
Chinese Journal OF Rice Science ›› 2022, Vol. 36 ›› Issue (6): 579-585.DOI: 10.16819/j.1001-7216.2022.220103
• Research Papers • Previous Articles Next Articles
MAO Hui1,#, PENG Yan2,#, MAO Bigang1,2, SHAO Ye2, ZHENG Wenjie1, HU Liming1, ZHOU Kai1, ZHAO Bingran1,2,*( )
)
Received:2022-01-06
Revised:2022-03-14
Online:2022-11-10
Published:2022-11-10
Contact:
ZHAO Bingran
About author:First author contact:# These authors contributed equally to this work
毛慧1,#, 彭彦2,#, 毛毕刚1,2, 韶也2, 郑文杰1, 胡黎明1, 周凯1, 赵炳然1,2,*( )
)
通讯作者:
赵炳然
作者简介:第一联系人:#共同第一作者
基金资助:MAO Hui, PENG Yan, MAO Bigang, SHAO Ye, ZHENG Wenjie, HU Liming, ZHOU Kai, ZHAO Bingran. Function and Effect Analysis of a New Gene Wx410 Regulating Amylose Synthesis in Rice[J]. Chinese Journal OF Rice Science, 2022, 36(6): 579-585.
毛慧, 彭彦, 毛毕刚, 韶也, 郑文杰, 胡黎明, 周凯, 赵炳然. 水稻直链淀粉合成调控新基因Wx410的功能与效应分析[J]. 中国水稻科学, 2022, 36(6): 579-585.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2022.220103
| 引物名称 Primer name | 正向引物序列 Forward primer sequence(5′-3′) | 反向引物序列 Reverse primer sequence(5′-3′) |
|---|---|---|
| Wx | WxF1: GCCGGAGGGCCGTTCGACGGCA | WxR1: TACTAAAATTGGTTGGATTCTGA |
| WxF2: GCCGAGTTGGTCAAAGGAA | WxR2: TCCAGCCTGCCGATGAACGCGATC | |
| WxF3: GGAACAGAAGGGCCCTGACG | WxR3: ATGGCATGGTATAATATGGAACAG | |
| KASP1 | F1: GAAGGTGACCAAGTTCATGCTTTCCAGGGCCTCAAGCCCC | R1: CGCTGGTCGTCACGCTGA |
| F2: GAAGGTCGGAGTCAACGGATTGTTCCAGGGCCTCAAGCCCA | ||
| KASP2 | F1: GAAGGTGACCAAGTTCATGCTGCGTTCATCGGCAGGCTGGA | R1: TTTGGCATATCGTGCAAGTGTGTCT |
| F2: GAAGGTCGGAGTCAACGGATTGCGTTCATCGGCAGGCTGGG | ||
| qWx | qWx-F: ACCTGACACTGGAGTTGATTAC | qWx-R: GTATGGGTTGTTGTTGAGGTTTAG |
| qActin | qActin-F: ACCTGACACTGGAGTTGATTAC | qActin-R: GTATGGGTTGTTGTTGAGGTTTAG |
| qHyg | qHyg-F: GCTTCTGCGGGCGATTTGTGT | qHyg-R: GGTCGCGGAGGCTATGGATGC |
Table 1. Sequence of primers used in the study.
| 引物名称 Primer name | 正向引物序列 Forward primer sequence(5′-3′) | 反向引物序列 Reverse primer sequence(5′-3′) |
|---|---|---|
| Wx | WxF1: GCCGGAGGGCCGTTCGACGGCA | WxR1: TACTAAAATTGGTTGGATTCTGA |
| WxF2: GCCGAGTTGGTCAAAGGAA | WxR2: TCCAGCCTGCCGATGAACGCGATC | |
| WxF3: GGAACAGAAGGGCCCTGACG | WxR3: ATGGCATGGTATAATATGGAACAG | |
| KASP1 | F1: GAAGGTGACCAAGTTCATGCTTTCCAGGGCCTCAAGCCCC | R1: CGCTGGTCGTCACGCTGA |
| F2: GAAGGTCGGAGTCAACGGATTGTTCCAGGGCCTCAAGCCCA | ||
| KASP2 | F1: GAAGGTGACCAAGTTCATGCTGCGTTCATCGGCAGGCTGGA | R1: TTTGGCATATCGTGCAAGTGTGTCT |
| F2: GAAGGTCGGAGTCAACGGATTGCGTTCATCGGCAGGCTGGG | ||
| qWx | qWx-F: ACCTGACACTGGAGTTGATTAC | qWx-R: GTATGGGTTGTTGTTGAGGTTTAG |
| qActin | qActin-F: ACCTGACACTGGAGTTGATTAC | qActin-R: GTATGGGTTGTTGTTGAGGTTTAG |
| qHyg | qHyg-F: GCTTCTGCGGGCGATTTGTGT | qHyg-R: GGTCGCGGAGGCTATGGATGC |
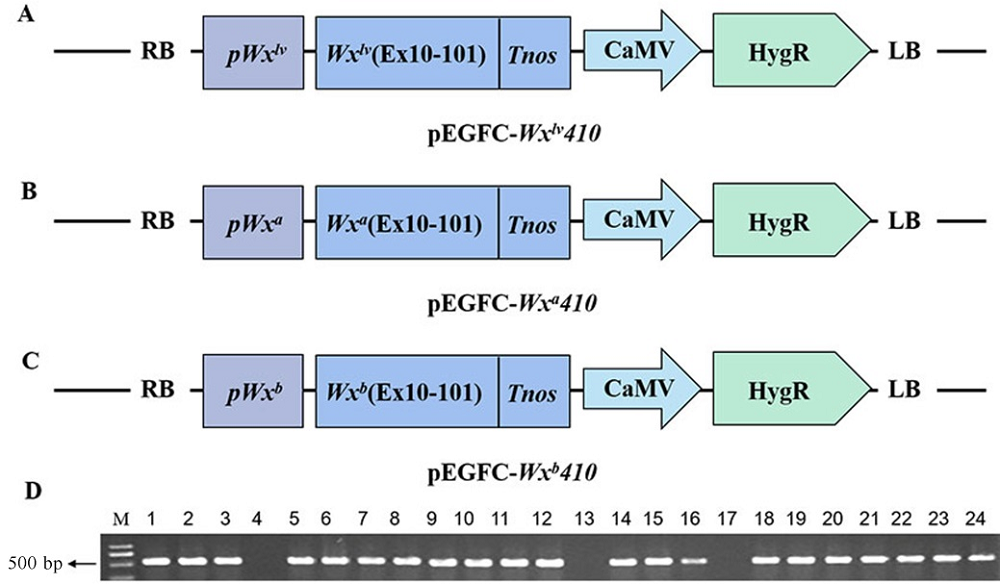
Fig. 1. Schematic diagram of Wx410 targeted mutant plant expression vector under different Wx allelic backgrounds and identification of T0 transgenic plants. M, DNA marker; Lanes 1-17, Hygromycin detection of different Wx410 transgenic lines; Lanes 18-24, Hygromycin test of control material.
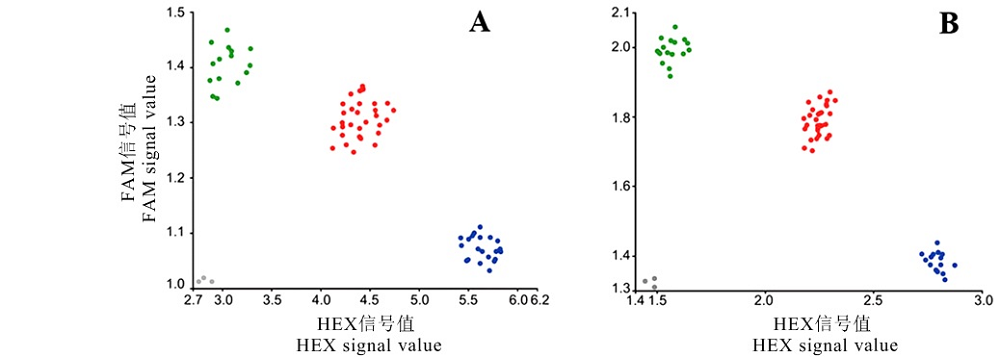
Fig. 2. KASP genotyping of transgenic T1 generation materials. A, Molecular marker KASP1 identifies transgenic T1 generation positive control plants (based on 23 bp nucleotide insertion in the second exon of Wx allele); B, Molecular marker KASP2 identifies homozygous plants of Wx410 targeted directed mutant transgenic line (based on SNP A-G at position 101 of exon 10 of Wx gene); Green, blue, red and gray dots represent transgenic receptors, homozygous lines, heterozygous materials and H2O, respectively.
| 材料 Material | 株高 Plant height /cm | 有效穗数 No. of effective panicles | 每穗总粒数 No. of grains per panicle | 结实率 Seed setting rate/% | 千粒重 1000-grain weight /g | 精米长宽比 Length to width ratio of milled rice |
|---|---|---|---|---|---|---|
| pEGFC | 132.7±0.6 a | 8.7±0.8 a | 82.3±4.2 a | 79.6±0.8 a | 36.1±0.3 a | 2.0±0.1 b |
| pEGFC-Wxlv | 131.7±1.5 a | 8.7±0.6 a | 81.3±3.2 a | 79.8±1.0 a | 36.0±0.8 a | 2.2±0.0 a |
| pEGFC-Wxlv410 | 131.0±2.0 a | 8.7±0.6 a | 80.0±4.6 a | 78.3±2.6 a | 36.6±0.3 a | 2.1±0.0 ab |
| pEGFC-Wxa | 132.0±1.0 a | 8.7±0.8 a | 82.0±3.0 a | 80.4±1.9 a | 36.2±0.3 a | 2.2±0.0 a |
| pEGFC-Wxa410 | 131.3±0.6 a | 8.3±0.6 a | 84.3±5.7 a | 80.4±2.0 a | 36.7±0.3 a | 2.1±0.0 ab |
| pEGFC-Wxb | 130.9±1.6 a | 8.7±0.6 a | 82.0±3.5 a | 78.9±2.6 a | 36.4±0.3 a | 2.2±0.0 a |
| pEGFC-Wxb410 | 129.3±1.5 a | 8.7±0.6 a | 78.0±4.4 a | 79.0±1.7 a | 35.8±0.6 a | 2.1±0.0 b |
Table 2. Comparison of agronomic traits between transgenic materials and control materials.
| 材料 Material | 株高 Plant height /cm | 有效穗数 No. of effective panicles | 每穗总粒数 No. of grains per panicle | 结实率 Seed setting rate/% | 千粒重 1000-grain weight /g | 精米长宽比 Length to width ratio of milled rice |
|---|---|---|---|---|---|---|
| pEGFC | 132.7±0.6 a | 8.7±0.8 a | 82.3±4.2 a | 79.6±0.8 a | 36.1±0.3 a | 2.0±0.1 b |
| pEGFC-Wxlv | 131.7±1.5 a | 8.7±0.6 a | 81.3±3.2 a | 79.8±1.0 a | 36.0±0.8 a | 2.2±0.0 a |
| pEGFC-Wxlv410 | 131.0±2.0 a | 8.7±0.6 a | 80.0±4.6 a | 78.3±2.6 a | 36.6±0.3 a | 2.1±0.0 ab |
| pEGFC-Wxa | 132.0±1.0 a | 8.7±0.8 a | 82.0±3.0 a | 80.4±1.9 a | 36.2±0.3 a | 2.2±0.0 a |
| pEGFC-Wxa410 | 131.3±0.6 a | 8.3±0.6 a | 84.3±5.7 a | 80.4±2.0 a | 36.7±0.3 a | 2.1±0.0 ab |
| pEGFC-Wxb | 130.9±1.6 a | 8.7±0.6 a | 82.0±3.5 a | 78.9±2.6 a | 36.4±0.3 a | 2.2±0.0 a |
| pEGFC-Wxb410 | 129.3±1.5 a | 8.7±0.6 a | 78.0±4.4 a | 79.0±1.7 a | 35.8±0.6 a | 2.1±0.0 b |
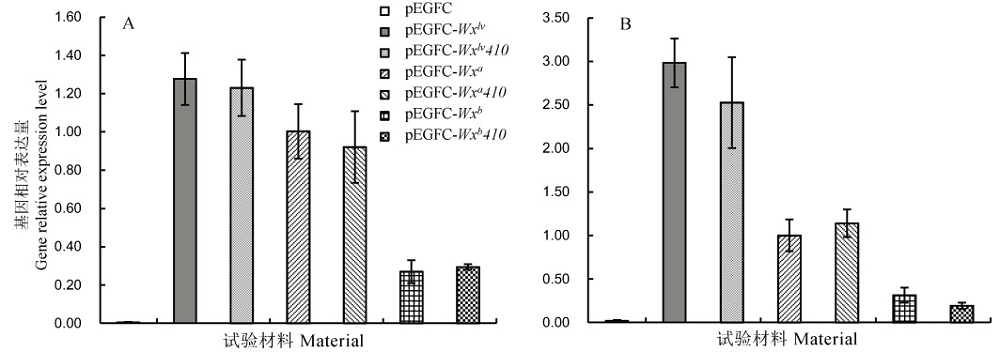
Fig. 4. Determination of relative expression level of Wx gene at different time after anthesis. A, 7 days after flowering; B, 14 days after flowering; Mean±SD, n=3. pEGFC, Negative control; pEGFC-Wxlv, pEGFC-Wxa and pEGFC-Wxb, Positive control; pEGFC-Wxlv410, pEGFC-Wxa410 and pEGFC-Wxb410, Wx410 targeted mutant transgenic lines under different Wx allelic backgrounds. The same below.

Fig. 5. OsGBSSⅠ activity in endosperm at different time after anthesis. A, Seven days after flowering; B, 14 days after flowering; Mean±SD, n=3; * and ** represent significant difference between Wx410 targeted mutant transgenic line and its positive control at 0.05 and 0.01 levels by t-test, respectively.
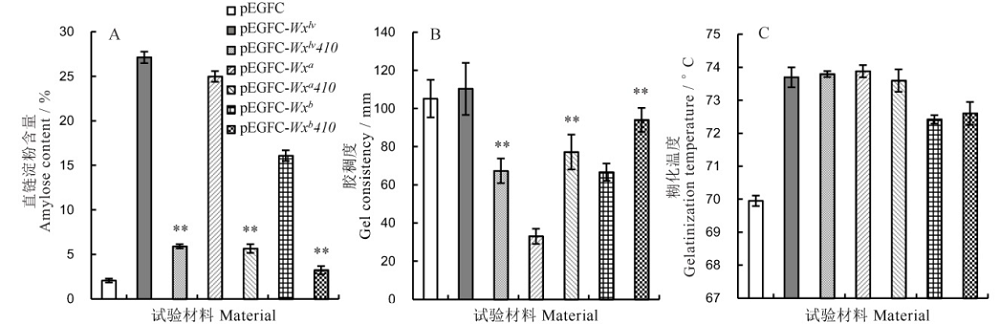
Fig. 6. Determination of endosperm physical and chemical properties of transgenic plants. Mean±SD, n=3; ** represent significant difference between Wx410 targeted mutant transgenic line and its positive control at 0.01 level by t-test.
| [1] | 方志强, 陆展华, 王石光, 刘维, 卢东柏, 王晓飞, 何秀英. 稻米品质性状研究进展与应用[J]. 广东农业科学, 2020, 47(5): 11-20. |
| Fang Z Q, Lu Z H, Wang S G, Liu W, Lu D B, Wang X F, He X Y. Research advances and applications of rice grain quality traits[J]. Guangdong Agricultural Sciences, 2020, 47(5): 11-20. (in Chinese with English abstract) | |
| [2] | Yang X H, Nong B X, Xia X Z, Zhang Z Q, Zeng Y, Liu K Q, Deng G F, Li D T. Rapid identification of a new gene influencing low amylose content in rice landraces (Oryza sativa L.) using genome-wide association study with specific-locus amplified fragment sequencing[J]. Genome, 2017, 60(6): 465-472. |
| [3] | Buléon A, Colonna P, Planchot V, Ball S. Starch granules: Structure and biosynthesis[J]. International Journal of Biological Macromolecules, 1998, 23(2): 85-112. |
| [4] | Tian Z X, Qian Q, Liu Q Q, Yan M X, Liu X F, Yan C J, Liu G F, Gao Z Y, Tang S Z, Zeng D L, Wang Y H, Yu J M, Gu M H, Li J Y. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(51): 21760-21765. |
| [5] | Juliano B O, Pascual C G. Differences in physicochemical properties of commercial rice from urban markets in West Africa[J]. Journal of Food Science and Technology, 2020, 57(4): 1505-1516. |
| [6] | Wang Z Y, Zheng F Q, Shen G Z, Gao J P, Snustad D P, Li M G, Zhang J L, Hong M M. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene[J]. The Plant Journal, 1995, 7(4): 613-622. |
| [7] | Gu M H, Liu Q Q, Yan C J, Tang S Z. Genetic variation and molecular improvement accelerating hybrid rice development// International Rice Research Institute. Grain quality of hybrid rice[M]. Los Banos (Philippines): International Rice Research Institute, 2010: 345-356. |
| [8] | Ball, S G, Wal M H, Visser R G. Progress in understanding the biosynthesis of amylose[J]. Trends in Plant Science, 1998, 3(12): 462-467. |
| [9] | 朱霁晖, 张昌泉, 顾铭洪, 刘巧泉. 水稻Wx基因的等位变异及育种利用研究进展[J]. 中国水稻科学, 2015, 29(4): 431-438. |
| Zhu J H, Zhang C Q, Gu M H, Liu Q Q. Progress in the Allelic variation of Wx gene and its application in rice breeding[J]. Chinese Journal of Rice Science, 2015, 29(4): 431-438. (in Chinese with English abstract) | |
| [10] | Suu T D, Hoai T T T, Hoa N T L, Loan H M, Yen D B,. Kumamaru T, Satoh H. Variation on grain quality in Vietnamese rice cultivars[J]. Journal of the Faculty of Agriculture, Kyushu University, 2012, 57(2): 365-371. |
| [11] | Sano Y, Katsumata M, Okuno K. Genetic studies of speciation in cultivated rice: 5. Inter-and intraspecific differentiation in the waxy gene expression of rice[J]. Euphytica, 1986, 35(1): 1-9. |
| [12] | Mikami I, Uwatoko N, Ikeda Y, Yamaguchi J, Hirano H Y, Suzuki Y, Sano Y. Allelic diversification at the Wx locus in landraces of Asian rice[J]. Theoretical and Applied Genetics, 2008, 116(7): 979-989. |
| [13] | Zhang C Q, Zhu J H, Chen S J, Fan X L, Li Q F, Lu Y, Wang M, Yu H X, Yi C D, Tang S Z, Gu M H, Liu Q Q. Wxlv, the ancestral allele of rice Waxy gene[J]. Molecular Plant, 2019, 12(8): 1157-1166. |
| [14] | Zhang C Q, Yang Y, Chen S J, Liu X J, Zhu J H, Zhou L H, Lu Y, Li Q F, Fan X L, Tang S Z, Gu M H, Liu Q Q. A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency[J]. Journal of Integrative Plant Biology, 2021, 63(5): 889-901. |
| [15] | Zhou H, Xia D, Zhao D, Li Y H, Li P B, Wu B, Gao G J, Zhang Q L, Wang G W, Xiao J H, Li X H, Yu S B, Lian X M, He Y Q. The origin of Wxla provides new insights into the improvement of grain quality in rice[J]. Journal of Integrative Plant Biology, 2021, 63(5): 878-888. |
| [16] | Sato H, Suzuki Y, Sakai M, Imbe T. Molecular characterization of Wxmq, a novel mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.)[J]. Breeding Science, 2002, 52(2): 131-135. |
| [17] | Mikami I, Aikawa M, Hirano H Y, Sano Y. Altered tissue-specific expression at the Wx gene of the opaque mutants in rice[J]. Euphytica, 1999, 105(2): 91-97. |
| [18] | Liu L L, Ma X D, Liu S J, Zhu C L, Jiang L, Wang Y H, Shen Y, Ren Y L, Dong H, Chen L M, Liu X, Zhao Z G, Zhai H Q, Wan J M. Identification and characterization of a novel Waxy allele from a Yunnan rice landrace[J]. Plant Molecular Biology, 2009, 71(6): 609-626. |
| [19] | Shao Y, Peng Y, Mao B G, Lü Q M, Yuan D Y, Liu X, Zhao B R. Allelic variations of the Wx locus in cultivated rice and their use in the development of hybrid rice in China[J]. PloS One, 2020, 15(5): e0232279. |
| [20] | Zhang C Q, Chen S J, Ren X Y, Lu Y, Liu D R, Cai X L, Li Q F, Gao J P, Liu Q Q. Molecular structure and physicochemical properties of starches from rice with different amylose contents resulting from modification of OSGBSSⅠ activity[J]. Journal of Agricultural and Food Chemistry, 2017, 65(10): 2222-2232. |
| [21] | Liu D R, Wang W, Cai X L. Modulation of amylose content by structure-based modification of OsGBSSⅠ activity in rice (Oryza sativa L.)[J]. Plant Biotechnology Journal, 2014, 12(9): 1297-1307. |
| [22] | 万映秀, 邓其明, 王世全, 刘明伟, 周华强, 李平. 水稻Wx基因的遗传多态性及其与主要米质指标的相关性分析[J]. 中国水稻科学, 2006, 20(6): 603-609. |
| Wan Y X, Deng Q M, Wang S Q, Liu M W, Zhou H Q, Li P. Genetic polymorphism of Wx gene and its correlation with major grain quality traits in rice[J]. Chinese Journal of Rice Science, 2006, 20(6): 603-609. (in Chinese with English abstract) | |
| [23] | Zhang J S, Zhang H, Botella J R, Zhu J K. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties[J]. Journal of Integrative Plant Biology, 2018, 60(5): 369-375. |
| [24] | Huang L C, Li Q F, Zhang C C, Chu R, Gu Z W, Tan H Y, Zhao D S, Fan X L, Liu Q Q. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system[J]. Plant Biotechnology Journal, 2020, 18(11): 2164-2166. |
| [25] | Zeng D C, Liu T L, Ma X L, Wang B, Zheng Z Y, Zhang Y L, Xie X R, Yang B W, Zhao Z, Zhu Q L, Liu Y G. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5' UTR-intron editing improves grain quality in rice[J]. Plant Biotechnology Journal, 2020, 18(12): 2385-2387. |
| [26] | Monsur M B, Cao N, Wei X J, Xie L H, Jiao G A, Tang S Q, Nese S, Shao G N, Hu P S. Improved eating and cooking quality of indica rice cultivar YK17 via adenine base editing of Wxa allele of granule-bound starch synthase I (GBSS I)[J]. Rice Science, 2021, 28: 427-430. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||