
Chinese Journal OF Rice Science ›› 2022, Vol. 36 ›› Issue (1): 27-34.DOI: 10.16819/j.1001-7216.2022.210302
• Research Papers • Previous Articles Next Articles
YANG Jinyu1,#, BAI Chen2,3,#, DING Xiaohui2,3, SHEN Hongfang1, WANG Lei1, YING Jiezheng1,*( ), E Zhiguo1,*(
), E Zhiguo1,*( )
)
Received:2021-03-03
Revised:2021-04-19
Online:2022-01-10
Published:2022-01-10
Contact:
YING Jiezheng, E Zhiguo
About author:First author contact:#These authors contributed equally to the work;
杨晋宇1,#, 白琛2,3,#, 丁小惠2,3, 申红芳1, 王磊1, 应杰政1,*( ), 鄂志国1,*(
), 鄂志国1,*( )
)
通讯作者:
应杰政,鄂志国
作者简介:第一联系人:#共同第一作者;
基金资助:YANG Jinyu, BAI Chen, DING Xiaohui, SHEN Hongfang, WANG Lei, YING Jiezheng, E Zhiguo. Genetic Analysis and Gene Mapping of a Male Sterile Mutant ms7 in Rice[J]. Chinese Journal OF Rice Science, 2022, 36(1): 27-34.
杨晋宇, 白琛, 丁小惠, 申红芳, 王磊, 应杰政, 鄂志国. 水稻雄性核不育突变体ms7的遗传分析及基因定位[J]. 中国水稻科学, 2022, 36(1): 27-34.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2022.210302

Fig. 1. Phenotype of ms7 and its wild type Zhonghui 161. A, ms7 and its wild type Zhonghui 161 (WT) at grain filling stage; B, Panicle of ms7 and WT at the harvest stage; C, Spikelet of ms7 and WT; D, I2-KI staining of ms7 and WT pollens.
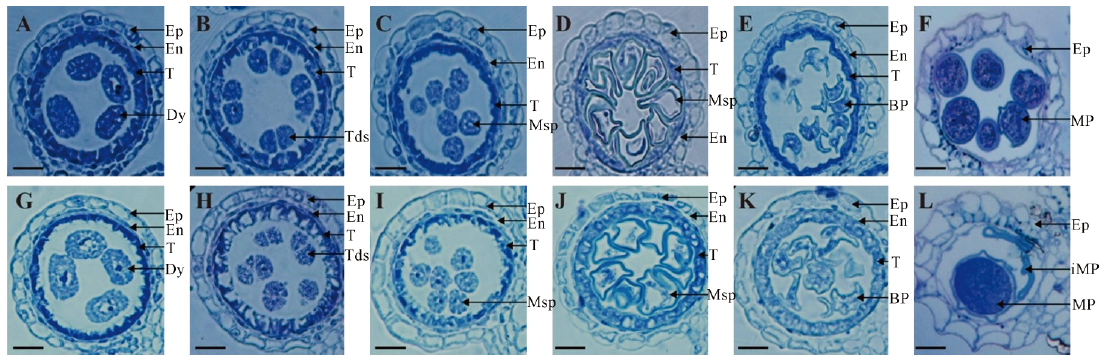
Fig. 2. Contrast of slice of anther between the ms7 mutant and its wild type. A-F, Wild-type anthers from stages 8a to 12; G-L, ms7 anthers from stages 8a to 12. Ep, Epidermis; En, Endothecium; T, Tapetum; Dy, Dyad cell; Tds, Tetrads; Msp, Microspore; BP, Biceullar pollen; MP, Mature pollen; iMP, Immature pollen. Bar=20 μm.
| 群体Group | 可育植株数No. of fertile plants | 不育植株数 No. of sterile plants | 卡方值 χ2 |
|---|---|---|---|
| F2-32 | 240 | 72 | 0.6154 |
| F2-33 | 232 | 73 | 0.1847 |
| F2-34 | 313 | 106 | 0.0199 |
| F2-35 | 273 | 76 | 1.9341 |
Table 1 Genetic analysis of the male-sterile trait in ms7.
| 群体Group | 可育植株数No. of fertile plants | 不育植株数 No. of sterile plants | 卡方值 χ2 |
|---|---|---|---|
| F2-32 | 240 | 72 | 0.6154 |
| F2-33 | 232 | 73 | 0.1847 |
| F2-34 | 313 | 106 | 0.0199 |
| F2-35 | 273 | 76 | 1.9341 |
| 样品 Sample | SNP位点数 SNP number | 转换的SNP数 Transition number | 颠换的SNP数 Transversion number | Ti/Tv | 杂合SNP数 Heterozygosity number | 纯合SNP数 Homozygosity number |
|---|---|---|---|---|---|---|
| 16F4Mix | 3 383 944 | 2 408 546 | 969 917 | 2.48 | 2 098 068 | 1 285 876 |
| 16S4 | 3 383 383 | 2 408 846 | 968 983 | 2.49 | 2 069 857 | 1 313 526 |
| IR9667 | 2 714 614 | 1 929 831 | 781 109 | 2.47 | 1 517 729 | 1 196 885 |
| ms7 | 3 014 721 | 2 147 993 | 863 049 | 2.49 | 549 661 | 2 465 060 |
Table 2. Statistics of SNP data.
| 样品 Sample | SNP位点数 SNP number | 转换的SNP数 Transition number | 颠换的SNP数 Transversion number | Ti/Tv | 杂合SNP数 Heterozygosity number | 纯合SNP数 Homozygosity number |
|---|---|---|---|---|---|---|
| 16F4Mix | 3 383 944 | 2 408 546 | 969 917 | 2.48 | 2 098 068 | 1 285 876 |
| 16S4 | 3 383 383 | 2 408 846 | 968 983 | 2.49 | 2 069 857 | 1 313 526 |
| IR9667 | 2 714 614 | 1 929 831 | 781 109 | 2.47 | 1 517 729 | 1 196 885 |
| ms7 | 3 014 721 | 2 147 993 | 863 049 | 2.49 | 549 661 | 2 465 060 |
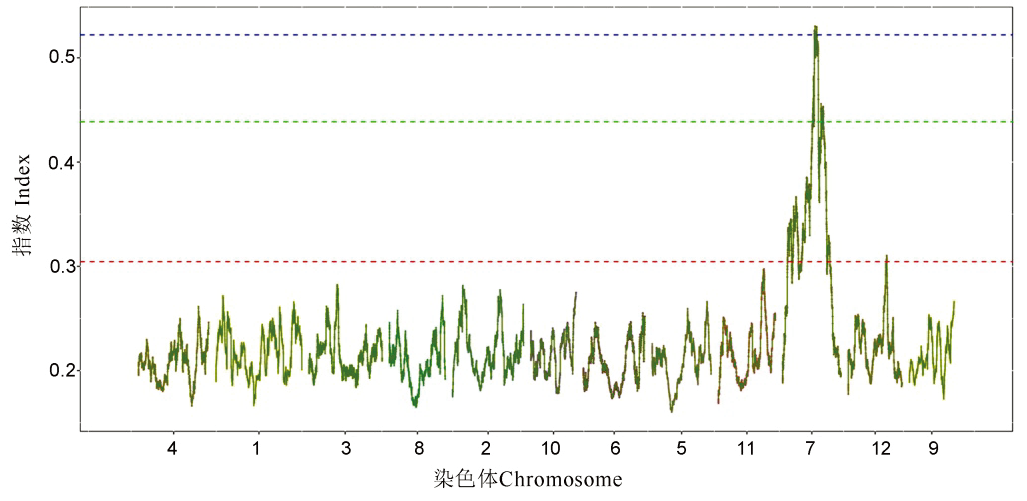
Fig. 3. Distribution of SNP-index on the chromosomes. The curve is the index value corresponding to the variation site on the chromosomes; the confidence interval thresholds from bottom to top are 95%, 99% and 99.9%, respectively.
| 引物名称 Primer name | 差异位点 Position | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|---|
| YD7036 | 16 913 833 | CCAGGGGATCTAAACAGG | TTGGACCATCACATAAGC |
| BSA11 | 16 969 331 | TGCCAATGTCGATTCCAAGAG | TCCAGCCTCTGTATACTTGCA |
| BSA6 | 17 002 663 | CCCTCACCTTCTGCAAGCTA | CGGTCTTTGAGGAGGTATCGA |
| BSA36 | 17 512 757 | AACAGAAAGCAACCAACCGC | TTCTTGGAGGTCAGGCAAGA |
| YD7013 | 17 777 239 | TGGTCTGCACTGCCTTAA | GGATGGACGGATGGGATA |
| YD7017 | 17 995 826 | ATCTTTCCCGATTGAGCT | CAGGCTTCATTCAAGACAA |
| YD7045 | 18 137 137 | AGGCTAAGGCGAGGAGAA | GCATACCAGCAAGGACGA |
| BSA47 | 18 400 602 | TTCATTCTTCTACCTTTGCCATCGG | TCTCTCTCACACTGGTCTCCTCCTC |
Table 3. Sequences and physical position of the eight InDel markers.
| 引物名称 Primer name | 差异位点 Position | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|---|
| YD7036 | 16 913 833 | CCAGGGGATCTAAACAGG | TTGGACCATCACATAAGC |
| BSA11 | 16 969 331 | TGCCAATGTCGATTCCAAGAG | TCCAGCCTCTGTATACTTGCA |
| BSA6 | 17 002 663 | CCCTCACCTTCTGCAAGCTA | CGGTCTTTGAGGAGGTATCGA |
| BSA36 | 17 512 757 | AACAGAAAGCAACCAACCGC | TTCTTGGAGGTCAGGCAAGA |
| YD7013 | 17 777 239 | TGGTCTGCACTGCCTTAA | GGATGGACGGATGGGATA |
| YD7017 | 17 995 826 | ATCTTTCCCGATTGAGCT | CAGGCTTCATTCAAGACAA |
| YD7045 | 18 137 137 | AGGCTAAGGCGAGGAGAA | GCATACCAGCAAGGACGA |
| BSA47 | 18 400 602 | TTCATTCTTCTACCTTTGCCATCGG | TCTCTCTCACACTGGTCTCCTCCTC |
| [1] | 鄂志国, 程本义, 孙红伟, 汪玉军, 朱练峰, 林海, 王磊, 童汉华, 陈红旗. 近40年我国水稻育成品种分析[J]. 中国水稻科学, 2019,33(6):523-531. |
| E Z G, Cheng B, Sun H, Wang Y, Zhu L, Lin Hai, Wang L, Tong H, Chen H. Analysis on Chinese improved rice varieties in recent four decades[J]. Chinese Journal of Rice Science, 2019,33(6):523-531. (in Chinese with English abstract) | |
| [2] | 范优荣, 曹晓风, 张启发. 光温敏雄性不育水稻的研究进展[J]. 科学通报, 2016,61(35):3822-3832. |
| Fan Y R, Cao X F, Zhang Q F. Progress on photoperiod thermo-sensitive genic male sterile rice[J]. Chinese Science Bulletin, 2016,61(35):3822-3832. (in Chinese with English abstract) | |
| [3] | Fan Y, Yang J, Mathioni S M, Yu J, Shen J, Yang X, Wang L, Zhang Q, Cai Z, Xu C, Li X, Xiao J, Meyers B C, Zhang Q,. PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016,113(52):15144. |
| [4] | Zhou H, Liu Q, Li J, Jiang D, Zhou L, Wu P, Lu S, Li F, Zhu L, Liu Z, Chen L, Liu Y G, Zhuang C. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA[J]. Cell Research, 2012,22(4):649-660. |
| [5] | Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice[J]. Proceedings of the National Academy of Sciences, 2012,109(7):2654-2659 |
| [6] | Zhou H, Zhou M, Yang Y, Li J, Zhu L, Jiang D, Dong J, Liu Q, Gu L, Zhou L, Feng M, Qin P, Hu X, Song C, Shi J, Song X, Ni E, Wu X, Deng Q, Liu Z, Chen M, Liu Y G, Cao X, Zhuang C. RNase Z S1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice [J]. Nature Communications, 2014,5:4884. |
| [7] | Pitnjam K, Chakhonkaen S, Toojinda T, Muangprom A. Identification of a deletion in tms2 and development of gene-based markers for selection[J]. Planta, 2008,228(5):813-822. |
| [8] | Yu B, Liu L, Wang T. Deficiency of very long chain alkanes biosynjournal causes humidity-sensitive male sterility via affecting pollen adhesion and hydration in rice[J]. Plant, Cell & Environment, 2019,42(12):3340. |
| [9] | Xue Z, Xu X, Zhou Y, Wang X, Zhang Y, Liu D, Zhao B, Duan L, Qi X. Deficiency of a triterpene pathway results in humidity-sensitive genic male sterility in rice[J]. Nature Communications, 2018,9:604. |
| [10] | Chen H, Zhang Z, Ni E, Lin J, Peng G, Huang J, Zhu L, Deng L, Yang F, Luo Q, Sun W, Liu Z, Zhuang C, Liu YG, Zhou H. HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynjournal and the humidity-sensitive genic male sterility in rice (Oryza sativa)[J]. New Phytologist, 2020,225(5):2077-2093. |
| [11] | 王多祥, 祝万万, 袁政, 张大兵. 水稻雄性发育功能基因的发掘及应用[J]. 生命科学, 2016,28(10):1180-1188. |
| Wang DX, Zhu WW, Yuan Z, Zhang DB. Functional research of rice male reproduction and its utilization in breeding[J]. Chinese Bulletin of Life Sciences, 2016,28(10):1180-1188. (in Chinese with English abstract) | |
| [12] | Li L, Li Y, Song S, Deng H, Li N, Fu X, Chen G, Yuan L. An anther development F-box (ADF) protein regulated by tapetum degeneration retardation (TDR) controls rice anther development[J]. Planta, 2015,241(1):157-166. |
| [13] | Lee S, Jung K H, An G, Chung Y Y. Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system[J]. Plant Molecular Biology, 2004,54(5):755-765. |
| [14] | Niu N, Liang W, Yang X, Jin W, Wilson Z A, Hu J, Zhang D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice[J]. Nature Communications, 2013,4 : 1445. |
| [15] | Shi J, Tan H, Yu X H, Liu Y, Liang W, Ranathunge K, Franke R B, Schreiber L, Wang Y, Kai G, Shanklin J, Ma H, Zhang D. Defective Pollen Wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase[J]. Plant Cell, 2011,23(6):2225-2246. |
| [16] | Mondol P C, Xu D, Duan L, Shi J, Wang C, Chen X, Chen M, Hu J, Liang W, Zhang D. Defective Pollen Wall 3 (DPW3), a novel alpha integrin-like protein, is required for pollen wall formation in rice[J]. New Phytologist, 2020,225(2):807-822. |
| [17] | Yang X, Liang W, Chen M, Zhang D, Zhao X, Shi J. Rice fatty acyl-CoA synthetase OsACOS12 is required for tapetum programmed cell death and male fertility[J]. Planta, 2017,246(1):105-122. |
| [18] | Xu D, Shi J, Rautengarten C, Yang L, Qian X, Uzair M, Zhu L, Luo Q, An G, Waßmann F, Schreiber L, Heazlewood J L, Scheller H V, Hu J, Zhang D, Liang W. Defective Pollen Wall 2 (DPW2) encodes an acyl transferase required for rice pollen development[J]. Plant Physiology, 2017,173(1):240-255. |
| [19] | Men X, Shi J, Liang W, Zhang Q, Lian G, Quan S, Zhu L, Luo Z, Chen M, Zhang D. Glycerol-3-phosphate acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice[J]. Journal of Experimental Botany, 2017,68(3):513-526. |
| [20] | Yu J, Meng Z, Liang W, Behera S, Kudla J, Tucker MR, Luo Z, Chen M, Xu D, Zhao G, Wang J, Zhang S, Kim YJ, Zhang D. A rice Ca2+ binding protein is required for tapetum function and pollen formation [J]. Plant Physiology, 2016,172(3):1772-1786. |
| [21] | Zheng S, Li J, Ma L, Wang H, Zhou H, Ni E, Jiang D, Liu Z, Zhuang C. OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers[J]. Proceedings of the National Academy of Sciences of USA, 2019,116(15):7549-7558. |
| [22] | Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB[J]. Plant Cell, 2009,21(5):1453. |
| [23] | Liu Z, Bao W, Liang W, Yin J, Zhang D. Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development[J]. Journal of Integrative Plant Biology, 2010,52(7):670-678. |
| [24] | Xiang X J, Sun L P, Yu P, Yang Z F, Zhang P P, Zhang Y X, Wu W X, Chen D B, Zhan X D, Khan R M, Abbas A, Cheng S H, Cao L Y. The MYB transcription factor Baymax1 plays a critical role in rice male fertility[J]. Theoretical and Applied Genetics, 2021,134(2):453-471. |
| [25] | Jung K H, Han M J, Lee Y S, Kim Y W, Hwang I, Kim M J, Kim Y K, Nahm B H, An G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development[J]. Plant Cell, 2005,17(10):2705-2722. |
| [26] | Ko S S, Li M J, Ku M S B, Ho Y C, Lin Y J, Chuang M H, Hsing H X, Lien Y C, Yang H T, Chang H C, Chan M T. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice[J]. Plant Cell, 2014,26(6):2486-2504. |
| [27] | Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W. The rice basic Helix-Loop-Helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell, 2014,26(4):1512-1524. |
| [28] | Li N, Zhang D S, Liu H S, Yin C S, Li X X, Liang W Q, Yuan Z, Xu B, Chu H W, Wang J, Wen T Q, Huang H, Luo D, Ma H, Zhang D B. The rice Tapetum Degeneration Retardation gene is required for tapetum degradation and anther development[J]. Plant Cell, 2006,18(11):2999-3014. |
| [29] | Ji C, Li H, Chen L, Xie M, Wang F, Chen Y, Liu YG. A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development[J]. Molecular Plant, 2013,6(5):1715-1718. |
| [30] | Cao H, Li X, Wang Z, Ding M, Sun Y, Dong F, Chen F, Liu L, Doughty J, Li Y, Liu Y X. Histone H2B monoubiquitination mediated by HISTONE MONOUBI-QUITINATION1 and HISTONE MONOUBIQUITI-NATION2 is involved in anther development by regulating tapetum degradation-related genes in rice[J]. Plant Physiology, 2015,168(4):1389. |
| [31] | Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson Z A, Zhang D. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice[J]. Plant Physiology, 2011,156(2):615-630. |
| [32] | Yang Z, Liu L, Sun L, Yu P, Zhang P, Abbas A, Xiang X, Wu W, Zhang Y, Cao L, Cheng S. OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice[J]. Plant Molecular Biology, 2019,99(1-2):175-191. |
| [33] | Yang Z, Sun L, Zhang P, Zhang Y, Yu P, Liu L, Abbas A, Xiang X, Wu W, Zhan X, Cao L, Cheng S. TDR INTERACTING PROTEIN 3, encoding a PHD-finger transcription factor, regulates Ubisch bodies and pollen wall formation in rice[J]. Plant Journal, 2019,99(5):844-861. |
| [34] | Li X, Gao X, Wei Y, Deng L, Ouyang Y, Chen G, Li X, Zhang Q, Wu C. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5’-triphosphate-dependent RNA helicases regulates tapetum degeneration[J]. Plant Cell, 2011,23(4):1416-1434. |
| [35] | Bai W, Wang P, Hong J, Kong W, Xiao Y, Yu X, Zheng H, You S, Lu J, Lei D, Wang C, Wang Q, Liu S, Liu X, Tian Y, Chen L, Jiang L, Zhao Z, Wu C, Wan J. Earlier degraded tapetum1 (EDT1) encodes an ATP-citrate lyase required for tapetum programmed cell death[J]. Plant Physiology, 2019,181(3):1223-1238. |
| [36] | Zhang P, Zhang Y, Sun L, Sinumporn S, Yang Z, Sun B, Xuan D, Li Z, Yu P, Wu W, Wang K, Cao L, Cheng S. The rice AAA-ATPase OsFIGNL1 is essential for male meiosis[J]. Frontiers in Plant Science, 2017,8:1639. |
| [37] | Tan H, Liang W, Hu J, Zhang D. MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice[J]. Developmental Cell, 2012,22(6):1127-1137. |
| [38] | Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009,25(14), 1754-1760. |
| [39] | McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo M A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome research, 2010,20(9):1297-1303. |
| [40] | Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009,25(16):2078-2079. |
| [41] | Zhang D, Luo X, Zhu L. Cytological analysis and genetic control of rice anther development[J]. Journal of Genetics and Genomics, 2011,38(9):379-390. |
| [42] | Wang C, Wang Y, Cheng Z, Zhao Z, Chen J, Sheng P, Yu Y, Ma W, Duan E, Wu F, Liu L, Qin R, Zhang X, Guo X, Wang J, Jiang L, Wan J. The role of OsMSH4 in male and female gamete development in rice meiosis[J]. Journal of Experimental Botany, 2016,67(5):1447-1459. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||