
Chinese Journal OF Rice Science ›› 2017, Vol. 31 ›› Issue (1): 1-12.DOI: 10.16819/j.1001-7216.2017.6132
• Orginal Article • Next Articles
Received:2016-09-30
Revised:2016-11-01
Online:2017-01-20
Published:2017-01-10
Contact:
Jinsong BAO
通讯作者:
包劲松
基金资助:CLC Number:
Yaling CHEN, Jinsong BAO. Progress in Structures, Functions and Interactions of Starch Synthesis Related Enzymes in Rice Endosperm[J]. Chinese Journal OF Rice Science, 2017, 31(1): 1-12.
陈雅玲, 包劲松. 水稻胚乳淀粉合成相关酶的结构、功能及其互作研究进展[J]. 中国水稻科学, 2017, 31(1): 1-12.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2017.6132
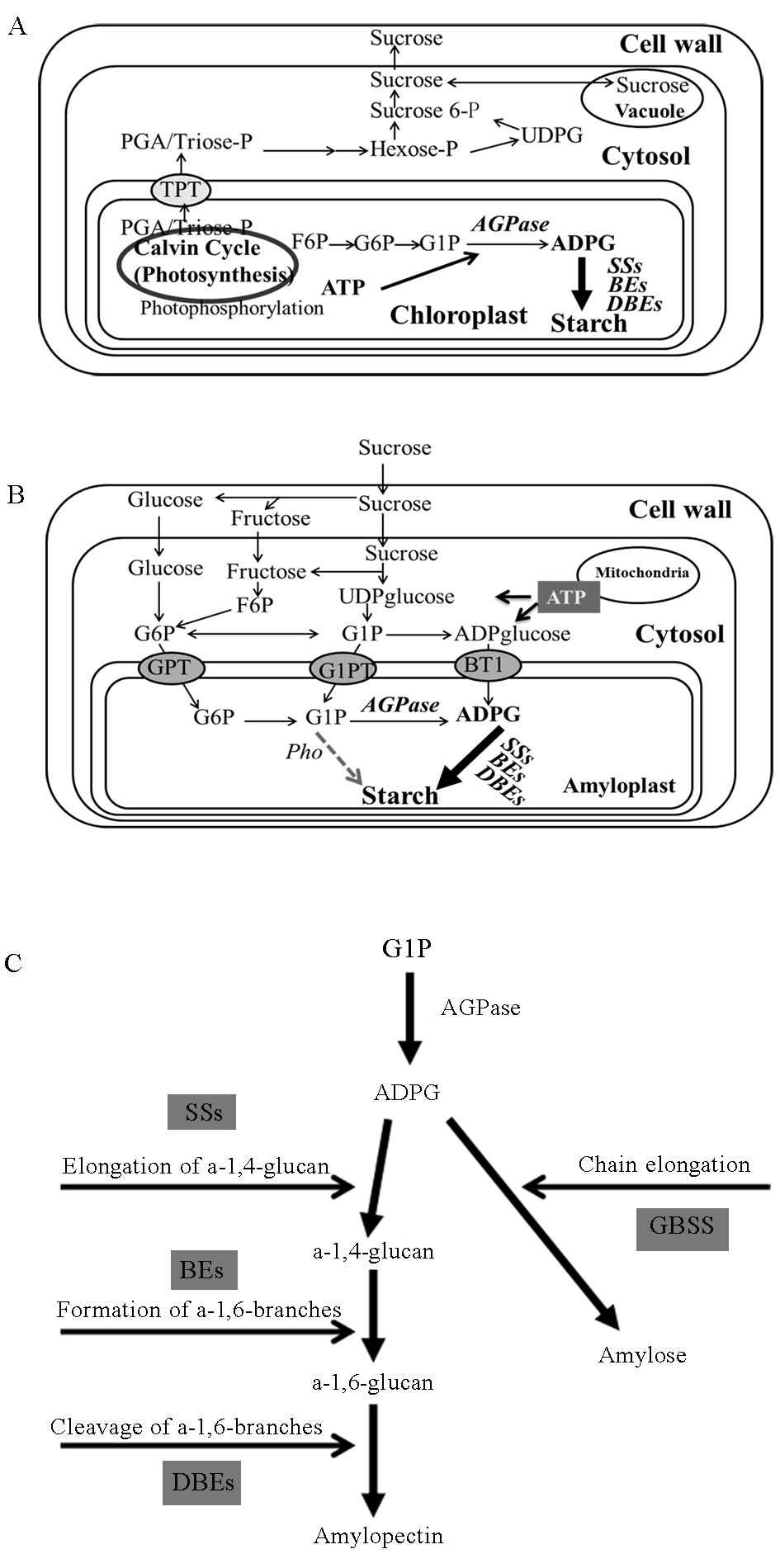
Fig. 1. Schematic representation of the starch biosynthetic pathway and the related metabolism in photosynthetic and non-photosynthetic tissues[7]. TPT, Triose phosphate translocator; GPT, G6P translocator; G1PT, Putative G1P transporter[8]; BT1, Brittle-1 protein. A, The synthesis of assimilatory starch in the leaf; B, The synthesis of reserve starch in storage tissues; C, The starch synthetic pathway from G1P.
| 酶 Enzyme | 基因 Gene | 染色体 Chromosome | 最高表达时期 Highest expression stage(DAF) | 突变体 Mutant | 功能 Function |
|---|---|---|---|---|---|
| AGPase | AGPLSU | 5,1,3,7 | 6~15 | osagpl2 | 酶的调控中心Regulation center of the enzyme |
| AGPSSU | 9,8 | 6 | osagps2 | 酶的活性中心 Activity center of the enzyme | |
| SSs | GBSSⅠ | 6 | 6,15 | waxy | 合成直连淀粉或形成超长链 Synthesis of amylose or formation of extra-long chain |
| SSⅠ | 6 | 10 | ssⅠ | 延伸DP 6-7的链形成DP 8-12的链 Formation of the DP 8-12 chain through elongated the DP 6-7 chain | |
| SSⅡa/SSⅡ-3 | 6 | 无明显表达高峰 No obvious peaks | Most japonica | 延伸DP≤10的短链形成 DP 12-24的中链 Formation of the DP 12-24 chain through elongated the DP≤10 short chain | |
| SSⅢa/SSⅢ-2 | 8 | 无明显表达高峰 No obvious peak | flo5 | 形成 DP>30的长链Formation of DP>30 long chain | |
| SSⅣa | 1 | 无明显表达高峰 No obvious peak | - | 涉及淀粉颗粒的形成及控制颗粒数量 Involve formation of starch granules and control the number of starch granules | |
| SSⅣb | 5 | 10 | ss4b | 涉及淀粉颗粒的形成及控制颗粒数量 Involve formation of starch granules and control the number of starch granules | |
| BEs | BEⅠ | 6 | 10 | sbe1 | 分支多聚葡萄糖链,形成B链簇状结构 Branch glucose chain and form the cluster structure of b chain |
| BEⅡb | 2 | 10 | ae | 分支多聚葡萄糖链,形成A链 Branch glucose chain and form A chain | |
| DBEs | ISA1 | 8 | 10 | sugary1 | 形成同源复合物,去除不当分支 Cleavage improper branch through homologous complexes of ISA1 |
| ISA2 | 5 | 10 | - | 与ISA1形成异源复合物 Form the heterogeneous complexes with ISA1 | |
| PUL | 4 | 10 | pul | 补偿ISA功能 Compensate the function of ISA |
Table 1 Function and expression pattern of starch synthesis genes in rice endosperm.
| 酶 Enzyme | 基因 Gene | 染色体 Chromosome | 最高表达时期 Highest expression stage(DAF) | 突变体 Mutant | 功能 Function |
|---|---|---|---|---|---|
| AGPase | AGPLSU | 5,1,3,7 | 6~15 | osagpl2 | 酶的调控中心Regulation center of the enzyme |
| AGPSSU | 9,8 | 6 | osagps2 | 酶的活性中心 Activity center of the enzyme | |
| SSs | GBSSⅠ | 6 | 6,15 | waxy | 合成直连淀粉或形成超长链 Synthesis of amylose or formation of extra-long chain |
| SSⅠ | 6 | 10 | ssⅠ | 延伸DP 6-7的链形成DP 8-12的链 Formation of the DP 8-12 chain through elongated the DP 6-7 chain | |
| SSⅡa/SSⅡ-3 | 6 | 无明显表达高峰 No obvious peaks | Most japonica | 延伸DP≤10的短链形成 DP 12-24的中链 Formation of the DP 12-24 chain through elongated the DP≤10 short chain | |
| SSⅢa/SSⅢ-2 | 8 | 无明显表达高峰 No obvious peak | flo5 | 形成 DP>30的长链Formation of DP>30 long chain | |
| SSⅣa | 1 | 无明显表达高峰 No obvious peak | - | 涉及淀粉颗粒的形成及控制颗粒数量 Involve formation of starch granules and control the number of starch granules | |
| SSⅣb | 5 | 10 | ss4b | 涉及淀粉颗粒的形成及控制颗粒数量 Involve formation of starch granules and control the number of starch granules | |
| BEs | BEⅠ | 6 | 10 | sbe1 | 分支多聚葡萄糖链,形成B链簇状结构 Branch glucose chain and form the cluster structure of b chain |
| BEⅡb | 2 | 10 | ae | 分支多聚葡萄糖链,形成A链 Branch glucose chain and form A chain | |
| DBEs | ISA1 | 8 | 10 | sugary1 | 形成同源复合物,去除不当分支 Cleavage improper branch through homologous complexes of ISA1 |
| ISA2 | 5 | 10 | - | 与ISA1形成异源复合物 Form the heterogeneous complexes with ISA1 | |
| PUL | 4 | 10 | pul | 补偿ISA功能 Compensate the function of ISA |
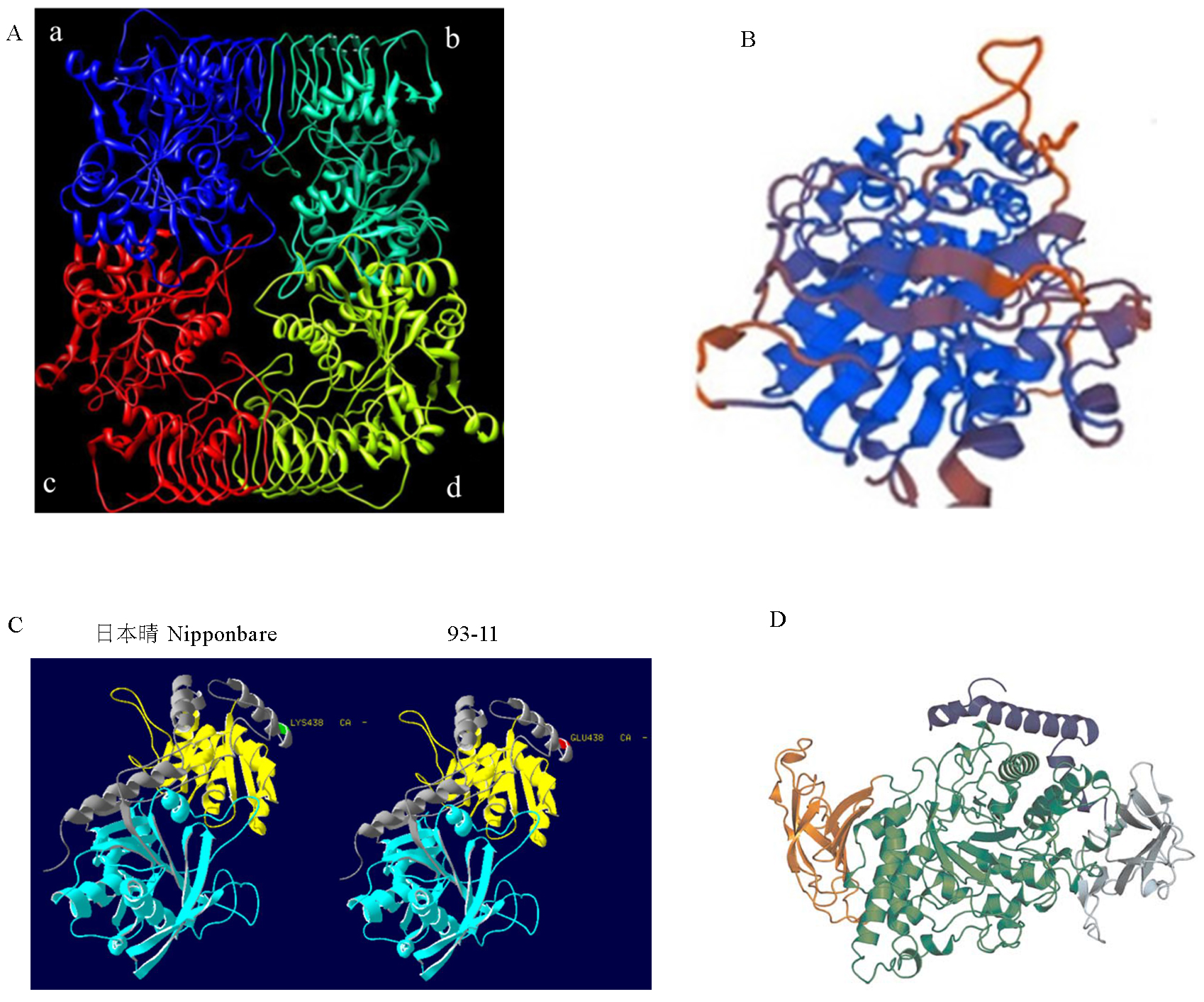
Fig. 2. 3D structure of starch synthesis related enzymes in rice. A, Molecular dynamics minimized structure of the rice AGPase[13]. Blue chain a, LS; Green chain c, LS; Cyan chain b, SS; Red chain d, SS. B, Homology model of GBSSⅠ, alpha helices are in blue, beta plated sheets in orange and coils in brown. C, Computational models of the 3D structure of SSⅠ of Nipponbare and 93-11. The blue color represents the starch synthase catalytic domain, and the yellow color represents the glycosyl transferase group. D, Structure of BEⅠ from Oryza sativa L. The purple color represents N-domain; The nattier blue represents CBM48; The blue-green color represents α-amylase; The orange represents C-domain.
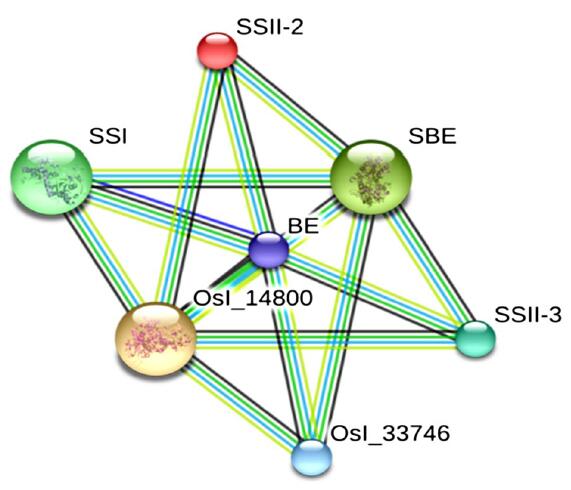
Fig. 3. Potential association networks of starch synthesis enzymes in rice endosperm. BE, Branching enzyme Ⅰ; BEⅡb, Branching enzyme Ⅱb; Osl-14800, Pullulanase; Osl-33746, Starch synthase Ⅱc.
| [1] | 朱德峰, 张玉屏, 陈惠哲, 向镜, 张义凯.中国水稻高产栽培技术创新与实践.中国农业科学, 2015, 48(17):3404-3414. |
| Zhu D F, Zhang Y P, Chen H Z, Xiang J, Zhang Y K.Innovation and practice of high-yield rice cultivation technology in China.Sci Agric Sin, 2015, 48(17): 3404-3414. (in Chinese with English abstract) | |
| [2] | Vandeputte G E, Delcour J A.From sucrose to starch granule to starch physical behaviour: A focus on rice starch.Carbohyd Polym, 2004, 58: 245-266. |
| [3] | Singh N, Kaur L, Sandhu K S,Nishinari K.Relationships between physicochemical, morphological, thermal, rheological properties of rice starches.Food Hydrocolloid, 2006, 20(4): 532-542. |
| [4] | 包劲松. 稻米淀粉品质遗传与改良研究进展. 分子植物育种, 2007, 5(6s): 1-20. |
| Bao J S.Progress in studies on inheritance and improvement of rice starch quality.Mol Plant Breeding, 2007, 5(6s): 1-20. (in Chinese with English abstract) | |
| [5] | Nakamura Y.Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue.Plant Cell Physiol, 2002, 43: 718-725. |
| [6] | Hannah L C, James M.The complexities of starch biosynthesis in cereal endosperms.Curr Opin Biotech, 2008, 19: 160-165. |
| [7] | Nakamura Y.Biosynthesis of reserve starch//Nakamura Y.Starch: Metabolism and Structure. Springer, Japan, 2015: 161-209. |
| [8] | Fettke J, Malinova I, Albrecht T, Hejazi M, Steup M.Glucose-1-phosphate transport into protoplasts and chloroplasts from leaves ofArabidopsis. Plant Physiol, 2011, 155(4): 1723-1734. |
| [9] | Tetlow I J, Davies E J, Vardy K A, Bowsher C G, Burrell M M, Emes M J.Subcellular localization of AD-Pglucose pyrophosphorylase in developing wheat endosperm and analysis of a plastidial isoform.J Exp Bot, 2003, 54: 715-725. |
| [10] | Bowsher C G, Scrase-Field E F A L, Esposito S, Emes M J, Tetlow I J. Characterization of ADP-glucose transport across the cereal endosperm amyloplast envelope.J Exp Bot, 2007, 58: 1321-1332. |
| [11] | Ohdan T, Francisco P B, Sawada T,Hirose T, Terao T, Satoh H, Nakamura Y.Expression profiling of genes involved in starch synthesis in sink and source organs of rice.J Exp Bot, 2005, 56(422): 3229-3244. |
| [12] | Lee S K, Hwang S K, Han M,Eom J S, Kang H G, Han Y, Choi S B, Cho M H, Bhoo S H, An G H, Hahn T R, Okita T W, Jeon J S.Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol Biol, 2007, 65(4): 531-546. |
| [13] | Dawar C, Jain S, Kumar S.Insight into the 3D structure of ADP-glucose pyrophosphorylase from rice (Oryza sativa L.). J Mol Model, 2013, 19(8): 3351-3367. |
| [14] | Seferoglu A B, Koper K, Can F B,Cevahir G, Kavakli I H.Enhanced heterotetrameric assembly of potato ADP-glucose pyrophosphorylase using reverse genetics.Plant Cell Physiol, 2014, pcu078. |
| [15] | Tuncel A, Cakir B, Hwang S K,Okita T W.The role of the large subunit in redox regulation of the rice endosperm ADP-glucose pyrophosphorylase.FEBS J, 2014, 281(21): 4951-4963. |
| [16] | Tang X J, Peng C, Zhang J,Cai Y, You X M, Kong F, Yan H G, Wang G X, Wang L, Jin J, Chen W W, Chen X G, Ma J, Wang P, Tiang L, Zhang W W, Wan J M.ADP-glucose pyrophosphorylase large subunit 2 is essential for storage substance accumulation and subunit interactions in rice endosperm.Plant Sci, 2016, 249: 70-83. |
| [17] | Tetlow I J, Morell M K, Emes M J.Recent developments in understanding the regulation of starch metabolism in higher plants.J Exp Bot, 2004, 55(406): 2131-2145. |
| [18] | Fujita N, Nakamura Y.Distinct and overlapping functions of starch synthase isoforms.Essent Rev Exp Biol, 2012a, 5: 115-140. |
| [19] | Denyer K.The isolation and characterization of novel low-amylose mutants ofPisum sativum L.Plant Cell Environ, 1995, 18(9):1019-1026. |
| [20] | Diall W M, Jiang H W, Chen Q S, Liu F, Wu P.Cloning and characterization of the granule-bound starch synthase Ⅱ gene in rice: Gene expression is regulated by the nitrogen level, sugar and circadian rhythm.Planta, 2003, 218(2): 261-268. |
| [21] | Wattoo J I, Iqbal M S, Arif M, Saleem Z, Shahid M N, Iqbal M.Homology modeling, functional annotation and comparative genome analysis of GBSS enzyme in rice and maize genomes.Int J Agric Biol, 2015, 17(5): 1061-1065. |
| [22] | Bligh H F J, Larkin P D, Roach P S,Jones C A, Fu H, Park W D. Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties.Plant Mol Biol, 1998, 38(3): 407-415. |
| [23] | Inukai T, Sako A, Hirano H Y,Sano Y.Analysis of intragenic recombination at wx in rice: Correlation between the molecular and genetic maps within the locus.Genome, 2000, 43(4): 589-596. |
| [24] | Mikami I, Aikawa M, Hirano H Y,Sano Y.Altered tissue-specific expression at theWx gene of the opaque mutants in rice. Euphytica, 1999, 105(2): 91-97. |
| [25] | Liu L L, Ma X D, Liu S J, Zhu C L, Jiang L, Wang Y H, Shen Y, Ren Y L, Dong H, Chen L M, Liu X, Zhao Z G, Zhai H Q, Wan JM.Identification and characterization of a novelWaxy allele from a Yunnan rice landrace. Plant Mol Biol, 2009, 71(6): 609-626. |
| [26] | Sato H, Suzuki Y, Sakai M, Imbe T.Molecular characterization of Wx-mq, a novel mutant gene for low-amylose content in endosperm of rice(Oryza sativa L.). Breeding Sci, 2002, 52(2): 131-135. |
| [27] | Larkin P D, Park W D.Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice(Oryza sativa L.). Mol Breeding, 2003, 12(4): 335-339. |
| [28] | Bergman C J, Delgado J T, McClung A M,Fjellstrom R G. An improved method for using a microsatellite in the rice waxy gene to determine amylose class. Cereal Chem, 2001, 78(3): 257-260. |
| [29] | 李枝桦, 陆春明, 卢宝荣, 王云月.云南传统栽培稻品种 waxy 基因序列分析.分子植物育种, 2011, 9(6):665-671. |
| Li Z H, Lu C M, Lu B R, Wang Y Y.Sequence analysis of waxy gene of yunnan rice landrace. Mol Plant Breeding, 2011, 9(6):665-671.(in Chinese with English abstract) | |
| [30] | Hanashiro I, Itoh K, Kuratomi Y, Yamazaki M, Igarashi T, Matsugasako J I, Takeda Y.Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice.Plant Cell Physiol, 2008, 49(6): 925-933. |
| [31] | Fujita N, Yoshida M, Kondo T,Saito K, Utsumi Y, Tokunaga T, Nishi H, Satoh J H, Park J L, Jane A, Miyao A, Hirochika Y, Nakamura Y.Characterization of SSⅢa-deficient mutants of rice: The function of SSⅢa and pleiotropic effects by SSⅢa deficiency in the rice endosperm. Plant Physiol, 2007, 144(4): 2009-2023. |
| [32] | Teng B, Zhang C, Zhang Y,Wu J, Li Z, Luo Z, Yang J.Comparison of amylopectin structure and activities of key starch synthesis enzymes in the grains of rice single-segment substitution lines with differentWx alleles. Plant Growth Reg, 2015, 77(2): 117-124. |
| [33] | Nakamura Y, Francisco P B, Hosaka Y,Sato A, Sawada T, Kubo A, Fujita N.Essential amino acids of starch synthase Ⅱa differentiate amylopectin structure and starch quality betweenjaponica and indica rice varieties. Plant Mol Biol, 2005, 58(2): 213-227. |
| [34] | Takemoto-Kuno Y, Suzuki K, Nakamura S,Satoh H, Ohtsubo K.Soluble starch synthase I effects differences in amylopectin structure betweenindica and japonica rice varieties. J Agric Food Chem, 2006, 54(24): 9234-9240. |
| [35] | Chen Y L, Bao J S.Underlying mechanisms of zymographic diversity in starch synthase I and pullulanase in rice-developing endosperm.J Agr Food Chem, 2016, 64(9): 2030-2037. |
| [36] | Cao H, James M G, Myers, A M.Purification and characterization of soluble starch synthases from maize endosperm.Arch BiochemI Biophy, 2000, 373(1): 135-146. |
| [37] | Commuri P D, Keeling P L.Chain-length specificities of maize starch synthase I enzyme: Studies of glucan affinity and catalytic properties.Plant J, 2001, 25: 475-486. |
| [38] | Fujita N, Yoshida M, Asakura N,Ohdan T, Miyao A, Hirochika H, Nakamura Y.Function and characterization of starch synthase I using mutants in rice.Plant Physiol, 2006, 140(3): 1070-1084. |
| [39] | Hirose T, Terao T.A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta, 2004, 220(1): 9-16. |
| [40] | Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y.Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties.Theor Appl Genet, 2002, 104(1): 1-8. |
| [41] | Jeon J S, Nayeon R, Tae-Ryong H,Harkamal W, Yasunori N.Starch biosynthesis in cereal endosperm.Plant Physiol Bioch, 2010, 48: 383-392. |
| [42] | Tetlow I J.Starch biosynthesis in developing seeds.Seed Sci Res, 2011, 21: 5-32. |
| [43] | Fujita N, Hanashiro I, Suzuki S, Higuchi T, Toyosawa Y, Utsumi Y, Itoh R, Aihara S, Nakamura Y.Elongated phytoglycogen chain length in transgenic rice endosperm expressing active starch synthase Ⅱa affects the altered solubility and crystallinity of the storage α-glucan.J Exp Bot, 2012b, 63(16): 5859-5872. |
| [44] | Wang K, Hasjim J, Wu A C, Li E, Henry R J, Gilbert R G.Roles of GBSSⅠ and SSⅡa in determining amylose fine structure.Carbohyd Polym, 2015, 127: 264-274. |
| [45] | Li Z, Li D, Du X,Wang H, Larroque O, Jenkins C L, Jobling S A, Morell M K.The barleyamo1 locus is tightly linked to the starch synthase Ⅲa gene and negatively regulates expression of granule-bound starch synthetic genes. J Exp Bot, 2011, 62(14): 5217-5231. |
| [46] | Lin Q, Huang B, Zhang M,Zhang X, Rivenbark J, Lappe R L, James M G, Myers A M, Hennen-Bierwagen T A. Functional interactions between starch synthase Ⅲ and isoamylase-type starch-debranching enzyme in maize endosperm.Plant Physiol, 2012, 158(2): 679-692. |
| [47] | Roldán I, Wattebled F, Mercedes Lucas M,Delvallé D, Planchot V, Jiménez S, Pérez R, Ball S, D’Hulst C, Mérida Á. The phenotype of soluble starch synthase Ⅳ defective mutants ofArabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J, 2007, 49(3): 492-504. |
| [48] | Toyosawa Y, Kawagoe Y, Matsushima R, Crofts N, Ogawa M, Fukuda M, Kumamaru T, Okazaki Y, Kusano M, Saito K, Toyooka K, Sato M, Ai Y F, Jane J L, Nakamura Y, Fujita N .Deficiency of starch synthase Ⅲa and Ⅳb alters starch granule morphology from polyhedral to spherical in rice endosperm.Plant Physiol, 2016, 170(3): 1255-1270. |
| [49] | Guan H, Li P, Imparl-Radosevich J, Preiss J, Keeling P.Comparing the properties ofEscherichia coli branching enzyme and maize branching enzyme. Arch Biochem Biophy, 1997, 342(1): 92-98. |
| [50] | Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Fujita N, Nakamura, Y.Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm.Plant Physiol, 2003, 133(3): 1111-1121. |
| [51] | Ryoo N, Yu C, Park C S, Baik M Y, Park I M, Cho M H, Bhoo S H, An G, Hahn T R, Jeon J S.Knockout of a starch synthase geneOsSSⅢa/Flo5 causes white-core floury endosperm in rice(Oryza sativa L.). Plant Cell Rep, 2007, 26(7): 1083-1095. |
| [52] | Nakamura Y, Utsumi Y, Sawada T, Aihara S, Utsumi C, Yoshida M, Kitamura S.Characterization of the reactions of starch branching enzymes from rice endosperm.Plant Cell Physiol, 2010, 51(5): 776-794. |
| [53] | Noguchi J, Chaen K, Vu N T,Akasaka T, Shimada H, Nakashima T, Nishi A, Satoh H, Omori T, Kakuta Y, Kimura M.Crystal structure of the branching enzyme I (BEI) fromOryza sativa L with implications for catalysis and substrate binding. Glycobiology, 2011, 21(8): 1108-1116. |
| [54] | Nishi A, Nakamura Y, Tanaka N,Satoh H.Biochemical and genetic analysis of the effects ofamylose extender mutation in rice endosperm. Plant Physiol, 2001, 127: 459-472. |
| [55] | Jiang H, Zhang J, Wang J,Xia M, Zhu S, Cheng B J.RNA interference-mediated silencing of the starch branching enzyme gene improves amylose content in rice . Genet Mol Res, 2013, doi. |
| [56] | Wong K S, Kubo A, Jane J L,Harada K, Satoh H, Nakamura Y.Structures and properties of amylopectin and phytoglycogen in the endosperm ofsugary-1 mutants of rice. J Cereal Sci, 2003, 37: 139-149. |
| [57] | Kubo A, Rahman S, Utsumi Y,Li Z, Mukai Y, Yamamoto M, Ugaki M, Harada K, Satoh H, Konik-Rose C, Morell M, Nakamura Y.Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase1 gene supports a direct role for isoamylase1 in amylopectin biosynthesis.Plant Physiol, 2005, 137(1): 43-56. |
| [58] | Utsumi Y, Utsumi C, Sawada T,Fujita N, Nakamura Y.Functional diversity of isoamylase oligomers: The ISA1 homooligomer is essential for amylopectin biosynthesis in rice endosperm.Plant Physiol, 2011, 156: 61-77. |
| [59] | 朱立楠, 刘海英, 孙璐璐, 孙涛, 郭雪冬, 朱方旭, 张忠臣, 金正勋.水稻灌浆过程中胚乳异淀粉酶基因家族表达特性及其与淀粉含量间关系分析. 中国水稻科学, 2015, 29(5): 528-534. |
| Zhu L N, Liu H Y, Sun L L, Sun T, Guo X D, Zhu F X, Zhang Z C, Jin Z X.Analysis of expression characteristics of isoamylase and the correlation with starch content during grain filling in rice.Chin J Rice Sci, 2015, 29(5): 528-534. (in Chinese with English abstract) | |
| [60] | Li Q F, Zhang G Y, Dong Z W,Yu H X, Gu M H, Sun S S M, Liu Q Q. Characterization of expression of theOsPUL gene encoding a pullulanase-type debranching enzyme during seed development and germination in rice. Plant Physiol Bioch, 2009, 47: 351-358. |
| [61] | Fujita N, Toyosawa Y, Utsumi Y,Higuchi T, Hanashiro I, Ikegami A, Akuzawa S, Yoshida M, Mori A, Inomata K, Itoh R, Miyao A, Hirochika H, Satoh H, Nakamura Y.Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. J Exp Bot, 2009, 60: 1009-1023. |
| [62] | Tian Z X, Qian Q, Liu Q,Yan M, Liu X, Yan C J, Liu G, Gao Z, Tang S, Zeng D, Wang Y, Yu J, Gu M, Li J.Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities.PNAS, 2009, 106(51): 21760-21765. |
| [63] | Yan C J, Tian Z X, Fang Y W,Yang Y C, Li J, Zeng S Y, Gu S L, Xu C W, Tang Z S, Gu M H.Genetic analysis of starch paste viscosity parameters in glutinous rice (Oryza sativa L.). Theor Appl Genet, 2011, 122: 63-76. |
| [64] | Kharabian-Masouleh A, Waters D L, Reinke R F, Ward R, Henry R J.SNP in starch biosynthesis genes associated with nutritional and functional properties of rice.Sci Rep, 2012, 2. |
| [65] | Yang F, Chen Y L, Tong C,Huang Y, Xu F F, Li K H, Corke H, Sun M, Bao J S.Association mapping of starch physicochemical properties with starch synthesis-related gene markers in nonwaxy rice (Oryza sativa L.). Mol Breeding, 2014, 34(4): 1747-1763. |
| [66] | Tetlow I J, Beisel K G, Cameron S,Makhmoudova A, Liu F, Bresolin N S, Wait R, Morell M K, Emes M J.Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes.Plant Physiol, 2008, 146(4): 1878-1891. |
| [67] | Liu F, Makhmoudova A, Lee E A,Wait R, Emes M J, Tetlow I J .The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts.J Exp Bot, 2009b, 60(15): 4423-4440. |
| [68] | Hennen-Bierwagen T A, Liu F, Marsh R S,Kim S, Gan Q, Tetlow I J, Emes M J, James M G, Myers A M. Starch biosynthetic enzymes from developing Zea mays endosperm associate in multisubunit complexes.Plant Physiol, 2008, 146:1892-1908. |
| [69] | Liu F, Ahmed Z, Lee E A,Donner E, Liu Q, Ahmed R, Morell, M K, Emes M J, Tetlow I J . Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein-protein interactions.J Exp Bot, 2012, 63(3): 1167-1183. |
| [70] | Bao J S.Towards understanding of the genetic and molecular basis of eating and cooking quality of rice.Cereal Food World, 2012, 57:148-156. |
| [71] | Nakamura Y, Ono M, Utsumi C,Steup M.Functional interaction between plastidial starch phosphorylase and starch branching enzymes from rice during the synthesis of branched maltodextrins.Plant Cell Physiol, 2012, 53(5): 869-878. |
| [72] | Nakamura Y, Aihara S, Crofts N,Sawada T, Fujita N.In vitro studies of enzymatic properties of starch synthases and interactions between starch synthase I and starch branching enzymes from rice.Plant Sci, 2014, 224: 1-8. |
| [73] | Crofts N, Abe N, Oitome N F,Matsushima R, Hayashi M, Tetlow, I J, Emes M J, Nakamura Y, Fujita N. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes.J Exp Bot, 2015, 66: 4469-4482. |
| [74] | She K C, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M, Tsuge T, Matsumoto K, Kudoh M, Itoh E, Kikuchi S, Kishimoto N, Yazaki J, Ando T, Yano M, Aoyama T, Sasaki T, Satoh H, Shimada H.A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality.Plant Cell, 2010, 22: 3280-3294. |
| [75] | Peng C, Wang Y, Liu F, Ren Y, Zhou K, Lv J, Zheng M, Zhao S L, Zhang L, Wang C M, Jiang L, Zhang X, Guo X P, Bao Y Q, Wan J M.FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm.Plant J, 2014, 77: 917-930. |
| [76] | Wang J C, Xu H, Zhu Y,Liu Q Q, Cai X L.OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm.J Exp Bot, 2013, 64: 3453-3466. |
| [77] | Tomita K, Horiuchih H, Terada K, Tanoi M, Kobayashi A, Kanda K, Tanaka I, Minobe T, Furuta H, Yamamoto A, Shinoyama H, Aoki K, Masaki N, Minami T, Sugimoto A, Kagashima C, Horiuchi K.New-hikari, a new rice cultivar.Bull Fukui Agric Exp, 2007, 44: 1-20. |
| [78] | 王才林, 陈摇涛, 张亚东, 朱镇, 赵凌, 林静.通过分子标记辅助选择培育优良食味水稻新品种.中国水稻科学, 2009, 23:25-30. |
| Wang C L, Chen T, Zhang Y D, Zhu Z, Zhao L, Lin J.Breeding of a new rice variety with good eating quality by marker assisted selection.Chin J Rice Sci, 2009, 23(1): 25-30. (in Chinese with English abstract) | |
| [79] | 王才林, 张亚东, 朱摇镇, 姚姝, 赵庆勇, 陈涛, 周丽慧, 赵凌.优良食味粳稻新品种南粳9108的选育与利用.江苏农业科学, 2013, 41:86-88. |
| Wang C L, Zhang Y D, Zhu Y Z, Yao S, Zhao Q Y, Chen T, Zhou L H, Zhao L.Breeding and utilization of a new good eating quality rice variety Nanjing 9108.Jiangsu Agric Sci, 2013, 41:86-88.(in Chinese with English abstract) | |
| [80] | 杨瑞芳, 白建江, 方军, 曾威, 朴钟泽, 李刚夑.分子标记辅助选择选育高抗性淀粉水稻新品种.核农学报, 2015, 29:2259-2267. |
| Yang R F, Bai J J, Fang J, Zeng W, Piao Z Z, Lee G S.Establishment of marker-assisted selection system for breeding rice varieties with high resistant starch content. J Nucl Agric Sci, 2015, 29(12): 2259-2267. (in Chinese with English abstract) | |
| [81] | Sacks W J, Deryng D, Foley J A, Ramankutty N.Crop planting dates: an analysis of global patterns.Global Ecol Bioge, 2010, 19(5): 607-620. |
| [82] | Liu Q, Wu X, Ma J, Li T, Zhou X, Guo T.Effects of high air temperature on rice grain quality and yield under field condition.Agron J, 2013, 105(2): 446-454. |
| [83] | Chen J, Tang L, Shi P, Yang B, Sun T, Cao W, Zhu Y.Effects of short-term high temperature on grain quality and starch granules of rice (Oryza sativa L.) at post-anthesis stage. Protoplasma, 2016: 1-9. |
| [84] | Liao J L, Zhou H W, Zhang H Y, Zhong P A, Huang Y J.Comparative proteomic analysis of differentially expressed proteins in the early milky stage of rice grains during high temperature stress.J Exp Bot, 2014, 65(2): 655-671. |
| [85] | Sreenivasulu N, Butardo V M, Misra G, Cuevas R P, Anacleto R, Kishor P B K. Designing climate-resilient rice with ideal grain quality suited for high-temperature stress.J Exp Bot, 2015, 66(7): 1737-1748. |
| [86] | Zhou H J, Wang L J, Liu G F, Meng X B, Jing Y H, Shu X L, Kong X L, Sun J, Yu H, Smith S M, Wu D X, Li J Y. (2016). Critical roles of soluble starch synthase SSⅢa and granule-bound starch synthase Waxy in synthesizing resistant starch in rice.P Natl Acad Sci, 2016, DOI: 10.1073/pnas.1615104113. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
