学, 2011, 25(1): 2: 25-30. |
| Yin D S, Xia M Y, Li J B, Wan B L, Zha Z P, Du X S, Qi H X. Development ofof STS marker linked to rice blast resistance gene Pi9 in marker-assisted selection breeding[J]. Chinese Journal of Rice Science, 2011, 25(1): 25-30. (in Chinese with English abstract) | |
| [5] | Zhu X Y, Chen S, Yang J Y, Zhou S C, Zeng L X, Han J L, Su J, Wang L, Pan Q H. The identification of Pi50(t), a new member of the rice blast resistance Pi2/Pi9 multigene family[J]. Theoretical and Applied Genetics, 2012, 124: 1295-1304. |
| [6] | Hulbert S H W, Webb C A S, Smith S M S, Sun Q. Resistance gene complexes: E: Evolution and utilization[J]. Annual Review of Phytopathology, 2001, 39:285-312. |
| [7] | 华丽霞, 汪文娟, 陈深, 汪聪颖, 曾烈先, 杨健源, 朱小源, 苏菁. 抗稻瘟病Pi2/9/z-t基因特异性分子标记的开发[J]. 中国水稻科学, 2015, 29(3): 3: 305-310. |
| Hua L X, Wang W J, Chen S, Wang C Y, Zeng L X, Yang J Y, Zhu X Y, Su J. Development of specific DNA markers for detecting the rice blast resistance gene alleles Pi2/9/z-t[J]. Chinese Journal of Rice Science, 2015, 29(3): 305-310. (in Chinese with English abstract) | |
| [8] | Deng Y W, Zhu X D, Shen Y, He Z H. Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9in a broad-spectrum resistant Chinese variety[J]. Theoretical and Applied Genetics, 2006, 113(4): 705-713. |
| [9] | Wang Z X, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes[J]. The Plant Journal, 1999, 19(1): 55-64. |
| [10] | Qu S L, Liu G Z, Zhou B, Bellizzi M Z, Zeng L D, Dai L H, Han B W, Wang G L. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice[J]. Genetics, 2006, 172(3): 1: 1901-1914. |
| [11] | Hassan B, Peng Y T, Li S, Yin X X, Chen C, Gulzar F, Zhou S X, Pu M, Ji Y P, Wang Y P, Zhao W S, Huang F, Peng Y L, Zhao Z X, Wang W M. Identification of the blast resistance genes in three elite restorer lines of hybrid rice[J]. Phytopathology Research, 2022, 4(1): 15. |
| [12] | Martin G B, Bogdanove A J, Sessa G. Understanding the functions of plant disease resistance proteins[J]. Annual Review of Plant Biology, 2003, 54: 23-61. |
| [13] | Belkhadir Y, Subramaniam R, Dangl J L. Plant disease resistance protein signaling: NBS-LRR proteins and their partners[J]. Current Opinion in Plant Biology, 2004, 7(4): 391-399. |
| [14] | Zhou B Q, Qu S H L, Liu G F D, Dolan M S, Sakai H L, Lu G D, Bellizzi M W, Wang G L. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea[J]. Molecular Plant-Microbe Interactions, 2006, 19( 11):1216-1228. |
| [15] | Tamura M, Tachida H. Evolution of the number of LRRs in plant disease resistance genes[J]. Molecular Genetics and Genomics, 2011, 285(5): 393-402. |
| [16] | Dodds P N L, Lawrence G J E, Ellis J G. Six amino acid changes confined to the leucine-rich repeat beta-strand/beta-turn motif determine the difference between the P and P2 rust resistance specificities in flax[J]. The Plant Cell, 2001, 13(1):163-178. |
| [17] | Wu J L, Fan Y Y, Li D B, Zheng K L, Leung H, Zhuang J Y. Genetic control of rice blast resistance in the durably resistant cultivar Gumei 2 against multiple isolates[J]. Theoretical and Applied Genetics, 2005, 111(1): 50-56. |
| [18] | Jeung J U, Kim B R, Cho Y C, Han S S, Moon H P, Lee Y T, Jena K K. A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice[J]. Theoretical and Applied Genetics, 2007, 115(8): 1163-1177. |
| [19] | Jiang N, Li Z Q, Wu J, Wang Y, Wu L Q, Wang S H, Wang D, Wen T, Liang Y, Sun P Y, Liu J L, Dai L Y, Wang Z L, Wang C, Luo M Z, Liu X L, Wang G L. Molecular mapping of the Pi2/9 allelic genePi2-2 conferring broad-spectrum resistance to Magnaporthe oryzae in the rice cultivar Jefferson[J]. Rice, 2012, 5(1): 29. |
| [20] | Su J, Wang W J, Han J L, Chen S, Wang C Y, Zeng L X, Feng A Q, Yang J Y, Zhou B, Zhu X Y. Functional divergence of duplicated genes results in a novel blast resistance gene Pi50 at the Pi2/9 locus[J]. Theoretical and Applied Genetics, 2015, 128(11): 2213-2225. |
| [21] | Deng Y W, Zhai K R, Xie Z, Yang D Y, Zhu X D, Liu J Z, Wang X, Qin P, Yang Y Z, Zhang G M, Li Q, Zhang J F, Wu S Q, Milazzo J, Mao B Z, Wang E, Xie H A, Tharreau D, He Z H. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance[J]. Science, 2017, 355(6328): 962-965. |
| [22] | Xie Z Y, Yan B X S, Shou J Y T, Tang J W, Wang X Z, Zhai K R L, Liu J Y L, Li Q L, Luo M Z D, Deng Y W H, He Z H. A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice[J]. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences, 2019,374(1767): 20180308. |
| [23] | Lü Q M, Xu X, Shang J J, Jiang G H, Pang Z Q, Zhou Z Z, Wang J, Liu Y, Li T, Li X B, Xu J C, Cheng Z K, Zhao X F, Li S G, Zhu L H. Functional analysis of Pid3-A4, an ortholog of rice blast resistance gene Pid3 revealed by allele mining in common wild rice[J]. Phytopathology, 2013, 103: 594-599. |
| [24] | Zhou Y, Lei F, Wang Q, He W C, Yuan B, Yuan W Y. Identification of novel alleles of the rice blast-resistance gene Pi9 through sequence-based allele mining[J]. Rice, 2020, 13: 80. |
| [25] | Li L Y W, Wang L J, Jing J X L, Li Z Q L, Lin F H, Huang L F P, Pan Q H. The Pikm gene, c, conferring stable resistance to isolates of Magnaporthe oryzae,was finely mapped in a crossover-cold region on rice chromosome 11[J]. Molecular Breeding, 2007, 20:179-188. |
| [26] | Kovi B, Sakai T, Abe A, Kanzaki E, Terauchi R, Shimizu M. Isolation of Pikps, an allele of Pik, from the aus rice cultivar Shoni[J]. Genes and Genetic Systems, 2023, 97(5): 229-235. |
| [27] | 黄衍焱, 李燕, 王贺, 王文明. 水稻小种特异性抗稻瘟病基因的等位性变异研究进展[J]. 植物病理学报, 2023, 53(5):753-768. |
| Huang Y Y L, Li Y W, Wang H W, Wang W M, Allelic variation in the race-specific blast resistance genes in rice[J]. Acta Phytopathologica Sinica, 2023, 53(5):753-768. (in Chinese with English abstract) | |
| [28] | Rybka K M, Miyamoto M A, Ando I S, Saito A K, Kawasaki S. High resolution mapping of the indica-derived rice blast resistance genes:II. Pi-ta2 and Pi-ta and a consideration of their origin[J]. Molecular Plant-Microbe Interactions, 1997, 10(4):517-524. |
| [29] | Liu X Q, Yang Q Z, Lin F, Hua L X, Wang C T, Wang L, Pan Q H. Identification and fine mapping of Pi39(t), a major gene conferring the broad-spectrum resistance to Magnaporthe oryzae[J]. Molecular Genetics and Genomics, 2007, 278(4): 403-410. |
| [30] | 吴俊, 刘雄伦, 戴良英, 王国梁. 水稻广谱抗稻瘟病基因研究进展[J]. 生命科学, 2007, 19(2):233-238. |
| Wu J, Liu X L, Dai L Y, Wang G L. Advances on the identification and characterization of broad-spectrum blast resistance genes in rice[J]. Chinese Bulletin of Life Science, 2007, 19(2): 233-238. (in Chinese with English abstract) | |
| [31] | Sallaud C, Lorieux M, Roumen E, Tharreau D, Berruyer R, Svestasrani P, Garsmeur O, Ghesquiere A, Notteghem J L. Identification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy[J]. Theoretical and Applied Genetics, 2003, 106: 794-803. |
| [32] | Liu B, Zhang S H, Zhu X Y, Yang Q Y, Wu S Z, Mei M T, Mauleon R, Leach J, Mew T, Leung H. Candidate defense genes as predictors of quantitative blast resistance in rice[J]. Molecular Plant-Microbe Interactions, 2004, 17: 1146-1152. |
| [33] | 韦新宇, 许旭明, 张锐, 陈美莲, 马彬林, 邹文广, 杨旺兴, 卓伟, 王宗华, 梁康迳. 籼粳交新种质康丰A对稻瘟病抗性的遗传[J]. 植物遗传资源学报, 2014, 15(5):1133-1137. |
| Wei X Y, Xu X M, Zhang R, Chen M L, Ma B L, Zou W G, Yang W X, Zhuo W, Wang Z H, Liang K J. Inheritance of blast resistance in new germplasm Kangfeng A from indica-japonica crosses[J]. Journal of Plant Genetic Resources, 2014, 15(5):1133-1137. (in Chinese with English abstract) | |
| [34] | Wei X Y Z, Zeng Y H Z, Zhang R H, Huang J H Y, Yang W X Z, Zou W G X, Xu X M. Fine mapping and identification of the rice blast-resistance locus Pi-kf2(t) as a new member of the Pi2/Pi9 multigene family[J]. Molecular Breeding, 2019,39:108. |
| [35] | Bonman J M, Khush G S, N, Nelson R J. Breeding rice for resistance to pests[J]. Annual Review of Phytopathology, 1992, 30:507-528. |
| [36] | 韦新宇, 曾跃辉, 杨旺兴, 肖长春, 候新坡, 黄建鸿, 邹文广, 许旭明. 利用CRISPR-Cas9技术编辑Badh2基因创制优质香型籼稻三系不育系[J]. . 作物学报, 2023, 49(8):2144-2159. |
| Wei X Y, Zeng Y H, Yang W X, Xiao C C, Hou X P, Huang J H, Zou W G, Xu X M. Development of high-quality fragrant indica CMS line by editing Badh2 gene using CRISPR-Cas9 technology in rice (Oryza sativa L.)[J]. Acta Agronomica Sinica, 2023, 49(8):2144-2159. (in Chinese with English abstract) | |
| [37] | Zeng Y H, Wei X Y, Xiao C C, Zhang R, Huang J H, Xu X M. Fine mapping and identification of a novel albino gene OsAL50 that is required for chlorophyll biosynthesis and chloroplast development in rice (Oryza sativa L.)[J]. Plant Growth Regulation, 2024, 103(2):389-407. |
| [38] | 曾跃辉, 韦新宇, 黄建鸿, 肖长春, 张锐, 尚伟, 许旭明. 不同来源特种稻香味和黑色种皮基因的鉴定与遗传特性分析[J]. 植物遗传资源学报, 2021, 22(4):951-962. |
| Zeng Y H, Wei X Y, Huang J H, Xiao C C, Zhang R, Shang W, Xu X M. Identification and genetic analysis of the genes for fragrance and black pericarp in special rice from different regions[J]. Journal of Plant Genetic Resources, 2021, 22(4):951-962. (in Chinese with English abstract) | |
| [39] | 陈深, 苏菁, 华丽霞, 汪文娟, 汪聪颖, 杨健源, 曾烈先, 朱小源. 水稻恢复系华占抗稻瘟病遗传分析及基因鉴定[J]. 植物病理学报, 2015, 45(6):598-605. |
| Chen S, Su J, Hua L X, Wang W J, Wang C Y, Yang J Y, Zeng L X, Zhu X Y. Genetic analysis and gene identification of restorer line Huazhan against rice blast[J]. Acta Phytopathologica Sinica, 2015, 45(6):598-605. (in Chinese with English abstract) | |
| [40] | Ellis J, Dodds P, Pryor T. The generation of plant disease resistance gene specificities[J]. Trends in Plant Science, 2000, 5(9):373-379. |
| [1] |
TANG Chenghan, CHEN Huizhe, HUAI Yan, SUN Liang, ZHANG Yuping, XIANG Jing, ZHANG Yikai, WANG Zhigang, XU Yiwen, WANG Yaliang.
Response of Machine-transplanting Quality and Yield Formation of Hybrid Rice Pot-mat Seedlings to Pot Depth [J]. Chinese Journal OF Rice Science, 2025, 39(4): 491-500. |
| [2] |
ZHU Peng, LING Xitie, WANG Jinyan, ZHANG Baolong, YANG Yuwen, XU Ke, QIU Shi.
Effects of Weed Control Methods on Grain Yield and Quality of Herbicide-resistant Rice Under Direct Seeding [J]. Chinese Journal OF Rice Science, 2025, 39(4): 501-515. |
| [3] |
DONG Liqiang, ZHANG Yikai, YANG Tiexin, FENG Yingying, MA Liang, LIANG Xiao, ZHANG Yuping, LI Yuedong.
Effect of Dense Sowing Nursery on Seedling Quality and Picking Characteristics for Mechanized Transplanting inNorthern japonica Rice [J]. Chinese Journal OF Rice Science, 2025, 39(4): 516-528. |
| [4] |
ZHOU Yang, YE Fan, LIU Lijun.
Research Progress of Typical Plant Growth-promoting Microorganism Enhancing Salt Stress Resistance in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(4): 529-542. |
| [5] |
ZHU Jianping, LI Xia, LI Wenqi, XU Yang, WANG Fangquan, TAO Yajun, JIANG Yanjie, CHEN Zhihui, FAN Fangjun, YANG Jie, YANG Jie.
Phenotypic Analysis and Gene Mapping of a Floury Endosperm Mutant we1 in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(4): 543-551. |
| [6] | HUANG Fudeng, WU Chunyan, HAO Yuanyuan, HAN Yifei, ZHANG Xiaobin, SUN Huifeng, PAN Gang . Transcriptome Analysis of Top Second Leaf Sheath of Rice Under Different Nitrogen Fertilizer Levels [J]. Chinese Journal OF Rice Science, 2025, 39(4): 563-571. |
| [7] | LU Yezi, QIU Jiehua, JIANG Nan, KOU Yanjun, SHI Huanbin. Research Progress in Effectors of Magnaporthe oryzae [J]. Chinese Journal OF Rice Science, 2025, 39(3): 287-294. |
| [8] | WANG Chaorui, ZHOU Yukun, WEN Ya, ZHANG Ying, FA Xiaotong, XIAO Zhilin, ZHANG Hao. Effects of Straw Returning Methods on Soil Characteristics and Greenhouse Gas Emissions in Paddy Fields and Their Regulation Through Water-fertilizer Interactions [J]. Chinese Journal OF Rice Science, 2025, 39(3): 295-305. |
| [9] | WANG Yaxuan, WANG Xinfeng, YANG Houhong, LIU Fang, XIAO Jing, CAI Yubiao, WEI Qi, FU Qiang, WAN Pinjun. Recent Advances in Mechanisms of Adaptation of Planthoppers to Rice Resistance [J]. Chinese Journal OF Rice Science, 2025, 39(3): 306-321. |
| [10] | HUANG Tao, WEI Zhaogen, CHENG Qi, CHENG Ze, LIU Xin, WANG Guangda, HU Keming, XIE Wenya, CHEN Zongxiang, FENG Zhiming, ZUO Shimin. Gene Cloning and Broad-spectrum Disease Resistance Analysis of Rice Lesion Mimic Mutant lm52 [J]. Chinese Journal OF Rice Science, 2025, 39(3): 322-330. |
| [11] | MA Shunting, HU Yungao, GAO Fangyuan, LIU Liping, MOU Changling, LÜ Jianqun, SU Xiangwen, LIU Song, LIANG Yuyu, REN Guangjun, GUO Hongming. Functional Study of Rice Eukaryotic Translation Initiation Factor OseIF6.2 in Grain Size Regulation [J]. Chinese Journal OF Rice Science, 2025, 39(3): 331-342. |
| [12] | ZHANG Bintao, LIU Congcong, GUO Mingliang, YANG Shaohua, WU Shiqiang, GUO Longbiao, ZHU Yiwang. Evaluation of Blast Resistance and Identification of Superior Haplotype of OsDR8 in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(3): 343-351. |
| [13] | LI Wenqi, XU Yang, WANG Fangquan, ZHU Jianping, TAO Yajun, LI Xia, FAN Fangjun, JIANG Yanjie, CHEN Zhihui, YANG Jie. Development and Application of KASP Marker for Broad-Spectrum Resistance Gene PigmR to Rice Blast [J]. Chinese Journal OF Rice Science, 2025, 39(3): 365-372. |
| [14] | WEI Huanhe, WANG Lulu, MA Weiyi, ZHANG Xiang, ZUO Boyuan, GENG Xiaoyu, ZHU Wang, ZHU Jizou, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, DAI Qigen. Grain-filling Characteristics and Its Relationship with Grain Yield Formation of japonica Rice Nanjing 9108 Under Combined Salinity-drought Stress [J]. Chinese Journal OF Rice Science, 2025, 39(3): 373-386. |
| [15] | ZHANG Haiwei, GU Xinyi, CHEN Mingshuai, LI Fukang, SHI Yuecheng, YANG Ting, JIANG Shuochen. Effects of Nitrogen Type of Basal Fertilizer on Growth, Grain Yield and Nitrogen Use Efficiency of Ratooning Rice [J]. Chinese Journal OF Rice Science, 2025, 39(3): 387-398. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||