
中国水稻科学 ›› 2026, Vol. 40 ›› Issue (1): 1-17.DOI: 10.16819/j.1001-7216.2026.240901
• 综述与专论 • 下一篇
李兴沂1,2, 陈玲2, 邵建韬1, 肖素勤2, 李金璐2, 付惠仙3, 殷富有2, 张建红4, 程在全2, 刘丽2,*( )
)
收稿日期:2024-09-01
修回日期:2024-11-10
出版日期:2026-01-10
发布日期:2026-01-21
通讯作者:
*email: liuliyaas@163.com基金资助:
LI Xingyi1,2, CHEN Ling2, SHAO Jiantao1, XIAO Suqin2, LI Jinlu2, FU Huixian3, YIN Fuyou2, ZHANG Jianhong4, CHENG Zaiquan2, LIU Li2,*( )
)
Received:2024-09-01
Revised:2024-11-10
Online:2026-01-10
Published:2026-01-21
摘要:
高产和优质是水稻育种的两大主要目标,产量与品质协同改良是水稻育种和功能基因组研究面临的重大挑战。本文从水稻淀粉合成的遗传调控、产量形成的分子生物学基础、产量与淀粉品质协同调控的分子机理及相关基因的克隆与功能研究等方面,总结水稻产量与淀粉品质协同调控的分子遗传研究进展,分析现有研究存在的问题,初步提出未来解决水稻产量与品质难以兼得关键问题的遗传改良策略,为利用分子育种技术协调产量与品质均衡,培育既高产又优质的水稻新品种提供信息参考。
李兴沂, 陈玲, 邵建韬, 肖素勤, 李金璐, 付惠仙, 殷富有, 张建红, 程在全, 刘丽. 水稻产量与淀粉品质协同调控的分子遗传研究进展[J]. 中国水稻科学, 2026, 40(1): 1-17.
LI Xingyi, CHEN Ling, SHAO Jiantao, XIAO Suqin, LI Jinlu, FU Huixian, YIN Fuyou, ZHANG Jianhong, CHENG Zaiquan, LIU Li. Progress in Molecular Genetic Research on Co-regulation of Grain Yield and Starch Quality in Rice[J]. Chinese Journal OF Rice Science, 2026, 40(1): 1-17.
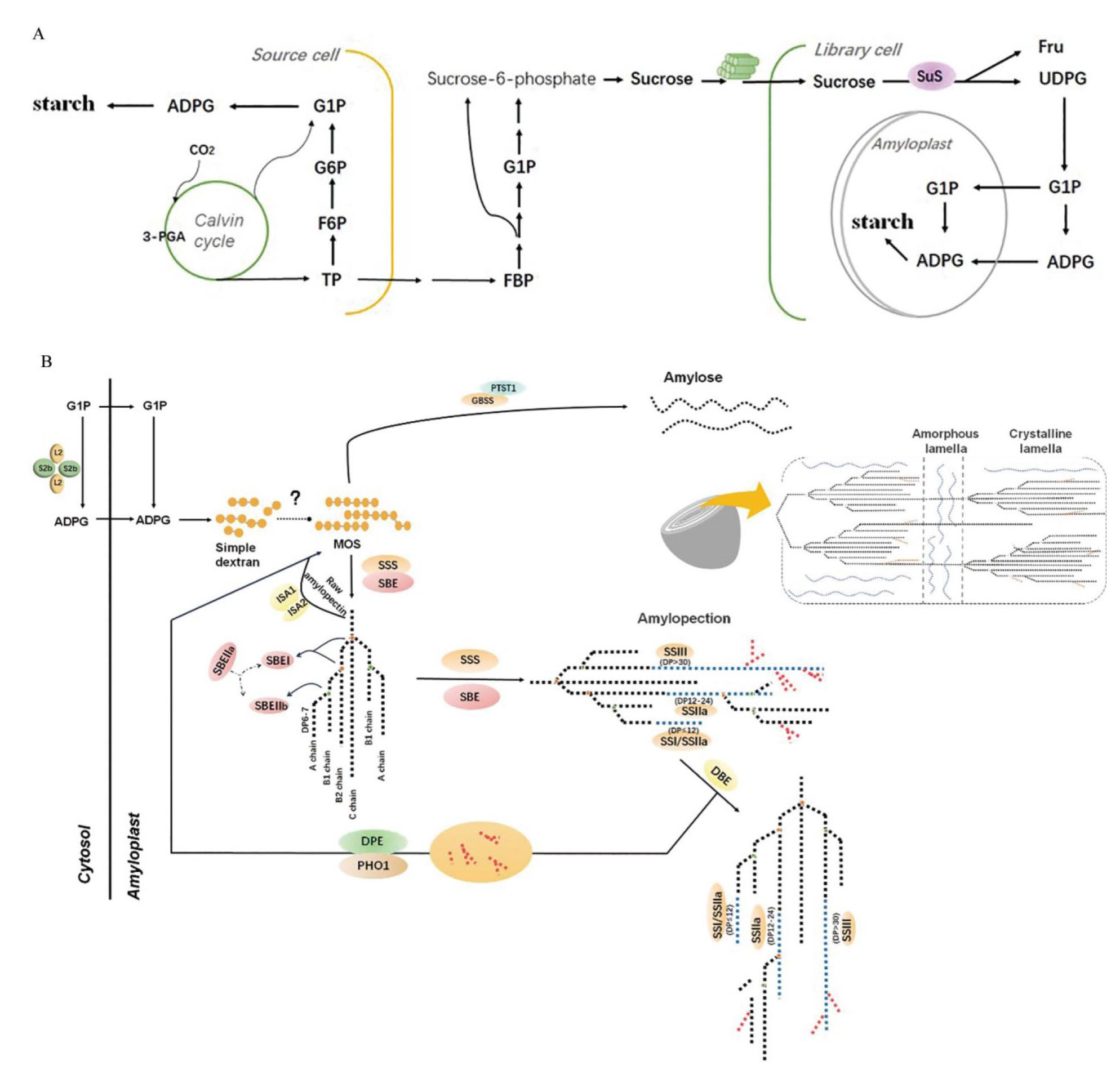
图1 淀粉的生物合成 A:源细胞(Source cell)中瞬时淀粉的合成及库细胞(Sink cell)中储藏淀粉的合成。SuS:蔗糖合酶;3-PGA:3-磷酸甘油酸;TP:磷酸丙糖;F6P:6-磷酸果糖。G6P:6-磷酸葡萄糖;G1P:1-磷酸葡萄糖;ADPG:腺苷二磷酸葡萄糖;FBP:1,6-二磷酸果糖;UDPG:二磷酸尿苷葡糖;Fru:果糖;B:淀粉合成途径中主要酶类与蛋白的功能及作用。AGPase由2种亚基(两个大亚基AGPL和两个小亚基AGPS)组成的异源四聚体;SBE:淀粉分支酶;GBSS:颗粒结合淀粉合成酶;SS/SSS:可溶性淀粉合成酶;DBE:脱分支酶;DPE:歧化酶;PHO:淀粉/α-葡聚糖磷酸化酶;Amylose:直链淀粉;Amylopectin:支链淀粉。
Fig. 1. Biosynthesis of starch A, Starch is synthesized as transient starch in source cells and as storage starch in library cells. SuS, Sucrose synthase; 3-PGA, 3-phosphoglyceric acid; TP, Triose phosphate; F6P, Fructose 6-phosphate; G6P, Glucose 6-phosphate; G1P, Glucose 1-phosphate; ADPG, Adenosine diphosphate glucose; FBP, Fructose 1,6-bisphosphate; UDPG, Uridine diphosphate glucose; Fru, Fructose. B, The main enzymes involving in starch synthesis and their functions in the pathway. AGPase is a heterotetramer composed of two structurally distinct subunits (two large regulatory subunits AGPL and two small catalytic subunits AGPS); SBE, Starch branching enzyme; GBSS, Granule-bound starch synthase; SS/SSS, Soluble starch synthase; DBE, Debranching enzyme; DPE, Dismutase; PHO, Starch/α-glucan phosphorylase.
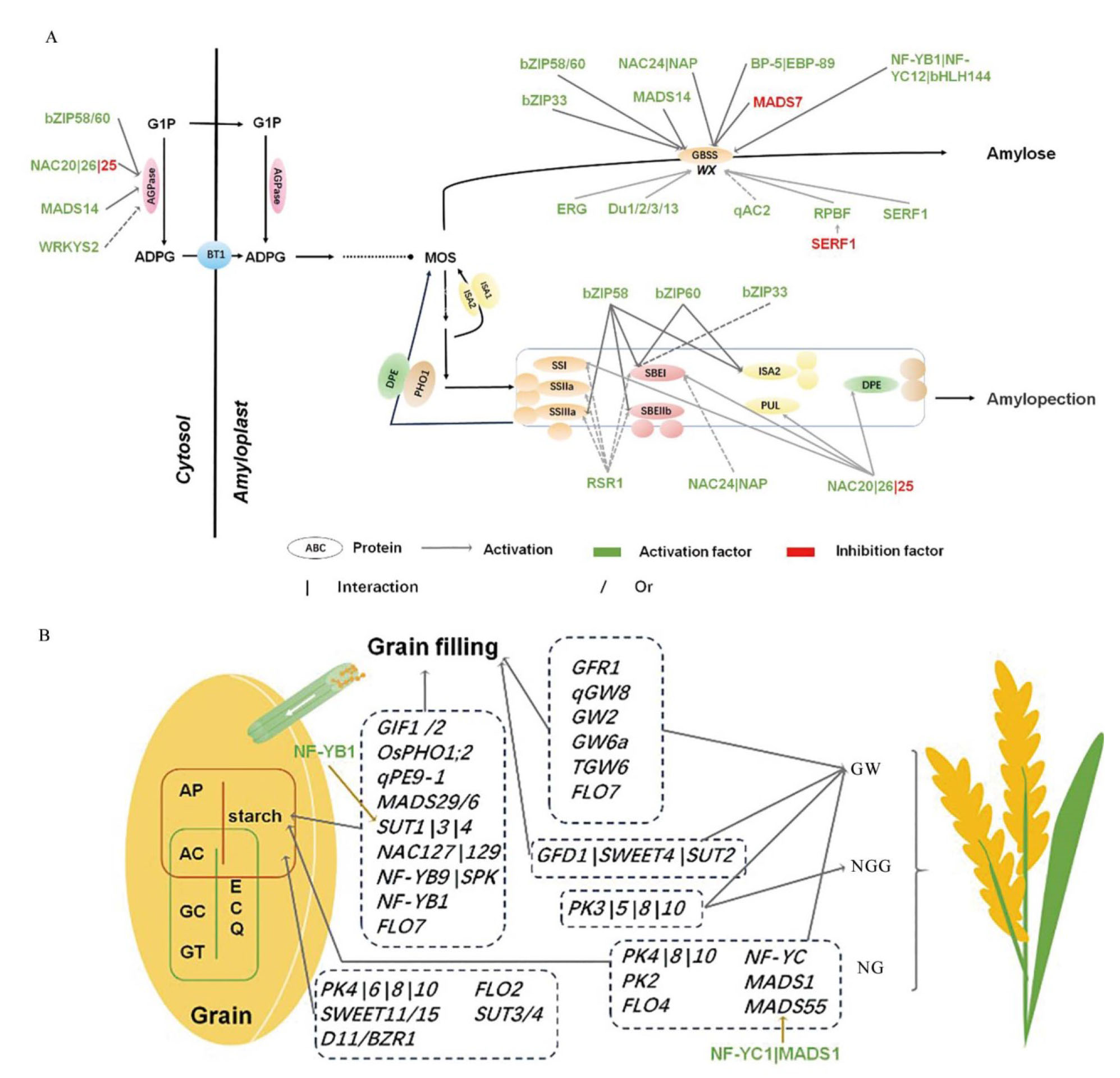
图2 已报道的淀粉及产量相关因子调控图 A:淀粉合成途径中,多种转录因子调控淀粉合成酶;B:已报道的淀粉及产量调控基因及转录因子。KW:籽重;NGS:穗粒数;NS:穗数。
Fig. 2. Network of reported regulatory factors of starch and grain yield in rice A, Multiple transcription factors regulating enzymes in the starch synthesis pathway; B, Genes and transcription factors regulating starch quality and grain yield. GW, Grain weight; NGG, Number of grain per panicles; NG, Number of panicles.
| 性状 Trait | 基因 Gene | 编码产物 Encoded protein/Enzyme | 基因效应 Gene effect | 参考文献 Reference | |
|---|---|---|---|---|---|
| 有效穗数 Effective panicle number | GS5 | 丝氨酸羧肽酶 | 正调控粒宽和粒重 | [ | |
| HTH5 | 磷酸吡哆醛结合蛋白 | 正调控结实率 | [ | ||
| qHd1 | Squamosa启动子 | 正调控有效穗 | [ | ||
| LAX2-4 | 核蛋白 | 负调控有效穗和穗粒数 | [ | ||
| OsDCL3b | Dicer酶 | 正调控有效穗 | [ | ||
| 穗粒数 Spikelets per panicle | GNP1 | GA20氧化酶 | 正调控穗粒数 | [ | |
| OsKRN2 | WD40蛋白 | 负调控穗粒数和产量 | [ | ||
| An-1 | bHLH蛋白 | 正调控穗粒数 | [ | ||
| COG1 | 转录因子OsMADS17 | 正调控穗粒数和粒重 | [ | ||
| NOG1 | 烯酰辅酶A水合酶 | 正调控穗粒数 | [ | ||
| LF1 | 转录因子HD-ZIP Ⅲ | 正调控穗粒数 | [ | ||
| 千粒重 1000-grain weight | GS3 | Gγ亚基 | 负调控粒长和粒重 | [ | |
| GSN1 | 双特异性磷酸酶 | 负调控粒型和穗粒数 | [ | ||
| TGW2 | 细胞数调控因子OsCNR1 | 负调控粒宽和粒重 | [ | ||
| DG1 | 毒性物外排转运蛋白 | 正调控籽粒灌浆 | [ | ||
| GLW7 | 转录因子OsSPL13 | 正调控粒长和粒重 | [ | ||
| GAD1 | 短分泌信号肽 | 正调控粒长、穗粒数 | [ | ||
| GW5 | 钙调素结合蛋白 | 正调控粒宽和粒重 | [ | ||
| GLW10 | 油菜素内酯信号激酶 | 正调控粒长和粒重 | [ | ||
| 淀粉合成Starch biosynthesis | Chalk5 | 液泡膜质子焦磷酸酶 | 负调控垩白 | [ | |
| Wx | 淀粉颗粒合成酶 | 负调控直链淀粉 | [ | ||
| WCR1 | F-box蛋白 | 负调控垩白 | [ | ||
| SSIIIb | 淀粉颗粒合成酶 | 与ssIIIa协同调控抗性淀粉 | [ | ||
| OsLESV | 淀粉结合蛋白 | 正调控储藏淀粉 | [ | ||
表1 已克隆的水稻产量与品质重要基因
Table 1. Major cloned genes related to yield and quality in rice
| 性状 Trait | 基因 Gene | 编码产物 Encoded protein/Enzyme | 基因效应 Gene effect | 参考文献 Reference | |
|---|---|---|---|---|---|
| 有效穗数 Effective panicle number | GS5 | 丝氨酸羧肽酶 | 正调控粒宽和粒重 | [ | |
| HTH5 | 磷酸吡哆醛结合蛋白 | 正调控结实率 | [ | ||
| qHd1 | Squamosa启动子 | 正调控有效穗 | [ | ||
| LAX2-4 | 核蛋白 | 负调控有效穗和穗粒数 | [ | ||
| OsDCL3b | Dicer酶 | 正调控有效穗 | [ | ||
| 穗粒数 Spikelets per panicle | GNP1 | GA20氧化酶 | 正调控穗粒数 | [ | |
| OsKRN2 | WD40蛋白 | 负调控穗粒数和产量 | [ | ||
| An-1 | bHLH蛋白 | 正调控穗粒数 | [ | ||
| COG1 | 转录因子OsMADS17 | 正调控穗粒数和粒重 | [ | ||
| NOG1 | 烯酰辅酶A水合酶 | 正调控穗粒数 | [ | ||
| LF1 | 转录因子HD-ZIP Ⅲ | 正调控穗粒数 | [ | ||
| 千粒重 1000-grain weight | GS3 | Gγ亚基 | 负调控粒长和粒重 | [ | |
| GSN1 | 双特异性磷酸酶 | 负调控粒型和穗粒数 | [ | ||
| TGW2 | 细胞数调控因子OsCNR1 | 负调控粒宽和粒重 | [ | ||
| DG1 | 毒性物外排转运蛋白 | 正调控籽粒灌浆 | [ | ||
| GLW7 | 转录因子OsSPL13 | 正调控粒长和粒重 | [ | ||
| GAD1 | 短分泌信号肽 | 正调控粒长、穗粒数 | [ | ||
| GW5 | 钙调素结合蛋白 | 正调控粒宽和粒重 | [ | ||
| GLW10 | 油菜素内酯信号激酶 | 正调控粒长和粒重 | [ | ||
| 淀粉合成Starch biosynthesis | Chalk5 | 液泡膜质子焦磷酸酶 | 负调控垩白 | [ | |
| Wx | 淀粉颗粒合成酶 | 负调控直链淀粉 | [ | ||
| WCR1 | F-box蛋白 | 负调控垩白 | [ | ||
| SSIIIb | 淀粉颗粒合成酶 | 与ssIIIa协同调控抗性淀粉 | [ | ||
| OsLESV | 淀粉结合蛋白 | 正调控储藏淀粉 | [ | ||
| [1] | Ren D Y, Ding C Q, Qian Q. Molecular bases of rice grain size and quality for optimized productivity[J]. Science Bulletin, 2023, 68(3): 314-350. |
| [2] | 田志喜, 严长杰, 钱前, 严松, 谢会兰, 王芳, 徐洁芬, 刘贵富, 王永红, 刘巧泉, 汤述翥, 李家洋, 顾铭洪. 水稻淀粉合成相关基因分子标记的建立[J]. 科学通报, 2010, 55(26): 2591-2601. |
| Tian Z X, Yan C J, Qian Q, Yan S, Xie H L, Xu J F, Liu G F, Wang Y H, Liu Q Q, Tang S Z, Li J Y, Gu M H. Establishment of molecular markers of genes related to starch synthesis in rice[J]. Scientific Bulletin, 2010, 55(26): 2591-2601. (in Chinese with English abstract) | |
| [3] | Shi Q B, Xia Y, Xue N, Wang Q B, Tao Q, Li M J, Xu D, Wang X F, Kong F Y, Zhang H S, Li G. Modulation of starch synthesis in Arabidopsis via phytochrome B-mediated light signal transduction[J]. Journal of Integrative Plant Biology, 2024, 66(5): 973-985. |
| [4] | 张青, 孟杉杉, 陈梓春, 王远江, 韦存虚, 王娟. 谷物胚乳淀粉合成相关酶的调控机制研究进展[J]. 植物生理学报, 2021, 57(1): 1-11. |
| Zhang Q, Meng S S, Chen Z C, Wang Y J, Wei C X, Wang J. Research progress on the regulatory mechanisms of starch synthesis related enzymes in grain endosperm[J]. Journal of Plant Physiology, 2021, 57(1): 1-11. (in Chinese with English abstract) | |
| [5] | Preiss J, Ball K, Hutney J, Smith-White B, Li L, Okita T W. Regulatory mechanisms involved in the biosynthesis of starch[J]. Pure and Applied Chemistry, 1991, 63(4): 535-544. |
| [6] | Brown I. Complex carbohydrates and resistant starch[J]. Nutrition Reviews, 1996, 54(11): S115. |
| [7] | Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y. Function and characterization of starch synthase I using mutants in rice[J]. Plant Physiology, 2006, 140(3): 1070-1084. |
| [8] | Commuri P D, Keeling P L. Chain-length specificities of maize starch synthase I enzyme: Studies of glucan affinity and catalytic properties[J]. The Plant Journal, 2001, 25(5): 475-486. |
| [9] | Umemoto T, Aoki N. Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme[J]. Functional Plant Biology, 2005, 32(9): 763-768. |
| [10] | Ohdan T, Francisco P B, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice[J]. Journal of Experimental Botany, 2005, 56(422): 3229-3244. |
| [11] | Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park J H, Jane J L, Miyao A, Hirochika H, Nakamura Y. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm[J]. Plant Physiology, 2007, 144(4): 2009-2023. |
| [12] | Seung D, Lu K J, Stettler M, Streb S, Zeeman S C. Degradation of glucan primers in the absence of starch synthase 4 disrupts starch granule initiation in Arabidopsis[J]. Journal of Biological Chemistry, 2016, 291(39): 20718-20728. |
| [13] | Szydlowski N, Ragel P, Raynaud S, Lucas M M, Roldán I, Montero M, Muñoz F J, Ovecka M, Bahaji A, Planchot V, Romero J P, D'Hulst C, Mérida Á. Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases[J]. Plant Cell, 2009, 21(8): 2443-2457. |
| [14] | Roldán I, Wattebled F, Lucas M M, Delvallé D, Planchot V, Jiménez S, Pérez R, Ball S, D'hulst C, Mérida Á. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation[J]. Plant Journal, 2007, 49(3): 492-504. |
| [15] | Zhang G, Cheng Z, Zhang X, Guo X, Su N, Jiang L, Mao L, Wan J. Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L.) uncovers interactive effects on the physicochemical properties of starch[J]. Genome, 2011, 54(6): 448-459. |
| [16] | Toyosawa Y, Kawagoe Y, Matsushima R, Crofts N, Ogawa M, Fukuda M, Kumamaru T, Okazaki Y, Kusano M, Saito K, Toyooka K, Sato M, Ai Y, Jane J L, Nakamura Y, Fujita N. Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm[J]. Plant Physiology, 2016, 170(3): 1255-1270. |
| [17] | Bao J S, Corke H, Sun M. Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.)[J]. Theoretical and Applied Genetics, 2006, 113: 1171-1183. |
| [18] | Nakamura Y, Aihara S, Crofts N, Sawada T, Fujita N. In viro studies of enzymatic properties of starch synthases and interactions between starch synthase I and starch branching enzymes from rice[J]. Plant Science, 2014, 224: 1-8. |
| [19] | Gurunathan S, Ramadoss B R, Mudili V, Siddaiah C, Kalagatur N K, Bapul J R K, Alqarawi A A, Hashem A, Allah E F A. Single nucleotide polymorphisms in starch biosynthetic genes associated with increased resistant starch concentration in rice mutant[J]. Frontiers in Genetics, 2019, 10: 946. |
| [20] | Pang Y H, Zhou X, Chen Y L, Bao J S. Comparative phosphoproteomic analysis of the developing seeds in two indica rice (Oryza sativa L.) cultivars with different starch quality[J]. Journal of Agricultural and Food Chemistry, 2018, 66(11): 3030-3037. |
| [21] | 贺晓鹏, 朱昌兰, 刘玲珑, 王方, 傅军如, 江玲, 张文伟, 刘宜柏, 万建民. 不同水稻品种支链淀粉结构的差异及其与淀粉理化特性的关系[J]. 作物学报, 2010, 36(2): 276-284. |
| He X P, Zhu C L, Liu L L, He X P, Zhu C L, Liu L L, Wang F, Fu J R, Jiang L, Zhang W W, Liu Y B, Wan J M. Difference of amylopectin structure among various rice genotypes differing in grain qualities and its relation to starch physicochemical properties[J]. Acta Agronomica Sinica, 2010, 36(2): 276-284. (in Chinese with English abstract). | |
| [22] | Sawada T, Francisco P B J, Aihara S, Utsumi Y, Yoshida M, Oyama Y, Tsuzuki M, Satoh H, Nakamura Y. Chlorella starch branching enzyme II (BEII) can complement the function of BEIIb in rice endosperm[J]. Plant and Cell Physiology, 2009, 50: 1062-1074. |
| [23] | Qu J Z, Xu S T, Zhang Z Q, Chen G Z, Zhong Y Y, Liu L S, Zhang R H, Xue J Q, Guo D W. Evolutionary, structural and expression analysis of core genes involved in starch synthesis[J]. Scientific Reports, 2018, 8(1): 12736. |
| [24] | Abe N, Asai H, Yago H, Oitome N F, Itoh R, Crofts N, Nakamura Y, Fujita N. Relationships between starch synthase I and branching enzyme isozymes determined using double mutant rice lines[J]. BMC Plant Biology, 2014, 14: 1-12. |
| [25] | Jiang H, Dian W, Wu P. Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme[J]. Phytochemistry, 2003, 63(1): 53-59. |
| [26] | Smith A M, Denyer K, Martin C R. What controls the amount and structure of starch in storage organs?[J]. Plant Physiology, 1995, 107(3): 673-677. |
| [27] | Wei K S, Cheng F M, Zhang Q F, Liu K G. Temperature stress at grain filling stage mediates expression of three isoform genes encoding starch branching enzymes in rice endosperm[J]. Rice Science, 2009, 16(3): 187-193. |
| [28] | Utsumi Y, Utsumi C, Sawada T, Fujita N, Nakamura Y. Functional diversity of isoamylase oligomers: The ISA1 homo-oligomer is essential for amylopectin biosynthesis in rice endosperm[J]. Plant Physiology, 2011, 156(1): 61-77. |
| [29] | Utsumi Y, Nakamura Y. Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the isoamylase1-isoamylase2 hetero-oligomer from rice endosperm[J]. Planta, 2006, 225: 75-87. |
| [30] | Goren A, Ashlock D, Tetlow I J. Starch formation inside plastids of higher plants[J]. Protoplasma, 2018, 255: 1855-1876. |
| [31] | Zeeman S C, Kossmann J, Smith A M. Starch: its metabolism, evolution, and biotechnological modification in plants[J]. Annual Review of Plant Biology, 2010, 61(1): 209-234. |
| [32] | Li Q F, Zhang G Y, Dong Z W, Yu H X, Gu M H, Sun S S M, Liu Q Q. Characterization of expression of the OsPUL gene encoding a pullulanase-type debranching enzyme during seed development and germination in rice[J]. Plant Physiology and Biochemistry, 2009, 47(5): 351-358. |
| [33] | Fujita N, Toyosawa Y, Utsumi Y, Higuchi T, Hanashiro I, Ikegami A, Akuzawa S, Yoshida M, Mori A, Inomata K, Itoh R, Miyao A, Hirochika H, Satoh H, Nakamura Y. Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm[J]. Journal of Experimental Botany, 2009, 60(3): 1009-1023. |
| [34] | Takaha T, Critchley J, Okada S, Smith S M. Normal starch content and composition in tubers of antisense potato plants lacking D-enzyme (4-α-glucanotransferase)[J]. Planta, 1998, 205: 445-451. |
| [35] | Seung D, Boudet J, Monroe J, Schreier T B, David L C, Abt M, Lu K J, Zanella M, Zeeman S C. Homologs of PROTEIN TARGETING TO STARCH control starch granule initiation in Arabidopsis leaves[J]. The Plant Cell, 2017, 29(7): 1657-1677. |
| [36] | Yan H G, Zhang W W, Wang Y H, Jin J, Xu H C, Fu Y S, Shan Z Z, Wang X, Teng X, Li X, Wang Y X, Hu X Q, Zhang W X, Zhu C Y, Zhang X, Zhang Y, Wang R Q, Zhang J, Cai Y, You X M, Chen J, Ge X Y, Wang L, Xu J H, Jiang L, Liu S J, Lei C L, Zhang X, Wang H Y, Ren YL, Wan J M. Rice LIKE EARLY STARVATION1 cooperates with FLOURY ENDOSPERM6 to modulate starch biosynthesis and endosperm development[J]. The Plant Cell, 2024, 36(5): 1892-1912. |
| [37] | Wang W, Wei X J, Gao AJ, Chen W Q, Wu Y W, Sheng Z H, Hu S K, Xie L H, Wang J Y, Tang S Q, Hu P S. GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield[J]. Journal of Integrative Plant Biology, 2020, 62(7): 948-966. |
| [38] | Anacleto R, Badoni S, Parween S, Butardo V M, Misra G, Cuevas R P, Kuhlmann M, Trinidad T P, Mallillin A C, Acuin C, Bird A R, Morell M K, Sreenivasulu N. Integrating a genome-wide association study with a large-scale transcriptome analysis to predict genetic regions influencing the glycaemic index and texture in rice[J]. Plant Biotechnology Journal, 2019, 17(7): 1261-1275. |
| [39] | Igarashi H, Ito H, Shimada T, Kang D J, Hamada S. A novel rice dull gene, LowAC1, encodes an RNA recognition motif protein affecting Waxyb pre-mRNA splicing[J]. Plant Physiology and Biochemistry, 2021, 162: 100-109. |
| [40] | Huang L, Tan H, Zhang C, Li Q F, Liu Q Q. Starch biosynthesis in cereal endosperms: An updated review over the last decade[J]. Plant Communications, 2021, 2(5): 100237. |
| [41] | Jin S K, Xu L N, Leng Y J, Yang Q Q, Wang S L, Jia S W, Song T, Wang R A, Tao T, Liu Q Q, Cai X L, Gao J P. The OsNAC24-OsNAP protein complex activates OsGBSSI and OsSBEI expression to fine-tune starch biosynthesis in rice endosperm[J]. Plant Biotechnology Journal, 2023, 21(11): 2224-2240. |
| [42] | 吴云飞, 张勇, 王磊磊, 余徐润, 熊飞. 水稻籽粒淀粉品质的影响因素及其机制研究进展[J]. 中国农学报, 2021, 37(6): 1-8. |
| Wu Y F, Zhang Y, Wang L L, Yu X R, Xiong F. Research progress on influencing factors and mechanisms of grain starch quality in rice[J]. Chinese Journal of Agronomy, 2021, 37(6): 1-8. (in Chinese with English abstract) | |
| [43] | Liu Z, Jiang S, Jiang L, Li W, Tang Y, He W, Wang M, Xing J, Cui Y, Lin Q, Yu F, Wang L. Transcription factor OsSGL is a regulator of starch synthesis and grain quality in rice[J]. Journal of Experimental Botany, 2022, 73(11): 3417-3430. |
| [44] | Zhu Y, Cai X L, Wang Z Y, Hong M M. An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene[J]. Journal of Biological Chemistry, 2003, 278(48): 47803-47811. |
| [45] | Wang J C, Xu H, Zhu Y, Liu Q, Cai X L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm[J]. Journal of Experimental Botany, 2013, 64(11): 3453-3466. |
| [46] | Cai Y, Xie D, Wang Z, Hong M M. Interaction of rice bZIP protein REB with the 5′-upstream region of both rice sbe1 gene and waxy gene[J]. Chinese Science Bulletin, 2002, 47: 310-314. |
| [47] | Feng T, Wang L, Li L, Liu Y, Chong K, Theißen G, Meng Z. OsMADS14 and NF-YB1 cooperate in the direct activation of OsAGPL2 and Waxy during starch synthesis in rice endosperm[J]. New Phytologist, 2022, 234(1): 77-92. |
| [48] | Zhang H, Xu H, Feng M, Zhu Y. Suppression of OsMADS7 in rice endosperm stabilizes amylose content under high temperature stress[J]. Plant Biotechnology Journal, 2018, 16(1): 18-26. |
| [49] | Cao R, Zhao S, Jiao G, Duan Y, Ma L, Dong N, Lu F, Zhu M, Shao G, Hu S, Sheng Z, Zhang J, Tang S, Wei X, Hu P. OPAQUE3, encoding a transmembrane bZIP transcription factor, regulates endosperm storage protein and starch biosynthesis in rice[J]. Plant Communications, 2022, 3(6): 100463. |
| [50] | Wang J, Chen Z, Zhang Q, Meng S, Wei C. The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis[J]. Plant Physiology, 2020, 184(4): 1775-1791. |
| [51] | Wang J, Zhang H, Wang Y, Meng S, Liu Q, Li Q, Zhao Z, Liu Q, Wei C. Regulatory loops between rice transcription factors OsNAC25 and OsNAC20/26 balance starch synthesis[J]. Plant Physiology, 2024, 195(2): 1365-1381. |
| [52] | Bebllo K, Hou Y, Zhao J, Jiao G A, Wu Y W, Li Z Y, Wang Y F, Tong X H, Wang W, Yuan W Y, Wei X J, Zhang J. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.)[J]. Plant Biotechnology Journal, 2019, 17(7): 1222-1235. |
| [53] | Geigenberger P. Regulation of starch biosynthesis in response to a fluctuating environment[J]. Plant Physiology, 2011, 155(4): 1566-1577. |
| [54] | Crofts N, Abe N, Oitome N F, Matsushima R, Hayashi M, Tetlow I J, Emes M J, Nakamura Y, Fujita N. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes[J]. Journal of Experimental Botany, 2015, 66(15): 4469-4482. |
| [55] | Schmidt R, Schippers J H M, Mieulet D, Watanabe M, Hoefgen R, Guiderdoni E, Roeber B M. SALT-RESPONSIVE ERF1 is a negative regulator of grain filling and gibberellin-mediated seedling establishment in rice[J]. Molecular Plant, 2014, 7(2): 404-421. |
| [56] | Honma Y, Adhikari P B, Kuwata K, Kagenishi T, Yokawa K, Notaguchi M, Kurotani K, Toda E, Uehara K B, Liu X, Zhu S, Wu X, Kasahara R D. High-quality sugar production by osgcs1 rice[J]. Communications Biology, 2020, 3(1): 617. |
| [57] | Fu F F, Xue H W. Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator[J]. Plant Physiology, 2010, 154(2): 927-938. |
| [58] | Pang Y H, Hu Y, Bao J S. Comparative phosphoproteomic analysis reveals the response of starch metabolism to high-temperature stress in rice endosperm[J]. International Journal of Molecular Sciences, 2021, 22(19): 10546. |
| [59] | Ying Y, Pang Y, Bao J. Comparative ubiquitome analysis reveals diverse functions of ubiquitination in rice seed development under high-temperature stress[J]. Seed Biology, 2023, 2(1): 69. |
| [60] | Meng X, Mujahid H, Zhang Y, Peng X, Redoña D E, Wang C, Peng Z. Comprehensive analysis of the lysine succinylome and protein co-modifications in developing rice seeds[J]. Molecular & Cellular Proteomics, 2019, 18(12): 2359-2372. |
| [61] | Nakamura Y, Ono M, Utsumi C, Steup M. Functional interaction between plastidial starch phosphorylase and starch branching enzymes from rice during the synthesis of branched maltodextrins[J]. Plant and Cell Physiology, 2012, 53(5): 869-878. |
| [62] | Chen Y, Bao J. Underlying mechanisms of zymographic diversity in starch synthase I and pullulanase in rice-developing endosperm[J]. Journal of Agricultural and Food Chemistry, 2016, 64(9): 2030-2037. |
| [63] | Hwang S K, Koper K, Satoh H, Okital T W. Rice endosperm starch phosphorylase (Pho1) assembles with disproportionating enzyme (Dpe1) to form a protein complex that enhances synthesis of malto-oligosaccharides[J]. Journal of Biological Chemistry, 2016, 291(38): 19994-20007. |
| [64] | Hwang S K, Koper K, Okita T W. The plastid phosphorylase as a multiple-role player in plant metabolism[J]. Plant Science, 2020, 290: 110303. |
| [65] | Hwang S K, Singh S, Cakir B, Satoh H, Okita T W. The plastidial starch phosphorylase from rice endosperm: Catalytic properties at low temperature[J]. Planta, 2016, 243: 999-1009. |
| [66] | Chao S F, Cai Y C, Feng B B, Jiao G A, Sheng Z H, Luo J, Tang S Q, Wang J L, Hu P S, Wei X J. Editing of rice isoamylase gene ISA1 provides insights into its function in starch formation[J]. Rice Science, 2019, 26(2): 77-87. |
| [67] | Dong N, Jiao G, Cao R, Li S F, Zhao S L, Duan Y Q, Ma L Y, Li X W, Lu F F, Wang H, Wang S W, Shao G E, Sheng Z H, Hu S K, Tang S Q, Wei X J, Hu P S. OsLESV and OsESV1 promote transitory and storage starch biosynthesis to determine rice grain quality and yield[J]. Plant Communications, 2024, 5(7): 100893. |
| [68] | Tian Z, Qian Q, Liu Q, Yan M, Liu X, Yan C, Liu G, Gao Z, Tang S, Zeng D, Wang Y, Yu J, Gu M, Li J. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities[J]. Proceedings of the National Academy of Sciences of the Uniced States, 2009, 106(51): 21760-21765. |
| [69] | Gong D, Zhang X, He F, Chen Y, Li R, Yao J P, Zhang M L, Zheng W J, Yu G X. Genetic improvements in rice grain quality: A review of elite genes and their applications in molecular breeding[J]. Agronomy, 2023, 13(5): 1375. |
| [70] | Inukai T, Sako A, Hirano H Y, Sano Y. Analysis of intragenic recombination at wx in rice: Correlation between the molecular and genetic maps within the locus[J]. Genome, 2000, 43(4): 589-596. |
| [71] | Bligh H F J, Larkin P D, Roach P S, Jones C A, Fu H, Park W D. Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties[J]. Plant Molecular Biology, 1998, 38: 407-415. |
| [72] | Larkin P D, Park W D. Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.)[J]. Molecular Breeding, 2003, 12: 335-339. |
| [73] | Bergman C J, Delgado J T, McClung A M, Fjellstrom R G. An improved method for using a microsatellite in the rice waxy gene to determine amylose class[J]. Cereal Chemistry, 2001, 78(3): 257-260. |
| [74] | Liu L, Ma X, Liu S, Zhu C, Jiang L, Wang Y, Shen Y, Ren Y, Dong H, Chen L, Liu X, Zhao Z, Zhai H, Wan J. Identification and characterization of a novel Waxy allele from a Yunnan rice landrace[J]. Plant Molecular Biology, 2009, 71: 609-626. |
| [75] | Sato H, Suzuki Y, Sakai M, Imbe T. Molecular characterization of Wxmq, a novel mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.)[J]. Breeding Science, 2002, 52(2): 131-135. |
| [76] | Lin L, Li Z, Ning M, Zou Z H, Zhang L, Liu Q Q, Wei C X. A mutant allele of the Wx gene encoding granule-bound starch synthase I results in extremely low amylose content in rice[J]. Plant Physiology, 2024: 196(4): 2296-2299. |
| [77] | Cai Y, Zhang W, Fu Y, Shan Z Z, Xu J H, Wang P, Kong F, Jin J, Yan H G, Ge X Y, Wang Y X, You X M, Chen J, Li X, Chen W W, Chen X G, Ma J, Tang X J, Zhang J, Bao Y Q, Jiang L, Wang H Y, Wan J M. Du13 encodes a C2H2 zinc-finger protein that regulates Wxb pre-mRNA splicing and microRNA biogenesis in rice endosperm[J]. Plant Biotechnology Journal, 2022, 20(7): 1387-1401. |
| [78] | Yao S, Zhang Y D, Lu K, Wang C L. Progress in functions, allelic variations and interactions of soluble starch synthases genes SSⅡa and SSⅢa in rice[J]. Chinese Journal of Rice Science, 2022, 36(3): 227. |
| [79] | Yu S, Ma Y, Menager L, Sun D W. Physicochemical properties of starch and flour from different rice cultivars[J]. Food and Bioprocess Technology, 2012, 5: 626-637. |
| [80] | Wang E, Wang J, Zhu X, Hao W, Wang L Y, Li Q, Zhang L X, He W, Lu B R, Lin H X, Ma H, Zhang G Q, He Z H. Control of rice grain-filling and yield by a gene with a potential signature of domestication[J]. Nature Genetics, 2008, 40(11): 1370-1374. |
| [81] | Wei X, Jiao G, Lin H, Sheng Z H, Shao G N, Xie L H, Tang S Q, Xu Q G, Hu P S. GRAIN INCOMPLETE FILLING2 regulates grain filling and starch synthesis during rice caryopsis development[J]. Journal of Integrative Plant Biology, 2017, 59(2): 134-153. |
| [82] | Ma B, Zhang L, Gao Q, Wang J M, Li X Y, Wang H, Liu Y, Lin H, Liu J Y, Wang X, Li Q, Deng Y W, Tang W H, Luan S, He Z H. A plasma membrane transporter coordinates phosphate reallocation and grain filling in cereals[J]. Nature Genetics, 2021, 53(6): 906-915. |
| [83] | Dong N N, Chen L N, Ahmad S, Cai YC, Duan Y Q, Li X W, Liu Y Q, Jiao G A, Xie L H, Hu S K, Shrng Z H, Shao G N, Wang L, Tang S Q, Wei X J, Hu P S. Genome-wide analysis and functional characterization of pyruvate kinase (PK) gene family modulating rice yield and quality[J]. International Journal of Molecular Sciences, 2022, 23(23): 15357. |
| [84] | Cai Y, Li S, Jiao G, Sheng Z H, Wu Y W, Shao G N, Xie L H, Peng C, Xu J F, Tang S Q, Wei X J, Hu P S. OsPK2 encodes a plastidic pyruvate kinase involved in rice endosperm starch synthesis, compound granule formation and grain filling[J]. Plant Biotechnology Journal, 2018, 16(11): 1878-1891. |
| [85] | Wang S, Wu K, Yuan Q, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D. Control of grain size, shape and quality by OsSPL16 in rice[J]. Nature Genetics, 2012, 44(8): 950-954. |
| [86] | Song X J, Huang W, Shi M, Zhu M Z, Lin H X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase[J]. Nature Genetics, 2007, 39(5): 623-630. |
| [87] | Chen K, Łyskowski A, Jaremko Ł, Jaremko M. Genetic and molecular factors determining grain weight in rice[J]. Frontiers in Plant Science, 2021, 12: 605799. |
| [88] | Huang X Z, Qian Q, Liu Z B, Sun H Y, He S Y, Luo D, Xia G M, Chu C C, Li J Y, Fu X D. Natural variation at the DEP1 locus enhances grain yield in rice[J]. Nature Genetics, 2009, 41(4): 494-497. |
| [89] | Wang L, Wang D, Yang Z, Jiang S, Qu J N, He W, Liu Z M, Xing J J, Ma Y C, Lin Q L, Yu F. Roles of FERONIA-like receptor genes in regulating grain size and quality in rice[J]. Science China Life Sciences, 2021, 64: 294-310. |
| [90] | Liu E, Zeng S, Zhu S, Liu Y, Wu G C, Zhao K M, Liu X L, Liu Q M, Dong Z Y, Dang X J, Xiu H, Li D L, Hu X X, Hong D L. Favorable alleles of GRAIN-FILLING RATE1 increase the grain-filling rate and yield of rice[J]. Plant Physiology, 2019, 181(3): 1207-1222. |
| [91] | Chen J Y, Liu S L, Siao W, Wang S J. Hormone and sugar effects on rice sucrose transporter OsSUT1 expression in germinating embryos[J]. Acta Physiologiae Plantarum, 2010, 32: 749-756. |
| [92] | 张雅文, 包淑慧, 唐振家, 王小文, 杨芳, 张德春, 胡一兵. 蔗糖转运蛋白OsSUT5在水稻花粉发育及结实中的作用[J]. 中国农业科学, 2021, 54(16): 3369-3385. |
| Zhang Y W, Bao S H, Tang Z J, Wang X W, Yang F, Zhang D C, Hu Y B. Effects of sucrose transporter OsSUT5 on pollen development and fruiting in rice[J]. Chinese Journal of Agricultural Sciences, 2021, 54(16): 3369-3385. (in Chinese with English abstract) | |
| [93] | Scofield G N, Hirose T, Aoki N, Furbank R T. Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice[J]. Journal of Experimental Botany, 2007, 58(12): 3155-3169. |
| [94] | Chung P, Hsiao H H, Chen H J, Chang C W, Wang S J. Influence of temperature on the expression of the rice sucrose transporter 4 gene, OsSUT4, in germinating embryos and maturing pollen[J]. Acta Physiologiae Plantarum, 2014, 36: 217-229. |
| [95] | Hu Z, Tang Z J, Zhang Y M, Niu L P, Yang F, Zhang D C, Hu Y B. Rice SUT and SWEET transporters[J]. International Journal of Molecular Sciences, 2021, 22(20): 11198. |
| [96] | Li P, Wang L H, Liu H B, Yuan M. Impaired SWEET-mediated sugar transportation impacts starch metabolism in developing rice seeds[J]. The Crop Journal, 2022, 10(1): 98-108. |
| [97] | Ma L, Zhang D, Miao Q, Yang J, Xuan Y H, HuY B. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling[J]. Plant and Cell Physiology, 2017, 58(5): 863-873. |
| [98] | Yang J, Luo D, Yang B, Frommer W B, Eom J S. SWEET 11 and 15 as key players in seed filling in rice[J]. New Phytologist, 2018, 218(2): 604-615. |
| [99] | Sun C, Wang Y, Yang X, Tang L, Wan C M, Liu J Q, Chen C P, Zhang H S, He C C, Liu C Q, Wang Q, Zhang K, Zhang W F, Yang B, Li S C, Zhu J, Sun Y J, Li W T, Zhou W H, Wang P R, Deng X J. MATE transporter GFD1 cooperates with sugar transporters, mediates carbohydrate partitioning and controls grain‐filling duration, grain size and number in rice[J]. Plant Biotechnology Journal, 2023, 21(3): 621-634. |
| [100] | Durand M, Mainson D, Porchrion B, Maurousset L, Lemoine R, Pourtau N. Carbon source-sink relationship in Arabidopsis thaliana: The role of sucrose transporters[J]. Planta, 2018, 247: 587-611. |
| [101] | Li S F, Wei X J, Ren Y, Qiu J H, Jiao G A, Guo X P, Tang S Q, Wan J M, Hu P S. OsBT1 encodes an ADP-glucose transporter involved in starch synthesis and compound granule formation in rice endosperm[J]. Scientific Reports, 2017, 7(1): 40124. |
| [102] | Liu D R, Xu L N, Wang W, Jia S W, Jin S K, Gao J P. OsRRM, an RNA-binding protein, modulates sugar transport in rice (Oryza sativa L.)[J]. Frontiers in Plant Science, 2020, 11: 605276. |
| [103] | Chen S Y, Wang Z Y, Cai X L. OsRRM, a Spen-like rice gene expressed specifically in the endosperm[J]. Cell Research, 2007, 17(8): 713-721. |
| [104] | Zhu X, Liang W, Cui X, Chen M J, Yin C S, Luo Z J, Zhu J Y, Lucas W J, Wang Z Y, Zhang D B. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein[J]. The Plant Journal, 2015, 82(4): 570-581. |
| [105] | Zhang H, Liang W, Yang X, Luo X, Jiang N, Ma H, Zhang D B. Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development[J]. The Plant Cell, 2010, 22(10): 672-689. |
| [106] | She K C, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M, Tsuge T, Matsumoto K, Kudoh M, Itoh E, Kikuchi S, Kisgimoto N, Yazaki J, Ando T, Yano M, Aoyama T, Sasaki T, Satoh H, Shimada H. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality[J]. The Plant Cell, 2010, 22(10): 3280-3294. |
| [107] | Zhang L, Zhao L, Lin L, Zhao L X, Liu Q Q, Wei C X. A novel mutation of OsPPDKB, encoding pyruvate orthophosphate dikinase, affects metabolism and structure of starch in the rice endosperm[J]. International Journal of Molecular Sciences, 2018, 19(8): 2268. |
| [108] | Zhang L, Ren Y, Lu B, Yang C Y, Feng Z M, Liu Z, Chen J, Ma W W, Wang Y, Yu X W, Wang Y L, Zhang W W, Wang Y H, Liu S J, Wu F Q, Zhang X, Guo X P, Bao Y Q, Jiang L, Wan J M. FLOURY ENDOSPERM7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice[J]. Journal of Experimental Botany, 2016, 67(3): 633-647. |
| [109] | Cui Z, Wang X, Dai Y, Sharma S, Yun Q, Li Q, E Z, Chen Chen C. Transcription factor OsNF-YC1 regulates grain size by coordinating the transcriptional activation of OsMADS1 in Oryza sativa L[J]. The Plant Journal, 2024, 119(3): 1465-1480. |
| [110] | Khanday I, Yadav S R, Vijayraghavan U. Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways[J]. Plant Physiology, 2013, 161(4): 1970-1983. |
| [111] | Zhang J, Nallmilli B R, Mujahid H, Peng Z H. OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice(Oryza sativa)[J]. The Plant Journal, 2010, 64(4): 604-617. |
| [112] | Yang X, Wu F, Lin X, Du X Q, Chong K, Gramzow L D, Schilling S S, Becker A, Theißen G, Meng Z. Live and let die-the Bsister MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa)[J]. The Public Library of Science Biology, 2012, 7(12): e51435. |
| [113] | Ren Y, Huang Z, Jiang H, Wang Z, Wu F S, Xiong Y, Yao J L. A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling[J]. Journal of Experimental Botany, 2021, 72(8): 2947-2964. |
| [114] | Xiong Y, Ren Y, Li W, Wu F S, Yang W J, Huang X L, Yao J L. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice[J]. Journal of Experimental Botany, 2019, 70(15): 3765-3780. |
| [115] | Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice[J]. Proceedings of the National Academy of Sciences, 2010, 107(45): 19579-19584. |
| [116] | Li Y, Fan C, Xing Y, Zhang Q. Natural variation in GS5 plays an important role in regulating grain size and yield in rice[J]. Nature Genetics, 2011, 43(12): 1266-1269. |
| [117] | Cao Z, Tang H, Cai Y, Zeng B, Zhao J, Tang X, Lu M, Wang H, Zhu X, Wu X, Yuan L, Wan J. Natural variation of HTH5 from wild rice, Oryza rufipogon Griff., is involved in conferring high-temperature tolerance at the heading stage[J]. Plant Biotechnology Journal, 2022, 20(8): 1591-1605. |
| [118] | Chen J Y, Zhang H W, Zhang H L, Ying J, Ma L, Zhuang J Y. Natural variation at qHd1 affects heading date acceleration at high temperatures with pleiotropism for yield traits in rice[J]. BMC Plant Biology, 2018, 18: 1-11. |
| [119] | Dai D, Chen J, Du C, Liang M, Wu M, Mou T, Zhang H, Ma L. A2-MB chromosome inversion interrupted transcription of lax2-4 and generated pleiotropic phenotypes in rice[J]. Journal of Plant Growth Regulation, 2022, 41(6): 2328-2337. |
| [120] | Liao P F, Ouyang J X, Zhang J J, Yang L, Wang X, Peng X J, Wang D, Zhu Y L, Li S B. OsDCL3b affects grain yield and quality in rice[J]. Plant Molecular Biology, 2019, 99: 193-204. |
| [121] | Wang Y, Zhai L, Chen K, Shen C, Liang Y, Wang C, Zhao X, Shu W, Xu J. Natural sequence variations and combinations of GNP1 and NAL1 determine the grain number per panicle in rice[J]. Rice, 2020, 13: 1-15. |
| [122] | Chen W, Chen L, Zhang X, Yang N, Guo J H, Wang M, Ji S H, Zhao X Y, Yin P F, Cai L C, Xu J, Zhang L L, Han Y J, Xiao Y N, Xu G L, Wang Y B, Wang S H, Wu S, Yang F, Jackson D, Cheng J K, Chen S H, Sun C Q, Qin F, Tian F, Fernie A R, Li J S, Yan J B, YangX H. Convergent selection of a WD40 protein that enhances grain yield in maize and rice[J]. Science, 2022, 375(6587): eabg7985. |
| [123] | Luo J, Liu H, Zhou T, Gu B G, Huang X H, ShangY Y, Zhu J J, Li Y, Zhao Y, Wang Y C, Zhao Q, Wang A H, Wang Z Q, Su T, Wang Z X, Han B. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice[J]. The Plant Cell, 2013, 25(9): 3360-3376. |
| [124] | Li Y, Wu S, Huang Y, Ma X, Tan L, Liu F, Lv Q, Zhu Z, Hu M, Fu Y, Zhang K, Gu P, Xie D, Sun H, Sun C. OsMADS17 simultaneously increases grain number and grain weight in rice[J]. Nature Communications, 2023, 14(1): 3098. |
| [125] | Huo X, Wu S, Zhu Z, Liu F, Fu Y, Cai H, Sun X, Gu P, Xie D, Tan L, Sun C. NOG1 increases grain production in rice[J]. Nature Communications, 2017, 8(1): 1497. |
| [126] | Zhang T, You J, Zhang Y, Yao W, Chen W, Duan Q, Xiao W, Ye L, Zhou Y, X S, Ling Y, He G, Li Y. LF1 regulates the lateral organs polarity development in rice[J]. New Phytologist, 2021, 231(3): 1265-1277. |
| [127] | Sun S, Wang L, Mao H, Shao L, Li X, Xiao J, Ouyang Y, Zhang Q. A G-protein pathway determines grain size in rice[J]. Nature Communications, 2018, 9(1): 851. |
| [128] | Guo T, Chen K, Dong N Q, Shi C L, Ye W W, Gao J P, Shan J X, Lin H X. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice[J]. The Plant Cell, 2018, 30(4): 871-888. |
| [129] | Ruan B, Shang L, Zhang B, Hu J, Wang Y, Lin H, Zhang A, Liu C, Peng Y, Zhu L, Ren D, Shen L, Dong G, Zhang G, Zeng D, Guo L, Qian Q, Gao Z. Natural variation in the promoter of TGW2 determines grain width and weight in rice[J]. New Phytologist, 2020, 227(2): 629-640. |
| [130] | Qin P, Zhang G, Hu B, Wu J, Chen W L, Ren Z J, Liu Y L, Xie J T, Yuan H, Tu B, Ma B, Wang Y P, Ye L M, Li L G, Xiang C B, Li S G. Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism[J]. Science Advances, 2021, 7(3): eabc8873. |
| [131] | Yang Q, Zhu W, Tang X, Wu Y C, Liu G Q, Zhao D S, Liu Q Q, Zhang Y, Zhang T. Improving rice grain shape through upstream open reading frame editing-mediated translation regulation[J]. Plant Physiology, 2024: kiae557. |
| [132] | Jin J, Hua L, Zhu Z, Tan L, Zhao X, Zhang W, Liu F, Fu Y, Cai H, Sun X, Gu P, Xie D, Sun C. GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication[J]. The Plant Cell, 2016, 28(10): 2453-2463. |
| [133] | Liu J, Chen J, Zheng X, Wu F Q, Lin Q, Heng Y Q, Tian P, Cheng Z J, Yu X W, Zhou K N, Zhang X, Guo X P, Wang J L, Wang H Y, Wan J M. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice[J]. Nature Plants, 2017, 3(5): 1-7. |
| [134] | Yuan H, Xu Z, Chen W, Deng C Y, Liu Y, Yuan M, Gao P, Shi H, Tu B, Li T, Kang L Z, Ma B, Wang Y P, Wang J, Chen X W, Li S G, Qin P. OsBSK2, a putative brassinosteroid-signalling kinase, positively controls grain size in rice[J]. Journal of Experimental Botany, 2022, 73(16): 5529-5542. |
| [135] | Li Y, Fan C, Xing Y, Peng Y, Luo L J, Bao Y, Peng B, Xie W B, Wang G W, Li X H, Xiao J H, Xu C G, He Y Q. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice[J]. Nature Genetics, 2014, 46(4): 398-404. |
| [136] | Wu B, Yun P, Zhou H, Xia D, Gu Y C, Li P B, Yao J L, Zhou Z Q, Chen J X, Liu R J, Cheng S Y, Zhang H, Zheng Y Y, Lou G M, Chen P L, Wan S S, Zhou M S, Li Y H, Gao G J, Zhang Q L, Li X H, Lian X M, He Y Q. Natural variation in WHITE-CORE RATE 1 regulates redox homeostasis in rice endosperm to affect grain quality[J]. The Plant Cell, 2022, 34(5): 1912-1932. |
| [137] | Wang A, Jing Y, Cheng Q, Zhou H J, Wang L J, Gong W X, Kou L Q, Liu G F, Meng X B, Chen M J, Ma H Y, Shu X L, Yu H, Wu D X, Li J Y. Loss of function of SSIIIa and SSIIIb coordinately confers high RS content in cooked rice[J]. Proceedings of the National Academy of Sciences, 2023, 120(19): e2220622120. |
| [138] | Wu Y, Zhao Y, Yu J, Wu C C, Wang Q, Liu X Y, Gao X H, Wu K, Fu X D, Liu Q. Heterotrimeric G protein γ subunit DEP1synergistically regulates grain quality and yield by modulating the TTP (TON1-TRM-PP2A) complex in rice[J]. Journal of Genetics and Genomics, 2023, 50(7): 528-531. |
| [139] | Fang L, Ma L, Zhao S, Cao R J, Jiao G A, Hu P S, Wei X J. Alanine aminotransferase (OsAlaAT1) modulates nitrogen utilization, grain yield, and quality in rice[J]. Journal of Genetics and Genomics, 2022, 49(5):510-513. |
| [140] | Zhai L, Yan A, Shao K, Wang S, Wang Y, Chen Z H, Xu J L. Large Vascular Bundle Phloem Area4 enhances grain yield and quality in rice via source-sink-flow[J]. Plant Physiology, 2023, 191(1): 317-334. |
| [141] | Wang Z, Wei K, Xiong M, Wang J D, Zhang C Q, Fan X L, Huang L C, Zhao D S, Liu Q Q, Li Q F. Glucan, Water-Dikinase1(GWD1), an ideal biotechnological target for potential improving yield and quality in rice[J]. Plant Biotechnology Journal, 2021, 19(12): 2606-2618. |
| [142] | Zhai L Y, Wang F, Yan A, Liang C W, Wang S, Wang Y, Xu J L. Pleiotropic effect of GNP1 underlying grain number per panicle on sink, source and flow in rice[J]. Frontiers in Plant Science, 2020, 11: 933. |
| [143] | Yang J, Kim S R, Lee S K, Cohi H, Jeon J S, An G. Alanine aminotransferase1 (OsAlaAT1) plays an essential role in the regulation of starch storage in rice endosperm[J]. Plant Science, 2015, 240: 79-89. |
| [144] | Fan F, Liu M, Li N, Guo Y, Yuan H R, Si F F, Cheng M X, Chen G L, Cai M, Li N W, Zhang Y X, Yu Y, Pi L M, Yang H C, Yang F, Wang K, Li S Q. Gain-of-function allele of HPY1 coordinates source and sink to increase grain yield in rice[J]. Science Bulletin, 2023, 68(19): 2155-2159. |
| [1] | 岳轩宇, 谢文亚, 冯志明, 陈宗祥, 胡珂鸣, 左示敏. OsERF93参与调控水稻纹枯病抗性的研究[J]. 中国水稻科学, 2026, 40(1): 37-50. |
| [2] | 王轶欣, 林参, 马刘洋, 陈龙, 奉保华, 倪深, 魏祥进, 贺记外, 陈天晓. 谷丙转氨酶基因OsAlaAT4调控水稻氮素吸收和产量[J]. 中国水稻科学, 2026, 40(1): 51-60. |
| [3] | 黄奇娜, 姜鸿瑞, 杨婕, 于坤宇, 杨长登, 梁燕. 种子休眠基因Sdr4的生物信息学分析与分子标记开发和应用[J]. 中国水稻科学, 2026, 40(1): 61-71. |
| [4] | 程朝平, 何旎清, 白康呈, 林少俊, 黄凤凰, 刘军化, 程祖锌, 黄成志, 杨德卫. 聚合稻瘟病抗性基因Pigm-1和Pid2的水稻三系不育系福梦A的选育与利用[J]. 中国水稻科学, 2026, 40(1): 72-84. |
| [5] | 刘亚萍, 董译词, 郑君妍, 邱绚, 刘鹏程, 叶亚峰, 刘斌美, 陈析丰, 马伯军. 水稻类病变早衰突变体lmes7的鉴定与基因精细定位[J]. 中国水稻科学, 2026, 40(1): 85-94. |
| [6] | 肖无为, 张雨晴, 朱辰光, 田贵生, 蔡岳宏, 王飞, 熊栋梁, 黄见良, 彭少兵, 崔克辉. 促芽肥施用时期和施用量对再生稻产量和头季稻米品质的影响[J]. 中国水稻科学, 2026, 40(1): 118-130. |
| [7] | 谢世民, 周誉株, 薛晓迪, 朱广飞, 孙良, 陈建能. 水稻钵苗取栽协同作业式移栽机构设计与试验[J]. 中国水稻科学, 2026, 40(1): 131-144. |
| [8] | 陈玲, 林文英, 梁丽梅, 欧阳由男, 叶胜海, 季芝娟. 水稻开花习性及其在粳型三系不育系选育中的应用[J]. 中国水稻科学, 2025, 39(6): 731-743. |
| [9] | 王娟, 吴丽娟, 洪海波, 姚志文, 王磊, 鄂志国. 水稻泛素结合酶E2的生物学功能研究进展[J]. 中国水稻科学, 2025, 39(6): 744-750. |
| [10] | 陶士博, 许娜, 徐正进, 刘畅, 徐铨. 水稻发芽期耐冷基因Cold6的克隆[J]. 中国水稻科学, 2025, 39(6): 751-759. |
| [11] | 陈伟, 叶元妹, 赵剑华, 冯志明, 陈宗祥, 胡珂鸣, 左示敏. 利用CRISPR/Cas9技术改良南粳46抽穗期[J]. 中国水稻科学, 2025, 39(6): 760-770. |
| [12] | 侯桂花, 周立国, 雷建国, 陈虹, 聂元元. 水稻OsRDR5基因功能及作用机制初步解析[J]. 中国水稻科学, 2025, 39(6): 779-788. |
| [13] | 陆帅, 陶涛, 刘冉, 周文玉, 曹蕾, 杨青青, 张明秋, 任鑫哲, 杨芝笛, 徐福祥, 环海东, 龚远航, 张皓程, 金素奎, 蔡秀玲, 高继平, 冷语佳. 水稻长护颖小粒突变体lsg8的表型鉴定与基因克隆[J]. 中国水稻科学, 2025, 39(6): 813-824. |
| [14] | 邓欢, 刘亚培, 王春连, 郭威, 陈析丰, 纪志远. 水稻抗白叶枯病新基因Xa49(t)的定位分析[J]. 中国水稻科学, 2025, 39(6): 825-831. |
| [15] | 卞金龙, 任高磊, 裘实, 许方甫, 胡忠磊, 张洪程, 魏海燕. 不同机插方式下控混肥施用方式对淮北地区优质食味粳稻产量及氮素利用的影响[J]. 中国水稻科学, 2025, 39(6): 847-862. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||