
Chinese Journal OF Rice Science ›› 2022, Vol. 36 ›› Issue (1): 13-26.DOI: 10.16819/j.1001-7216.2022.210312
• Research Papers • Previous Articles Next Articles
ZHANG Taohui1,#, WANG Haiyu1,#, WAN Hua1, ZHANG Liping1, XIE Zhenwei1, CHEN Keyi1, HE Xiaodong1, ZHAO Zhigang1,*( ), WAN Jianmin1,2
), WAN Jianmin1,2
Received:2021-03-23
Revised:2021-04-22
Online:2022-01-10
Published:2022-01-10
Contact:
ZHAO Zhigang
About author:First author contact:#These authors contributed equally to this work;
张涛荟1,#, 王海宇1,#, 万华1, 张莉萍1, 谢振威1, 陈可毅1, 何晓栋1, 赵志刚1,*( ), 万建民1,2
), 万建民1,2
通讯作者:
赵志刚
作者简介:第一联系人:#共同第一作者;
基金资助:ZHANG Taohui, WANG Haiyu, WAN Hua, ZHANG Liping, XIE Zhenwei, CHEN Keyi, HE Xiaodong, ZHAO Zhigang, WAN Jianmin. Cytological Observation of a Female and Male Sterile Osfma2 Mutant in Rice and Its Map-based Cloning[J]. Chinese Journal OF Rice Science, 2022, 36(1): 13-26.
张涛荟, 王海宇, 万华, 张莉萍, 谢振威, 陈可毅, 何晓栋, 赵志刚, 万建民. 水稻雌雄不育突变体Osfma2的细胞学观察及基因图位克隆[J]. 中国水稻科学, 2022, 36(1): 13-26.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2022.210312
| 引物名称 | 正向引物序列 | 反向引物序列 | 实验目的 |
|---|---|---|---|
| Name | Forward sequence(5’-3’) | Reverse sequence(5’-3’) | Purpose |
| RM162 | GCCAGCAAAACCAGGGATCCGG | CCGCAAGCCGCACAAGACCTTG | 精细定位 Fine mapping |
| H971-5 | CCGCCCGCGTTTTATTTACT | GGCAACATTCTTCGGCCTC | 精细定位 Fine mapping |
| HF-2 | CGCCGCCAAGAGCTAATTAA | TGAGAGATCTCGATCGACTTCTC | 精细定位 Fine mapping |
| HF-3 | GCCTATGTTTAGCCGCGAAA | GGAAGGAGAAGTTGAGGGGG | 精细定位 Fine mapping |
| Hi-1 | CCGGACCGTGATTTCGTTAG | TCAATACTAAAATCTTCGCCCCT | 精细定位 Fine mapping |
| Hh-4 | CTTTCTCCCCGTCGATCCTT | GCCCCACGGAGCATATCTAG | 精细定位 Fine mapping |
| Hh-9 | GGCAGGAAGTCCAAAAAGCT | TCCATAAAGCAAGCTGATGCA | 精细定位 Fine mapping |
| He-6 | CGTAGGCGAGGTGGTACAAT | GAGAGGTCACTGGTCAGCTT | 精细定位 Fine mapping |
| RM5463 | ACCCTTGCAGACAACGTACC | GCATGCAGCTGCTGGTATAT | 精细定位 Fine mapping |
| QHG | AGCTCTGCAGCAACCAGATA | TGCTTTGGGCATTCCAGTTC | qRT-PCR |
Table 1 Fine-mapping and quantitative primer sequences used in the study.
| 引物名称 | 正向引物序列 | 反向引物序列 | 实验目的 |
|---|---|---|---|
| Name | Forward sequence(5’-3’) | Reverse sequence(5’-3’) | Purpose |
| RM162 | GCCAGCAAAACCAGGGATCCGG | CCGCAAGCCGCACAAGACCTTG | 精细定位 Fine mapping |
| H971-5 | CCGCCCGCGTTTTATTTACT | GGCAACATTCTTCGGCCTC | 精细定位 Fine mapping |
| HF-2 | CGCCGCCAAGAGCTAATTAA | TGAGAGATCTCGATCGACTTCTC | 精细定位 Fine mapping |
| HF-3 | GCCTATGTTTAGCCGCGAAA | GGAAGGAGAAGTTGAGGGGG | 精细定位 Fine mapping |
| Hi-1 | CCGGACCGTGATTTCGTTAG | TCAATACTAAAATCTTCGCCCCT | 精细定位 Fine mapping |
| Hh-4 | CTTTCTCCCCGTCGATCCTT | GCCCCACGGAGCATATCTAG | 精细定位 Fine mapping |
| Hh-9 | GGCAGGAAGTCCAAAAAGCT | TCCATAAAGCAAGCTGATGCA | 精细定位 Fine mapping |
| He-6 | CGTAGGCGAGGTGGTACAAT | GAGAGGTCACTGGTCAGCTT | 精细定位 Fine mapping |
| RM5463 | ACCCTTGCAGACAACGTACC | GCATGCAGCTGCTGGTATAT | 精细定位 Fine mapping |
| QHG | AGCTCTGCAGCAACCAGATA | TGCTTTGGGCATTCCAGTTC | qRT-PCR |
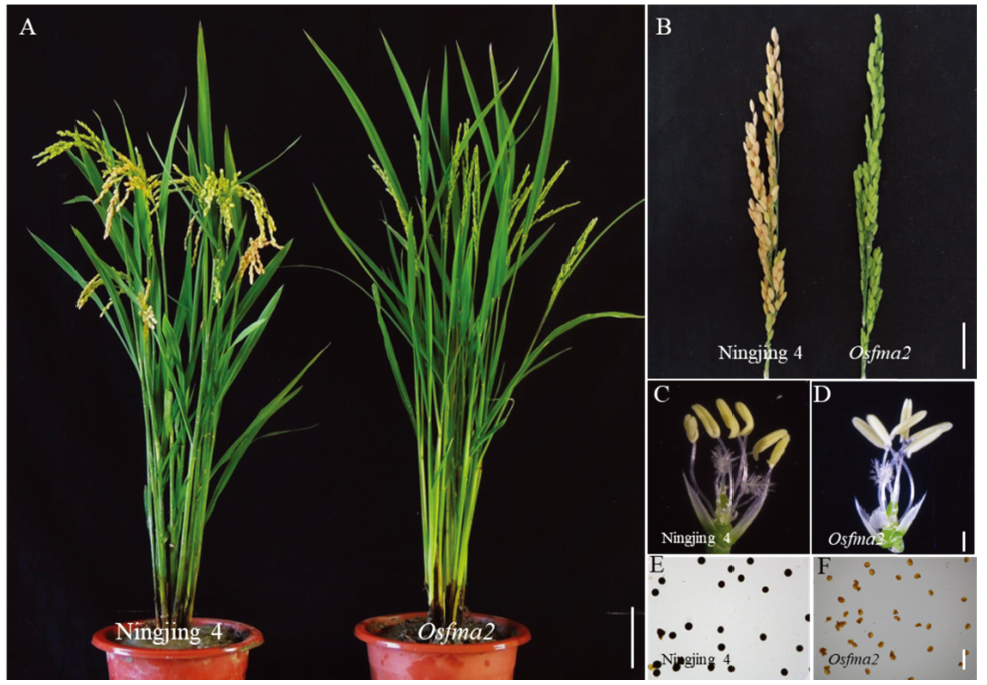
Fig. 1. Phenotypes and sterility of the wild type and the mutant Osfma2. A, Plant morphology of the wild type (Ningjing 4) and the Osfma2 mutant, bar=15 cm. B, Panicle of the wild type and the Osfma2 mutant, bar=4 cm. C, Anther of wild type, bar=600 μm. D, Anther of the mutant Osfma2, bar=600 μm. E, Pollen of Ningjing 4 stained with I2-KI, bar= 50 μm. F, Pollen of Osfma2 stained with I2-KI, bar= 50 μm.
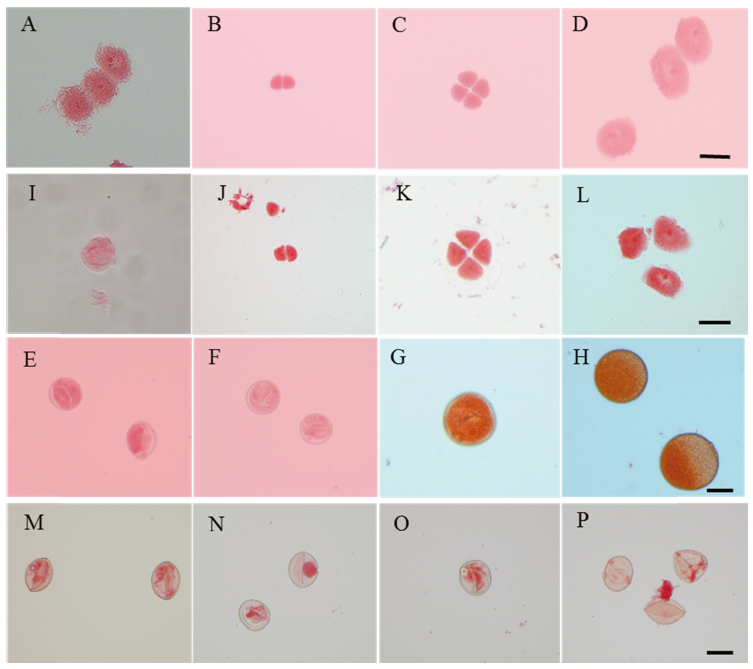
Fig. 2. Developmental process of pollen in the wild type and the mutant Osfma2. A-H, Wild type; I-P, Mutant Osfma2. A and I, Archesporial cell; B and J, Dyads; C and K, Tetrads; D and L, Early microspore; E and M, Late microspore; F and N, Early bicellular pollen; G and O, Late bicellular pollen; H and P, Mature pollen. Bar=10 μm.

Fig. 3. Transverse section of anthers of the wild type and the mutant Osfma2. A-F, Wild type; G-L, Mutant. A and G, Before the stage of meiosis; B and H, 8b stage of anther development; C and I, The 9th stage of anther development; D and J, The 10th stage of anther development; E and K, The 11th stage of anther development; F and L, Mature stages of anther development. E, Epidermis; En, Endothecium; ML, Middle layer; T, Tapetum; MMC, Microspore mother cell; Tds, Tetrads; Msp, Microspore parietal cell; BP, Bicellular pollen; Mp, Mature pollen. Bar=10 μm.
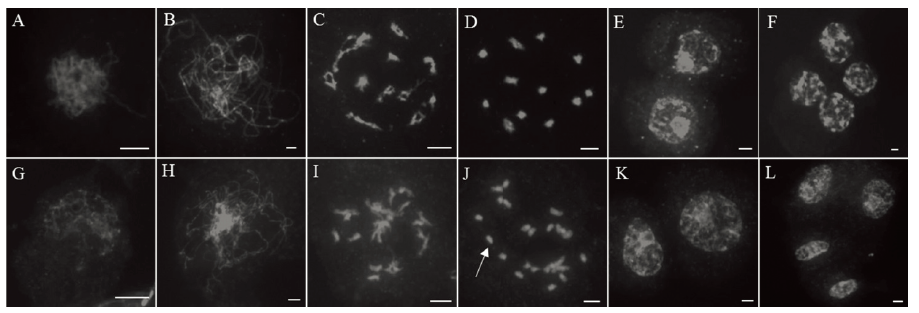
Fig. 4. Chromosome observation of archesporial cell of the wild type and the mutant Osfma2. A~F, Wild type; G~L, Mutant Osfma2. A and G, Zygotene; B and H, Pachytene; C and I, Diplotene; D and J, Diakinesis; E and K, Telophase I; F and L, Telophase II. The white arrows point to univalents. Bar=5μm.
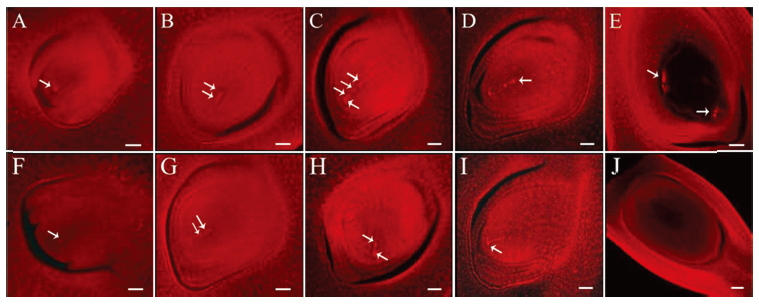
Fig. 5. Observation of embryo sac development of the wild type and the mutant Osfma2. A~E, Embryo sac development of wild type; F~J, Embryo sac development of mutant Osfma2. A and F, Archesporial cell stage; B and G, Dyad stage; C and H, Tetrad stage; D and I, Functional megaspore stage; E and J, Mature embryo stage. Bar=60 μm in E and J. Bar=30 μm in others.
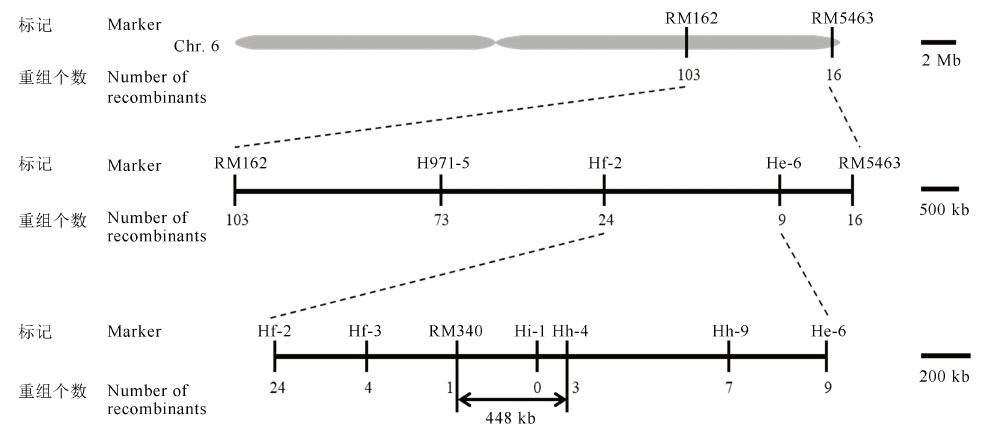
Fig. 6. OsFMA2 gene mapping on chromosome 6. The preliminary and fine mapping of OsFMA2. The OsFMA2 gene was ultimately located in the 448-kb interval between the molecular markers RM340 and Hh-4.
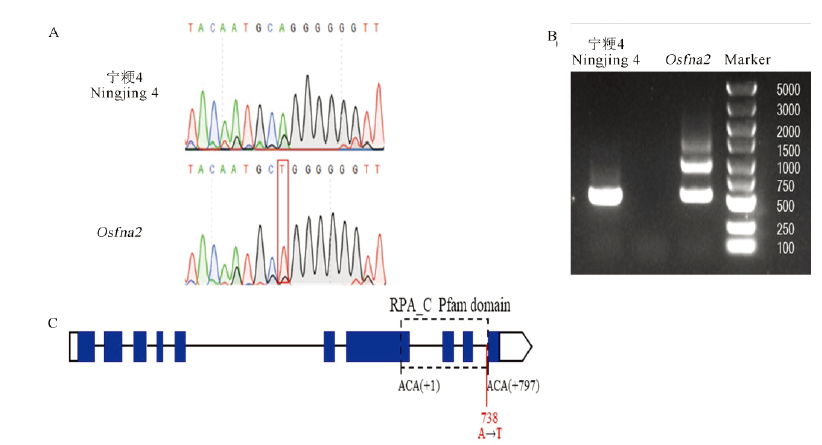
Fig. 8. Cloning and sequence analysis of the sterility gene OsFMA2. A, Sequencing differences between the wild type and the mutant; B, Differential splicing of the OsFMA2 gene in the wild type and the mutant; C, Gene structure of OsFMA2. The blue box represents the exon, the line represents the intron, and the dotted box represents the RPA_C conserved domain, which is calculated from the first base of the first amino acid to the last base of the last amino acid. The solid red line represents the mutation. The white boxes represent 5′ UTR and 3′ UTR.
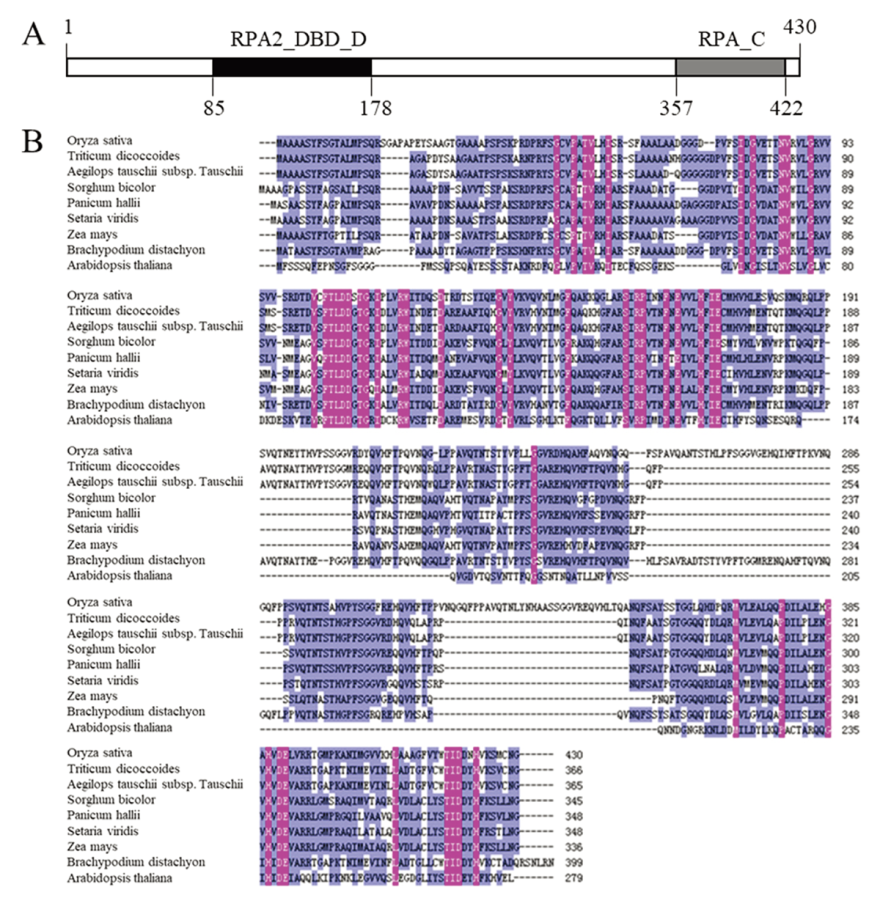
Fig. 9. Bio-information analysis of OsFMA2. A, Protein-structure prediction of OsFMA2. The numbers indicate the position of amino acid residue at the OsFMA2; B, Multiple sequence alignment between OsFMA2 and RPA protein of other species.
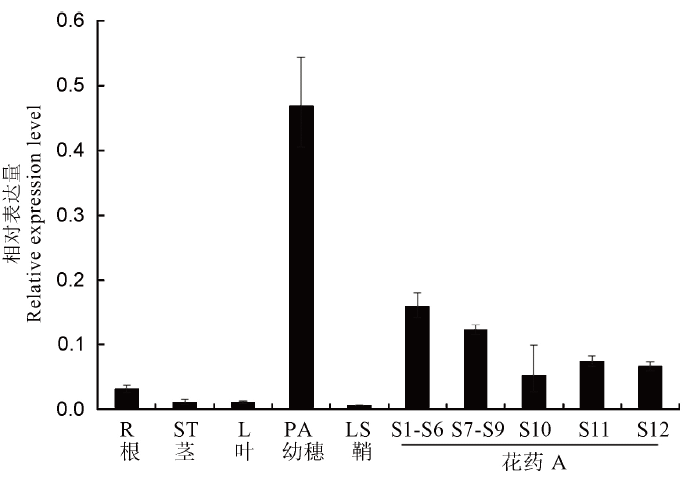
Fig. 11. Relative expression levels of OsFMA2 in different rice tissues. R, Root; ST, Stem; L, Leaf; PA, Panicle; LS, Leaf sheath; S1-S6, Microspore mother cell stage; S7-S9, Meiotic division of male gamete; S10, Mononucleate stage; S11, Binuclear stage; S12, Tri-nuclear stage. Osubiquitin was used as an internal control. Error bars show the SD (n=3).
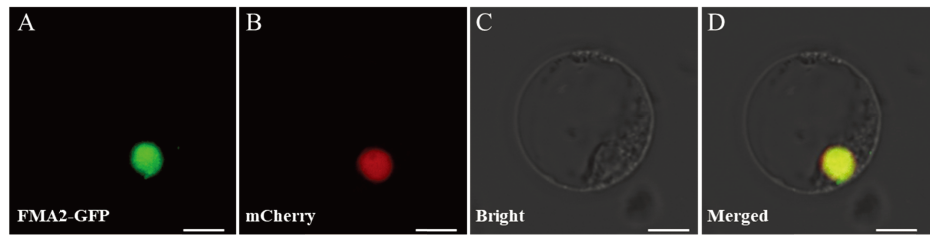
Fig. 12. Subcellular localization of OsFMA2 in rice protoplast. A, Subcellular localization of OsFMA2-GFP in rice protoplast; B, Localization of nuclear marker; C, Bright field of rice protoplast status; D, Merged image of GFP, nuclear marker and bright field. Bar=10 μm.
| [1] | 谭何新, 文铁桥, 张大兵. 水稻花粉发育的分子机理[J]. 植物学通报, 2007,24(3):330-339. |
| Tan H X, Wen T Q, Zhang D B. Molecular mechanisms of pollen development in Oryza sativa[J]. Chinese Bulletin of Botany, 2007,24(3):330-339. (in Chinese with English abstract) | |
| [2] | 马西青, 方才臣, 邓联武, 万向元. 水稻隐性核雄性不育基因研究进展及育种应用探讨[J]. 中国水稻科学, 2012,26(5):511-520. |
| Wan X Q, Fang C C, Deng L W, Wan X Y. Research progress and breeding application of recessive genic male sterility genes in rice[J]. Chinese Journal of Rice Science, 2012,26(5):511-520. (in Chinese with English abstract) | |
| [3] | 官文祥, 邓赟, 李小旭, 吴为人, 郑燕. 水稻雌性不育分子机理研究进展[J]. 分子植物育种, 2017,15(2):672-684. |
| Guan W X, Deng Y, Li X X, Wu W R, Zheng Y. Advances in research on molecular mechanism of female sterility in rice (Oryza sativa L.)[J]. Molecular Plant Breeding, 2017,15(2):672-684. (in Chinese with English abstract) | |
| [4] | 刘春宏, 方珊茹, 刘玉芹, 沈伟锋. 水稻雄性核不育基因的研究进展[J]. 台湾农业探索, 2012,19(1):71-75. |
| Liu C H, Fang S R, Liu Y Q, Shen W F. Research progress on genic male sterile genes in rice (Oryza sativa L)[J]. Taiwan Agricultural Research, 2012,19(1):71-75. (in Chinese with English abstract) | |
| [5] | Wang C, Liu Q, Shen Y, Hua Y, Wang J J, Lin J R, Wu M G, Sun T T, Cheng Z K, Mercier R. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes[J]. Nature Biotechnology, 2019,37(3):283-286. |
| [6] | Nonomur K, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N. The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis[J]. The Plant Cell, 2004,16(4):1008-1020. |
| [7] | Chang L, Ma H, Xue H W. Functional conservation of the meiotic genes SDS and RCK in male meiosis in the monocot rice[J]. Cell Research, 2009,19(6):768-782. |
| [8] | Yu H X, Wang M, Tang D, Wang K J, Chen F L, Gong Z Y, Gu M H, Cheng Z K. OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice[J]. Chromosoma, 2010,119(6):625-636. |
| [9] | Wang Y X, Copenhaver G. Meiotic recombination: mixing it up in plants[J]. Annual Review of Plant Biology, 2018,69(1):577-609. |
| [10] | Shingu Y, Tokai T, Agawa Y, Toyota K, Ahmed S, Kobayashi M, Komatsu A, Mikawa T, Yamamoto M, Wakasa K, Shibata T, Kusano K. The double-stranded break-forming activity of plant SPO11s and a novel rice SPO11 revealed by a Drosophila bioassay[J]. BMC Molecular Biology, 2012,13:1-16. |
| [11] | Zhang B W, Wang M, Tang D, Li Y F, Xu M, Gu M H, Cheng Z K, Yu H X. XRCC3 is essential for proper double-strand break repair and homologous recombination in rice meiosis[J]. Journal of Experimental Botany, 2015,66(19):5713-5725. |
| [12] | Deng Z Y, Wang T. OsDMC1 is required for homologous pairing in Oryza sativa[J]. Plant Molecular Biology, 2007,65(1-2):31-42. |
| [13] | Sheridan S, Yu X, Roth R, Heuser J, Sehorn M, Sung P, Egelman E, Bishop D. A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments[J]. Nucleic Acids Research, 2008,36(12):4057-4066. |
| [14] | Sakane I, Kamataki C, Takizawa Y, Nakashima M, Toki S, Ichikawa H, Ikawa S, Shibata T, Kurumizaka H. Filament formation and robust strand exchange activities of the rice DMC1A and DMC1B proteins[J]. Nucleic Acids Research, 2008,36(13):4266-4276. |
| [15] | Morozumi Y, Ino R, Ikawa S, Mimida N, Shimizu T, Toki S, Ichikawa H, Shibata T, Kurumizaka H. Homologous pairing activities of two rice RAD51 proteins, RAD51A1 and RAD51A2[J]. PloS ONE, 2013,8(10):e75451. |
| [16] | Vries S, Baart E, Dekker M, Siezen A, Rooij D, Boer P, Riele H. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis[J]. Gene & Development, 1999,13(5):523-531. |
| [17] | Mimitou E, Symington L. DNA end resection: Many nucleases make light work[J]. DNA Repair, 2009,8(9):983-995. |
| [18] | Youds J, Boulton S. The choice in meiosis defining the factors that influence crossover or non-crossover formation[J]. Journal of Cell Science, 2011,124(4):501-513. |
| [19] | Fairman M, Stillman B. Cellular factors required for multiple stages of SV40 DNA-replication in vitro[J]. The EMBO Journal, 1988,7(4):1211-1218. |
| [20] | Iftode C, Daniely Y, Borowiec J. Replication Protein A (RPA): The Eukaryotic SSB[J]. Critical Reviews in Biochemistry and Molecular Biology, 1999,34(3):141-180. |
| [21] | Osman K, Sanchez-Moran E, Mann S, Jones G, Franklin F. Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair[J]. The EMBO Journal, 2009,28(4):394-404. |
| [22] | Takashi Y, Kobayashi Y, Tanaka K, Tamura K. Arabidopsis replication protein A 70a is required for DNA damage response and telomere length homeostasis[J]. Plant Cell Physiology, 2009,50(11):1965-1976. |
| [23] | Aklilu B, Soderquist R, Culligan K. Genetic analysis of the replication protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication[J]. Nucleic Acids Research, 2014,42(5):3104-3118. |
| [24] | Chang Y X, Gong L, Yuan W Y, Li X W, Chen G X, Li X H, Zhang Q F, Wu C Y. Replication Protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice[J]. Plant Physiology, 2009,151(4):2162-2173. |
| [25] | Li X W, Chang Y X, Xin X D, Zhu C M, Li X H, Higgins J, Wu C Y. Replication protein A2c coupled with replication protein A1c regulates crossover formation during meiosis in rice[J]. The Plant Cell, 2013,25(10):3885-3899. |
| [26] | 冯九焕, 卢永根, 刘向东, 徐雪宾. 水稻花粉发育过程及其分期[J]. 中国水稻科学, 2001,15(1):22-29. |
| Feng J H, Lu Y G, Liu X D, Xu X B. Pollen development and its stages in rice (Oryza sativa L.)[J]. Chinese Journal of Rice Science, 2001,15(1):22-29. (in Chinese with English abstract) | |
| [27] | Zhang D B, Luo X, Zhu L. Cytological analysis and genetic control of rice anther development[J]. Journal of Genetics and Genomics, 2011,38(9):379-390. |
| [28] | Xia R, Wang J G, Liu C Y, Wang Y Q, Zhai J X, Liu J, Hong X H, Cao X F, Zhu J K, Gong Z Z. ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis[J]. The Plant Cell, 2006,18(1):85-103. |
| [29] | Belanger K, Griffith A, Baker H, Hansen J, Kovacs L, Seconi J, Strine A. The karyopherin Kap95 and the C-termini of Rfa1, Rfa2, and Rfa3 are necessary for efficient nuclear import of functional RPA complex proteins in Saccharomyces cerevisiae[J]. DNA and Cell Biology, 2011,30(9):641-651. |
| [30] | Keshav K F, Chen C, Dutta A. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex[J]. Molecular and Cellular Biology, 1995,15(6):3119-3128. |
| [31] | Zhang J, Han F P. Centromere pairing precedes meiotic chromosome pairing in plants[J]. Science China Life Sciences, 2017,60(11):1197-1202. |
| [32] | Simonet J, Zick D. Genes involved in caryogamy and meiosis in Podospora anserine[J]. Molecular Genetics Genomics, 1978,162(3):237-242. |
| [33] | Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M. Genome sequencing reveals agronomically important loci in rice using MutMap[J]. Nature Biotechnology, 30(2):174-178. |
| [34] | Ishibashi T, Kimura S, Sakaguchi K. A higher plant has three different types of RPA heterotrimeric complex[J]. Journal of Biochemistry, 2006,139(1):99-104. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||