
Chinese Journal OF Rice Science ›› 2025, Vol. 39 ›› Issue (3): 331-342.DOI: 10.16819/j.1001-7216.2025.2401010
• Research Papers • Previous Articles Next Articles
MA Shunting1, HU Yungao1, GAO Fangyuan2, LIU Liping2, MOU Changling2, LÜ Jianqun2, SU Xiangwen2, LIU Song2, LIANG Yuyu1, REN Guangjun2,*( ), GUO Hongming2,*(
), GUO Hongming2,*( )
)
Received:2024-10-25
Revised:2025-03-27
Online:2025-05-10
Published:2025-05-21
Contact:
*email: guangjun61@sina.com;hongmingguo552@163.com
马顺婷1, 胡运高1, 高方远2, 刘利平2, 牟昌铃2, 吕建群2, 苏相文2, 刘松2, 梁毓玉1, 任光俊2,*( ), 郭鸿鸣2,*(
), 郭鸿鸣2,*( )
)
通讯作者:
*email: guangjun61@sina.com;hongmingguo552@163.com
基金资助:MA Shunting, HU Yungao, GAO Fangyuan, LIU Liping, MOU Changling, LÜ Jianqun, SU Xiangwen, LIU Song, LIANG Yuyu, REN Guangjun, GUO Hongming. Functional Study of Rice Eukaryotic Translation Initiation Factor OseIF6.2 in Grain Size Regulation[J]. Chinese Journal OF Rice Science, 2025, 39(3): 331-342.
马顺婷, 胡运高, 高方远, 刘利平, 牟昌铃, 吕建群, 苏相文, 刘松, 梁毓玉, 任光俊, 郭鸿鸣. 水稻真核翻译起始因子OseIF6.2调控粒型的功能研究[J]. 中国水稻科学, 2025, 39(3): 331-342.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2025.2401010
| 引物名称 Primer name | 引物序列 Sequence (5'-3') | 用途 Purpose |
|---|---|---|
| sgRNA序列 | GCGAGATTGGAGTGTTCGCGAGG | OseIF6.2基因sgRNA序列 |
| eIF6.2-CDS-F | ATGATTTTTTTTTTCAAAAATCGGTTCTGCATTGCA | CDS 扩增 |
| eIF6.2-CDS-R | CTATAGGTAGCTGCTGACGACGAGCGA | CDS amplification |
| K21-F | AGTTTGTAACAGCTGCTGGGATTAC | 亚细胞定位载体 |
| K21-NOSR-R | CCATCTCATAAATAACGTCATGCATTACATGTT | Subcellular localization vector |
| eIF6.2-Cas9-F | AGGAGTTCTTGGTTGGCTGACTGCGA | 基因编辑材料鉴定 |
| eIF6.2-Cas9-R | ACGCACATCCTCCCGATGATCCT | Identification of gene editing materials |
| OsCas9-1000-F | GAGGAGACAATCACCCCCTGGAACT | Cas9蛋白鉴定 |
| OsCas9-1000-R | TTATCGGACTTGCCCCTGTTCTTGT | Identification of Cas9 protein |
| eIF6.1-2YC-767-F | ACGATAGTTAATTAACATGGCGACCCGTATTCAGTTTG | BiFC载体 |
| eIF6.1-2YC-767-R | GATCGACAGTTATGTGCACTAGTGGCGCGCCC | Bimolecular fluorescence complementation vector |
| eIF6.1-CDS-759-F | CCTTAATTAAGGATGGCGACCCGTATTCAGTTTGAGAAC | CDS 扩增 |
| eIF6.1-CDS-759-R | TTGGCGCGCCAACACATAACTGTCGATCAAAG | CDS amplification |
| eIF6.2-BD-798-F | GAGGACCTGCATATGATGATTTTTTTTTTCAAAAATCG | 酵母双杂载体 |
| eIF6.2-BD-798-R | GATCCCCGGGAATTCCTATAGGTAGCTG | Construction of yeast two-hybrid vector |
| eIF6.2-2YN-797-F | AC GATAGTTAATTAACATGATTTTTTTTTTCAAAAATCGG | BiFC载体构建 |
| eIF6.2-2YN-797-R | CACTAGTGGCGCGCCCTAGGTAGCTGC | Constructionof of bimolecular fluorescence complementation vector |
| Actin-qPCR-F | TGTATGCCAGTGGTCGTACCA | 内参基因定量引物 |
| Actin-qPCR-R | CCAGCAAGGTCGAGACGAA | Primers of internal reference gene |
| eIF6.2-qPCR-148-F | AGAGAACTTCTTCAGCGTGTTC | OseIF6.2 定量引物 |
| eIF6.2-qPCR-148-R | CTCTTGGTCTGTGGTGGTATG | qRT-PCR primers of OseIF6.2 |
| OsEXPA5-F | AAGGCTGTGGCTTGATTGACA | 细胞膨大相关基因定量引物 |
| OsEXPA5-R | TTAGGCCCAATTTTGCTATTTTG | qRT-PCR primers of genes related to cell expansion |
| OsEXPA8-F | GCGATGAGCCGCAACTG | 细胞膨大相关基因定量引物 |
| OsEXPA8-R | CTCTTCCATCCTATGCCACG | qRT-PCR primers of genes related to cell expansion |
| OsEXPA30-F | CCAAGTTCAGGGCGATGCAG | 细胞膨大相关基因定量引物 |
| OsEXPA30-R | ACGGTGATCGCCGGCGAGCC | qRT-PCR primers of genes related to cell expansion |
| OsEXPB3-F | CTTTGAGTGGTTGGAGTGGTGG | 细胞膨大相关基因定量引物 |
| OsEXPB3-R | GCAGCCTTCTTGGAGATGGAA | qRT-PCR primers of genes related to cell expansion |
Table 1. Primers used in this study
| 引物名称 Primer name | 引物序列 Sequence (5'-3') | 用途 Purpose |
|---|---|---|
| sgRNA序列 | GCGAGATTGGAGTGTTCGCGAGG | OseIF6.2基因sgRNA序列 |
| eIF6.2-CDS-F | ATGATTTTTTTTTTCAAAAATCGGTTCTGCATTGCA | CDS 扩增 |
| eIF6.2-CDS-R | CTATAGGTAGCTGCTGACGACGAGCGA | CDS amplification |
| K21-F | AGTTTGTAACAGCTGCTGGGATTAC | 亚细胞定位载体 |
| K21-NOSR-R | CCATCTCATAAATAACGTCATGCATTACATGTT | Subcellular localization vector |
| eIF6.2-Cas9-F | AGGAGTTCTTGGTTGGCTGACTGCGA | 基因编辑材料鉴定 |
| eIF6.2-Cas9-R | ACGCACATCCTCCCGATGATCCT | Identification of gene editing materials |
| OsCas9-1000-F | GAGGAGACAATCACCCCCTGGAACT | Cas9蛋白鉴定 |
| OsCas9-1000-R | TTATCGGACTTGCCCCTGTTCTTGT | Identification of Cas9 protein |
| eIF6.1-2YC-767-F | ACGATAGTTAATTAACATGGCGACCCGTATTCAGTTTG | BiFC载体 |
| eIF6.1-2YC-767-R | GATCGACAGTTATGTGCACTAGTGGCGCGCCC | Bimolecular fluorescence complementation vector |
| eIF6.1-CDS-759-F | CCTTAATTAAGGATGGCGACCCGTATTCAGTTTGAGAAC | CDS 扩增 |
| eIF6.1-CDS-759-R | TTGGCGCGCCAACACATAACTGTCGATCAAAG | CDS amplification |
| eIF6.2-BD-798-F | GAGGACCTGCATATGATGATTTTTTTTTTCAAAAATCG | 酵母双杂载体 |
| eIF6.2-BD-798-R | GATCCCCGGGAATTCCTATAGGTAGCTG | Construction of yeast two-hybrid vector |
| eIF6.2-2YN-797-F | AC GATAGTTAATTAACATGATTTTTTTTTTCAAAAATCGG | BiFC载体构建 |
| eIF6.2-2YN-797-R | CACTAGTGGCGCGCCCTAGGTAGCTGC | Constructionof of bimolecular fluorescence complementation vector |
| Actin-qPCR-F | TGTATGCCAGTGGTCGTACCA | 内参基因定量引物 |
| Actin-qPCR-R | CCAGCAAGGTCGAGACGAA | Primers of internal reference gene |
| eIF6.2-qPCR-148-F | AGAGAACTTCTTCAGCGTGTTC | OseIF6.2 定量引物 |
| eIF6.2-qPCR-148-R | CTCTTGGTCTGTGGTGGTATG | qRT-PCR primers of OseIF6.2 |
| OsEXPA5-F | AAGGCTGTGGCTTGATTGACA | 细胞膨大相关基因定量引物 |
| OsEXPA5-R | TTAGGCCCAATTTTGCTATTTTG | qRT-PCR primers of genes related to cell expansion |
| OsEXPA8-F | GCGATGAGCCGCAACTG | 细胞膨大相关基因定量引物 |
| OsEXPA8-R | CTCTTCCATCCTATGCCACG | qRT-PCR primers of genes related to cell expansion |
| OsEXPA30-F | CCAAGTTCAGGGCGATGCAG | 细胞膨大相关基因定量引物 |
| OsEXPA30-R | ACGGTGATCGCCGGCGAGCC | qRT-PCR primers of genes related to cell expansion |
| OsEXPB3-F | CTTTGAGTGGTTGGAGTGGTGG | 细胞膨大相关基因定量引物 |
| OsEXPB3-R | GCAGCCTTCTTGGAGATGGAA | qRT-PCR primers of genes related to cell expansion |
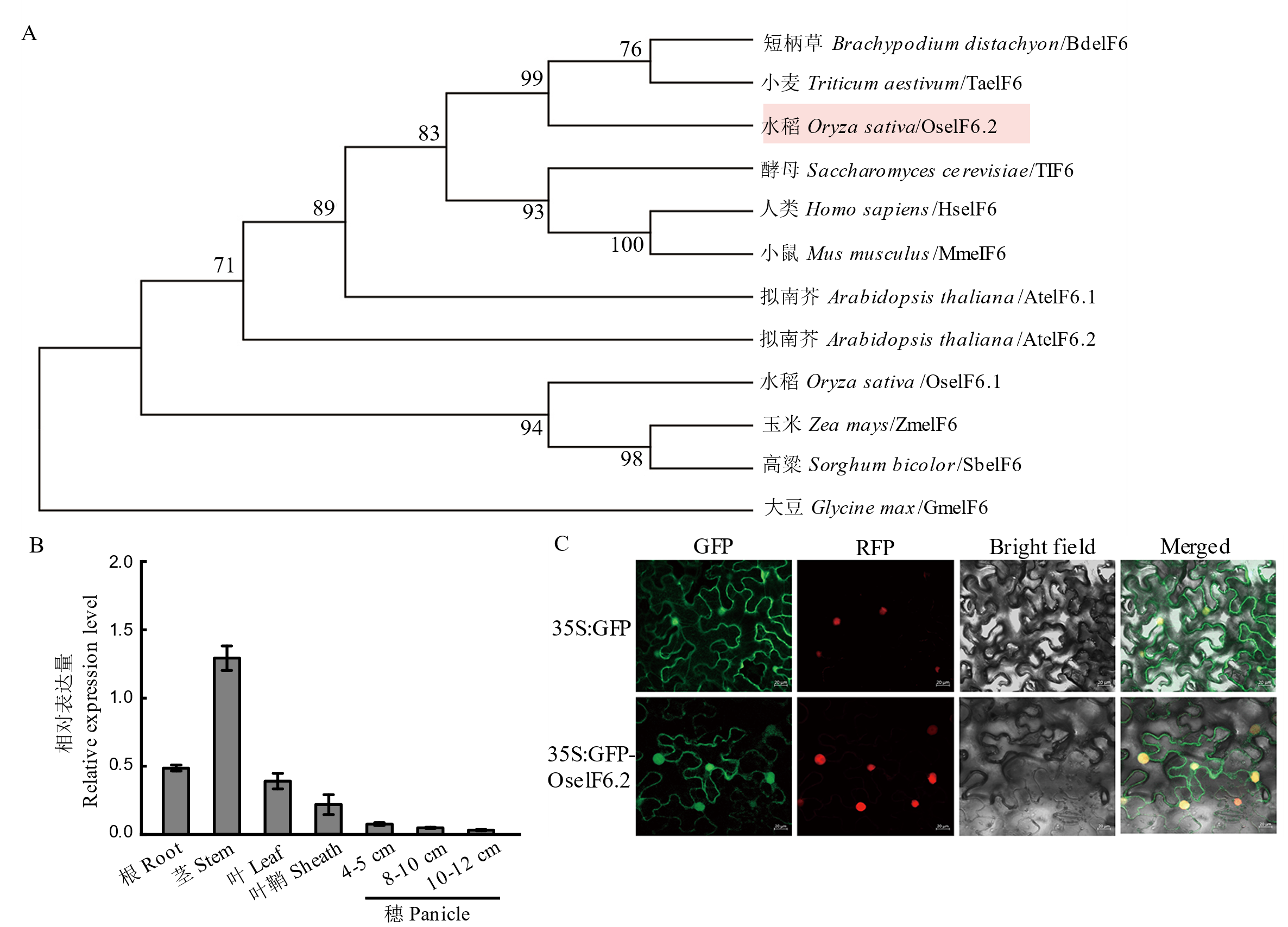
Fig. 1. Expression pattern and subcellular localization of OseIF6.2 A, Comparative phylogenetic analysis of eIF6 protein cross diverse eukaryotic species. B, Relative expression of OseIF6.2 in root, stem, leaf, and sheath of young seedlings and developing panicles at lengths of 4-5, 8-10, and 10-12 cm. OsActin served as the internal control. Values represent means ± SE from at least three independent experiments. C, Subcellular localization of OseIF6.2 in N. benthamiana leaves. YFP: Yellow fluorescent protein, RFP: Red fluorescent protein. Scale bars = 20 μm.

Fig. 2. Types and phenotypic analysis of OseIF6.2 mutants A, Mutation types of OseIF6.2. B, Plants and panicles of WT and oseif6.2 mutant at the mature stage. Scale bars = 10 cm for plant height and 1 cm for panicle length. C, Panicle length of WT and oseif6.2 mutant. D, Number of primary branches per panicle of WT and oseif6.2 mutant. E, Number of secondary branches of WT and oseif6.2 mutant. F, Seed setting rate of WT and oseif6.2 mutant. Values represent means ± SE from at least three independent experiments, Student's t-test: *P< 0.05, **P< 0.01, ***P< 0.001.

Fig. 3. Mutation of OseIF6.2 alters grain shape A, Mature grains of WT and oseif6.2 mutants. Scale bars =1 cm. B, Grain length of WT and oseif6.2 mutants; C, Grain width; D, Grain thickness; E, 1000-grain weight; F, Filled grains per panicle; G, Panicle number per plant; H, Grain yield per plant. Values represent means ± SE from at least three independent experiments, Student's t-test: *P < 0.05, **P < 0.01, ***P< 0.001.

Fig. 4. OseIF6.2 regulates grain size by affecting cell expansion A, Scanning electron microscopy images of the glume outer surfaces of WT and oseif6.2 mutants. Scale bars = 100 μm. Cell length (B) and width (C) of the outer epidermal of WT and oseif6.2 mutant lemmas. D, Cell area of the outer epidermal of WT and oseif6.2 mutant lemmas. Values represent means ± SE from at least three independent experiments. Student's t-test: *P < 0.05, **P < 0.01, ***P < 0.001.
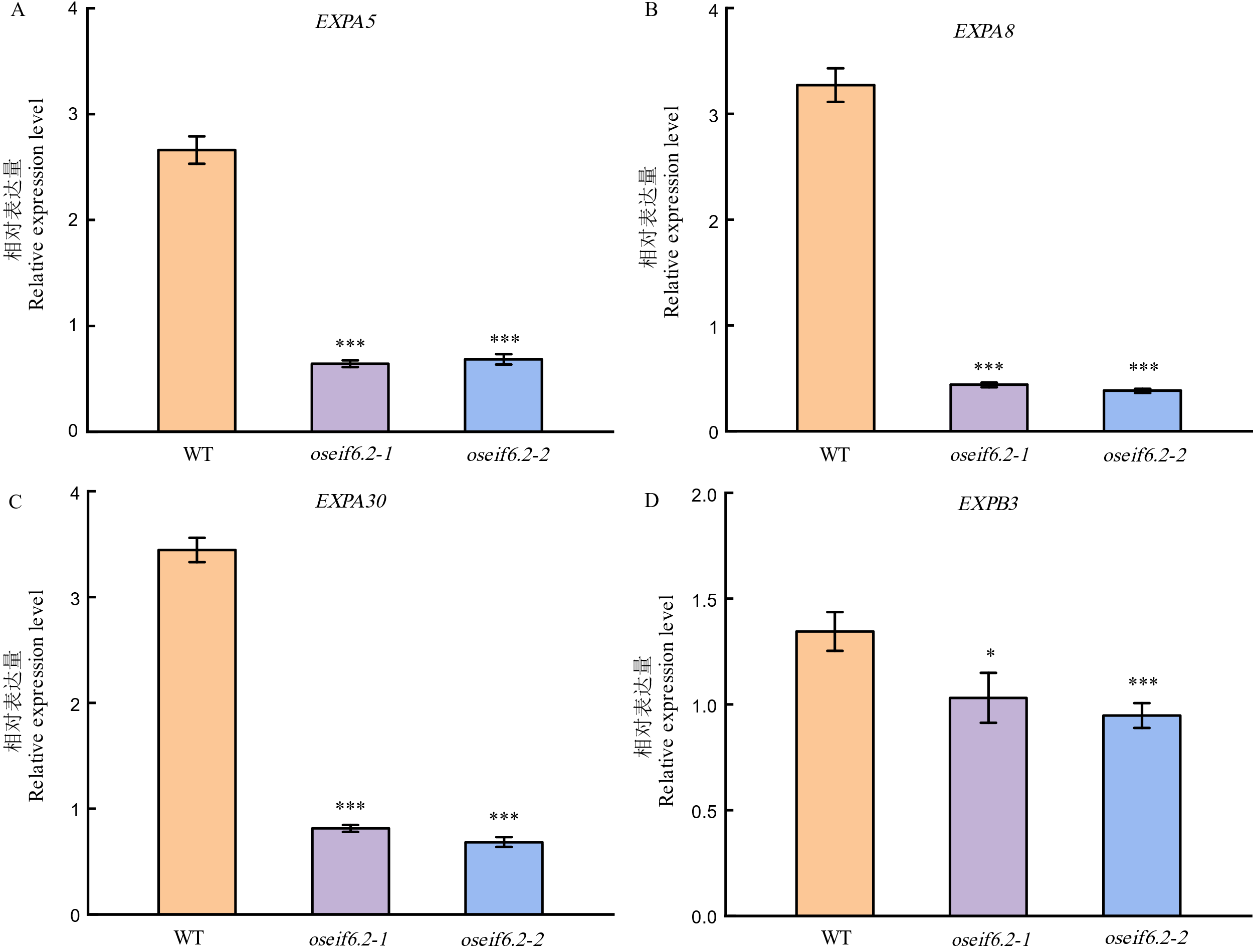
Fig. 5. Expression levels of genes related to cell expansion in WT and oseif6.2 mutant lines The values represent means ± SE derived from at least three independent experiments. Student's t-test: *P < 0.05, **P < 0.01, ***P < 0.001.

Fig. 6. OseIF6.2 physically interacts with OseIF6.1 A, OseIF6.2 interacts with OseIF6.1 in yeast cells. Transformed cells were cultured on DDO or QDO/X/A media. B, Bimolecular fluorescence complementation assays verifies the interaction between OseIF6.2 and OseIF6.1 in N. benthamiana. OseIF6.2-nYFP was coexpressed with OseIF6.1-cYFP in cells of N. benthamiana. Scale bars = 10 μm.
| [1] | Su S, Hong J, Chen X, Zhang C, Chen M, Luo Z, Chang S, Bai S, Liang W, Liu Q, Zhang D. Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice[J]. Plant Biotechnology Journal, 2021, 19(11): 2304-2318. |
| [2] | Merrick W C, Pavitt G D. Protein synthesis initiation in eukaryotic cells[J]. Cold Spring Harbor Perspectives in Biology, 2018, 10(12): a033092. |
| [3] | Jackson R J, Hellen C U T, Pestova T V. The mechanism of eukaryotic translation initiation and principles of its regulation[J]. Nature Reviews Molecular Cell Biology, 2010, 11(2): 113-127. |
| [4] | Guo J, Jin Z, Yang X, Li J F, Chen J G. Eukaryotic initiation factor 6, an evolutionarily conserved regulator of ribosome biogenesis and protein translation[J]. Plant Signaling & Behavior, 2011, 6(5): 766-771. |
| [5] | Aitken C E, Lorsch J R. A mechanistic overview of translation initiation in eukaryotes[J]. Nature structural & Molecular Biology, 2012, 19(6): 568-576. |
| [6] | Raabe K, Honys D, Michailidis C. The role of eukaryotic initiation factor 3 in plant translation regulation[J]. Plant Physiology and Biochemistry, 2019, 145: 75-83. |
| [7] | Castellano M, Merchante C. Peculiarities of the regulation of translation initiation in plants[J]. Current Opinion in Plant Biology, 2021, 63: 102073. |
| [8] | Singha D L, Maharana J, Panda D, Dehury B, Modi M K, Singh S. Understanding the thermal response of rice eukaryotic transcription factor eIF4A1 towards dynamic temperature stress: insights from expression profiling and molecular dynamics simulation[J]. Journal of Biomolecular Structure and Dynamics, 2021, 39(7): 2575-2584. |
| [9] | Ma L, Yang Y, Wang Y, Cheng K, Zhou X, Li J, Zhang J, Li R, Zhang L, Wang K, Zeng N, Gong Y, Zhu D, Deng Z, Qu G, Zhu B, Fu D, Luo Y, Zhu H. SlRBP1 promotes translational efficiency via SleIF4A2 to maintain chloroplast function in tomato[J]. The Plant Cell, 2022, 34(7): 2747-2764. |
| [10] | Rausell A, Kanhonou R, Yenush L, Serrano R, Ros R. The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants[J]. Plant Journal, 2003, 34: 257-267. |
| [11] | Sahoo R K, Gill S S, Tuteja N. Pea DNA helicase 45 promotes salinity stress tolerance in IR64 rice with improved yield[J]. Plant Signaling&Behavior, 2012, 7: 1042-104. |
| [12] | Zhang L, Liu X, Gaikwad K, Kou X, Wang F, Tian X, Xin M, Ni Z, Sun Q, Peng H, Vierling E. Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in Arabidopsis thaliana[J]. The Plant Cell, 2017, 29(8): 1952-1969. |
| [13] | Zheng T C, Zang L N, Dai L J, Yang C P, Qu G Z. Two novel eukaryotic translation initiation factor 5A genes from Populus simonii×P. nigra confer tolerance to abiotic stresses in Saccharomyces cerevisiae[J]. Journal Frontiers Research, 2017, 28: 453-46. |
| [14] | Li L S, Luo C, Huang F, Liu Z L, An Z Y, Dong L, He X H. Identification and characterization of the mango eIF gene family reveals MieIF1A-a, which confers tolerance to salt stress in transgenic Arabidopsis[J]. Scientia Horticulturae, 2019, 248: 274-281. |
| [15] | Wang L, Xu C, Wang C, Wang Y. Characterization of a eukaryotic translation initiation factor 5A homolog from Tamarix androssowii involved in plant abiotic stress tolerance[J]. BMC Plant Biology, 2012, 12(1): 118. |
| [16] | 王雷, 孙尧, 李瑶, 吴琼, 夏新莉. 罗布麻eIF-5A基因功能[J]. 北方园艺, 2017(23): 190-193. |
| Wang L, Sun Y, Li Y, Wu Q, XIia X L. Function of eIF-5A gene in Apocynum venetum L[J]. Norther Horticulture, 2017(23):190-193. | |
| [17] | Kenia S D, Mayra A L, Eloísa H L, Brenda N R, Marlon A P T, Jesús H J P, Marina G R, Tzvetanka D D. Arabidopsis thaliana eIF4E1 and eIF(iso)4E participate in cold response and promote translation of some stress-related mRNAs[J]. Frontiers in Plant Science, 2021, 12(12): 698585. |
| [18] | Wang W, Xu M, Liu X, Tu J. The rice eukaryotic translation initiation factor 3 subunit e (OseIF3e) influences organ size and pollen maturation[J]. Frontiers in Plant Science, 2016, 7: 1399. |
| [19] | Huang Y, Zheng P, Liu X, Chen H, Tu J. OseIF3h regulates plant growth and pollen development at translational level presumably through interaction with OsMTA2[J]. Plants, 2021, 10(6): 1101. |
| [20] | Kim T H, Kim B H, Yahalom A, Chamovitz D A, von Arnim A G. Translational regulation via 5' mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h[J]. The Plant Cell, 2004, 16(12): 3341-3356. |
| [21] | Kim B H, Cai X, Vaughn J N, vonArnim A G. On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation[J]. Genome Biology. 2007, 8: R60. |
| [22] | Li N, Li Y. Signaling pathways of seed size control in plants[J]. Current opinion in Plant Bitology, 2016, 33: 23-32. |
| [23] | Xia C, Wang Y J, Li W Q, Chen Y R, Deng Y, Zhang X Q, Chen L Q, Ye D. The Arabidopsis eukaryotic translation initiation factor 3, subunit F (AteIF3f), is required for pollen germination and embryogenesis[J]. The Plant Journal, 2010, 63(2): 189-202. |
| [24] | Feng H, Chen Q, Feng J, Zhang J, Yang X, Zuo J. Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death[J]. Plant Physiology, 2007, 144: 1531-1545. |
| [25] | Hopkins M T, Lampi Y, Wang T W, Liu Z, Thompson J E. Eukaryotic translation initiation factor 5A is involved in pathogen-induced cell death and development of disease symptoms in Arabidopsis[J]. Plant Physiology, 2008, 148: 479-489. |
| [26] | Wang T W, Lu L, Wang D, Thompson J E. Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eucaryotic translation initiation factor 5A from tomato[J]. Journal of Biological Chemistry, 2001, 276(20): 17541. |
| [27] | Lin L, Cao J, Du A, An Q, Chen X, Yuan S, Batool W, Shabbir A, Zhang D, Wang Z, Norvienyeku J. EIF3k domain-containing protein regulates conidiogenesis, appressorium turgor, virulence, stress tolerance, and physiological and pathogenic development of Magnaporthe oryzae Oryzae[J]. Frontiers in Plant Science, 2021, 12: 748120. |
| [28] | Zhang X, Yin Y, Su Y, Jia Z, Jiang L, Lu Y, Zheng H, Peng J, Rao S, Wu G, Chen J, Yan F. eIF4A, a target of siRNA derived from rice stripe virus, negatively regulates antiviral autophagy by interacting with ATG5 in Nicotiana benthamiana[J]. PLoS Pathogens, 2021, 17(9): e1009963. |
| [29] | Roy B, von Arnim A G. Translational regulation of cytoplasmic mRNAs[J]. The Arabidopsis book/American Society of Plant Biologists, 2013, 11: e0165. |
| [30] | Si K, Maitra U. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor[J]. Molecular and Cellular Biology, 1999, 19(2): 1416-1426. |
| [31] | Basu U, Si K, Warner J R, Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis[J]. Molecular and Cellular Biology, 2001, 21(5): 1453-1462. |
| [32] | Ceci M, Gaviraghi C, Gorrini C, Sala L A, Offenhäuser N, Marchisio P C, Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly[J]. Nature, 2003, 426(6966): 579-584. |
| [33] | Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation[J]. EMBO Reports, 2009, 10(5): 459-465. |
| [34] | Gandin V, Miluzio A, Barbieri A M, Beugnet A, Kiyokawa H, Marchisio P C, Biffo S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation[J]. Nature, 2008, 455(7213): 684-688. |
| [35] | Kausik S, Jayanta C, Jorge C H. Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6[J]. Proceedings of the National Academy of Sciences, 1997, 94(26): 14285-14290. |
| [36] | Sanvito F, Piatti S, Villa A, Bossi M, Lucchini G, Marchisio PC, Biffo S. The beta4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly.[J]. The Journal of Cell Biology, 1999, 144(5): 823-837. |
| [37] | Kapp L D, Lorsch J R. The molecular mechanics of eukaryotic translation[J]. Annual Review of Biochemistry, 2004, 73(1): 657-704. |
| [38] | Sonenberg N, Hinnebusch A G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets[J]. Cell, 2009, 136(4): 731-745. |
| [39] | Russell D W, Spremulli L L. Mechanism of action of the wheat germ ribosome dissociation factor: Interaction with the 60S subunit[J]. Archives of Biochemistry and Biophysics, 1980, 201(2): 518-526. |
| [40] | Kato Y, Konishi M, Shigyo M, Yoneyama T, Yanagisawa S. Characterization of plant eukaryotic translation initiation factor 6 (eIF6) genes: The essential role in embryogenesis and their differential expression in Arabidopsis and rice[J]. Biochemical and Biophysical Research Communications, 2010, 397(4): 673-678. |
| [41] | Guo H M, Lv J Q, Su X W, Chen L, Ren J S, Liu L P, Ren M X, Liu S, Dai M L, Ren G J, Gao F Y. Rice OseIF6. 1 encodes a eukaryotic translation initiation factor and is essential for the development of grain and anther[J]. Frontiers in Plant Science, 2024, 15: 1366986. |
| [42] | Sparkes I A, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants[J]. Nature Protocols, 2006, 1(4): 2019-2025. |
| [43] | Aslanidis C, Jong P J, Schmitz G. Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products[J]. Genome Research, 1994, 4(3): 172-177. |
| [44] | Raychaudhuri P, Stringer E A, Valenzuela D M, Maitra U. Ribosomal subunit antiassociation activity in rabbit reticulocyte lysates. Evidence for a low molecular weight ribosomal subunit antiassociation protein factor (Mr= 25,000)[J]. Journal of Biological Chemistry, 1984, 259(19): 11930-11935. |
| [45] | Xing Y, Zhang Q. Genetic and molecular bases of rice yield[J]. Annual Review of Plant Biology, 2010, 61:421-442. |
| [46] | 何秀英, 伍时照, 宋美芳, 林贤琛. 水稻籽粒发育和胚乳淀粉粒形成的研究[J]. 广东农业科学, 2000(2): 8-10. |
| He X Y, Wu S Z, Song M F, Lin X C. Research on rice grain development and endosperm starch grain formation[J]. Guangdong Agricultural Science, 2000, (2): 8-10. (in Chinese with English abstract) | |
| [47] | 黄海祥, 钱前. 水稻粒形遗传与长粒型优质粳稻育种进展[J]. 中国水稻科学, 2017, 31(6): 665-672. |
| Huang H X, Qian Q. Progress in Rice Grain Shape Genetics and Breeding of Long Grain High Quality Japonica Rice[J]. Chinese Journal of Rice Science, 2017, 31(6): 665-672. (in Chinese with English abstract) | |
| [48] | 况慧云, 张慧, 许寿增, 黄英金. 水稻产量基因设计育种及其发展前景[J]. 安徽农学通报, 2014, 20(8):42-45. |
| Kuang H Y, Zhang H, Xu S Z, Huang Y J. Design and breeding of rice yield genes and their development prospects[J]. Anhui Agricultural Bulletin, 2014, 20(8): 42-45. (in Chinese with English abstract) | |
| [49] | 李孝琼, 韦宇, 高国庆, 邓国富, 郭嗣斌. 水稻遗传图谱构建及粒形相关性状的QTL定位[J]. 南方农业学报, 2014, 45(7): 1154-1159. |
| Li X Q, Wei Y, Gao G Q, Deng G F, Guo S B. Construction of rice genetic map and QTL mapping of grain shape related traits[J]. Journal of Southern Agriculture, 2014, 45(7): 1154-1159. (in Chinese with English abstract) | |
| [50] | Wood L C, Ashby M N, Grunfeld C, Feingold K R. Cloning of murine translation initiation factor 6 and functional analysis of the homologous sequence YPR016c in Saccharomyces cerevisiae[J]. Journal of Biological Chemistry, 1999, 274(17): 11653-11659. |
| [51] | Guo H M, Cui Y C, Huang L J, Ge L, Xu X R, Xue D Y, Tang M, Zheng J S, Yi Y, Chen L. The RNA binding protein OsLa influences grain and anther development in rice[J]. The Plant Journal, 2022, 110(5): 1397-1414. |
| [1] |
TANG Chenghan, CHEN Huizhe, HUAI Yan, SUN Liang, ZHANG Yuping, XIANG Jing, ZHANG Yikai, WANG Zhigang, XU Yiwen, WANG Yaliang.
Response of Machine-transplanting Quality and Yield Formation of Hybrid Rice Pot-mat Seedlings to Pot Depth [J]. Chinese Journal OF Rice Science, 2025, 39(4): 491-500. |
| [2] |
ZHU Peng, LING Xitie, WANG Jinyan, ZHANG Baolong, YANG Yuwen, XU Ke, QIU Shi.
Effects of Weed Control Methods on Grain Yield and Quality of Herbicide-resistant Rice Under Direct Seeding [J]. Chinese Journal OF Rice Science, 2025, 39(4): 501-515. |
| [3] |
DONG Liqiang, ZHANG Yikai, YANG Tiexin, FENG Yingying, MA Liang, LIANG Xiao, ZHANG Yuping, LI Yuedong.
Effect of Dense Sowing Nursery on Seedling Quality and Picking Characteristics for Mechanized Transplanting inNorthern japonica Rice [J]. Chinese Journal OF Rice Science, 2025, 39(4): 516-528. |
| [4] |
ZHOU Yang, YE Fan, LIU Lijun.
Research Progress of Typical Plant Growth-promoting Microorganism Enhancing Salt Stress Resistance in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(4): 529-542. |
| [5] |
ZHU Jianping, LI Xia, LI Wenqi, XU Yang, WANG Fangquan, TAO Yajun, JIANG Yanjie, CHEN Zhihui, FAN Fangjun, YANG Jie, YANG Jie.
Phenotypic Analysis and Gene Mapping of a Floury Endosperm Mutant we1 in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(4): 543-551. |
| [6] | HUANG Fudeng, WU Chunyan, HAO Yuanyuan, HAN Yifei, ZHANG Xiaobin, SUN Huifeng, PAN Gang . Transcriptome Analysis of Top Second Leaf Sheath of Rice Under Different Nitrogen Fertilizer Levels [J]. Chinese Journal OF Rice Science, 2025, 39(4): 563-571. |
| [7] | LU Yezi, QIU Jiehua, JIANG Nan, KOU Yanjun, SHI Huanbin. Research Progress in Effectors of Magnaporthe oryzae [J]. Chinese Journal OF Rice Science, 2025, 39(3): 287-294. |
| [8] | WANG Chaorui, ZHOU Yukun, WEN Ya, ZHANG Ying, FA Xiaotong, XIAO Zhilin, ZHANG Hao. Effects of Straw Returning Methods on Soil Characteristics and Greenhouse Gas Emissions in Paddy Fields and Their Regulation Through Water-fertilizer Interactions [J]. Chinese Journal OF Rice Science, 2025, 39(3): 295-305. |
| [9] | WANG Yaxuan, WANG Xinfeng, YANG Houhong, LIU Fang, XIAO Jing, CAI Yubiao, WEI Qi, FU Qiang, WAN Pinjun. Recent Advances in Mechanisms of Adaptation of Planthoppers to Rice Resistance [J]. Chinese Journal OF Rice Science, 2025, 39(3): 306-321. |
| [10] | HUANG Tao, WEI Zhaogen, CHENG Qi, CHENG Ze, LIU Xin, WANG Guangda, HU Keming, XIE Wenya, CHEN Zongxiang, FENG Zhiming, ZUO Shimin. Gene Cloning and Broad-spectrum Disease Resistance Analysis of Rice Lesion Mimic Mutant lm52 [J]. Chinese Journal OF Rice Science, 2025, 39(3): 322-330. |
| [11] | ZHANG Bintao, LIU Congcong, GUO Mingliang, YANG Shaohua, WU Shiqiang, GUO Longbiao, ZHU Yiwang. Evaluation of Blast Resistance and Identification of Superior Haplotype of OsDR8 in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(3): 343-351. |
| [12] | WEI Xinyu, ZENG Yuehui, XIAO Changchun, HUANG Jianhong, RUAN Hongchun, YANG Wangxing, ZOU Wenguang, XU Xuming. Cloning and Functional Verification of Rice-Blast Resistance Gene Pi-kf2(t) in Kangfeng B [J]. Chinese Journal OF Rice Science, 2025, 39(3): 352-364. |
| [13] | LI Wenqi, XU Yang, WANG Fangquan, ZHU Jianping, TAO Yajun, LI Xia, FAN Fangjun, JIANG Yanjie, CHEN Zhihui, YANG Jie. Development and Application of KASP Marker for Broad-Spectrum Resistance Gene PigmR to Rice Blast [J]. Chinese Journal OF Rice Science, 2025, 39(3): 365-372. |
| [14] | WEI Huanhe, WANG Lulu, MA Weiyi, ZHANG Xiang, ZUO Boyuan, GENG Xiaoyu, ZHU Wang, ZHU Jizou, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, DAI Qigen. Grain-filling Characteristics and Its Relationship with Grain Yield Formation of japonica Rice Nanjing 9108 Under Combined Salinity-drought Stress [J]. Chinese Journal OF Rice Science, 2025, 39(3): 373-386. |
| [15] | ZHANG Haiwei, GU Xinyi, CHEN Mingshuai, LI Fukang, SHI Yuecheng, YANG Ting, JIANG Shuochen. Effects of Nitrogen Type of Basal Fertilizer on Growth, Grain Yield and Nitrogen Use Efficiency of Ratooning Rice [J]. Chinese Journal OF Rice Science, 2025, 39(3): 387-398. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||