
中国水稻科学 ›› 2023, Vol. 37 ›› Issue (3): 329-336.DOI: 10.16819/j.1001-7216.2023.220903
• 实验技术 • 上一篇
罗举1, 杨素文2, 贝文勇3, 余军伟4, 唐健1, 刘淑华1,*( )
)
收稿日期:2022-09-06
修回日期:2022-11-12
出版日期:2023-05-10
发布日期:2023-05-16
通讯作者:
*email: liushuhua@caas.cn
基金资助:
LUO Ju1, YANG Suwen2, BEI Wenyong3, YU Junwei4, TANG Jian1, LIU Shuhua1,*( )
)
Received:2022-09-06
Revised:2022-11-12
Online:2023-05-10
Published:2023-05-16
Contact:
*email: liushuhua@caas.cn
摘要:
【目的】褐飞虱(Nilaparvata lugens)是水稻重大害虫,灯光诱捕一直是褐飞虱监测的重要方法。然而,褐飞虱的两个同属近似种伪褐飞虱(N. muiri)和拟褐飞虱(N. bakeri)也具有扑灯行为,常被误判为褐飞虱。针对测报灯下褐飞虱属近似种形态鉴定费时费力、专业技能要求高,伪褐飞虱和拟褐飞虱常被误判为褐飞虱这一问题,拟建立一种褐飞虱属3种飞虱的快速分类鉴定方法。【方法】以条形码基因ITS1为靶标,筛选褐飞虱属通用引物及物种特异性探针,建立并优化直接多重Taqman qPCR检测体系(Direct Multiplex TaqMan quantitative PCR, dmTqPCR),并分析其特异性、灵敏度及实用性。【结果】本研究建立的3种飞虱dmTqPCR检测方法特异性强,可准确区分褐飞虱、伪褐飞虱和拟褐飞虱;灵敏度高,检出限可达10拷贝/反应;大样本检测结果表明,382个样本从样本获取到结果输出的整个过程可在3 h内完成,检出率及准确率均为100%。【结论】本研究建立的褐飞虱属近似种的dmTqPCR鉴定方法可以用于褐飞虱、伪褐飞虱和拟褐飞虱的快速分类鉴定,有利于灯下褐飞虱的精准测报。
罗举, 杨素文, 贝文勇, 余军伟, 唐健, 刘淑华. 直接多重TaqMan qPCR方法快速鉴定褐飞虱属3种飞虱[J]. 中国水稻科学, 2023, 37(3): 329-336.
LUO Ju, YANG Suwen, BEI Wenyong, YU Junwei, TANG Jian, LIU Shuhua. Direct Multiplex TaqMan qPCR Assay for Rapid Detection of Three Sibling Species from Nilaparvata Distant[J]. Chinese Journal OF Rice Science, 2023, 37(3): 329-336.

图1 褐飞虱及其两个近似种的ITS1基因多序列比对 Nl―褐飞虱;Nm―伪褐飞虱;Nb―拟褐飞虱。下划线序列代表引物位置,蓝色背景为探针位置。
Fig. 1. Multiple alignment of ITS1 genes from N. lugens and its two sibling species. Nl, N. lugens; Nm, N.muiri; Nb, N. bakeri. Primer sequences were underlined, and the probe sequences were highlighted with blue background.
| 引物/探针名称 Primer/Probe name | 引物/探针序列(5’-3’) Primer/Probe sequence (5’-3’) | 描述 Description |
|---|---|---|
| ITS1-F | CCAAGTGATCCACCACTCAGG | 褐飞虱属通用引物General primer |
| ITS1-R | GGATTCGTCTTGTCGGCTAGG | |
| Nl-ITS1-P | FAM-CCACCATACATTGTAATG-MGB | 褐飞虱探针N.1ugens-specific probe |
| Nm-ITS1-P | VIC-CTAGTCATTACACAATGTA-MGB | 伪褐飞虱探针N.muiri-specific probe |
| Nb-ITS1-P | ROX-CATTACATTGTATGTTGGC-MGB | 拟褐飞虱探针N.bakeri-specific probe |
表1 本研究用到的引物和探针
Table 1. Primers and probes used in the study.
| 引物/探针名称 Primer/Probe name | 引物/探针序列(5’-3’) Primer/Probe sequence (5’-3’) | 描述 Description |
|---|---|---|
| ITS1-F | CCAAGTGATCCACCACTCAGG | 褐飞虱属通用引物General primer |
| ITS1-R | GGATTCGTCTTGTCGGCTAGG | |
| Nl-ITS1-P | FAM-CCACCATACATTGTAATG-MGB | 褐飞虱探针N.1ugens-specific probe |
| Nm-ITS1-P | VIC-CTAGTCATTACACAATGTA-MGB | 伪褐飞虱探针N.muiri-specific probe |
| Nb-ITS1-P | ROX-CATTACATTGTATGTTGGC-MGB | 拟褐飞虱探针N.bakeri-specific probe |
| 地点 Location | 坐标Coordinates | 样本数量Number of specimen | |||
|---|---|---|---|---|---|
| 东经 Longitude (E) | 北纬 Latitude (N) | 褐飞虱 N. lugens | 伪褐飞虱 N. muiri | 拟褐飞虱 N. bakeri | |
| 广西Guangxi | 110.80 | 24.17 | 85 | 59 | 21 |
| 湖南Hunan | 111.73 | 27.25 | 75 | 27 | 43 |
| 浙江Zhejiang | 119.95 | 30.07 | 29 | 35 | 8 |
表2 样本收集地点
Table 2. Regions of specimen collection in China.
| 地点 Location | 坐标Coordinates | 样本数量Number of specimen | |||
|---|---|---|---|---|---|
| 东经 Longitude (E) | 北纬 Latitude (N) | 褐飞虱 N. lugens | 伪褐飞虱 N. muiri | 拟褐飞虱 N. bakeri | |
| 广西Guangxi | 110.80 | 24.17 | 85 | 59 | 21 |
| 湖南Hunan | 111.73 | 27.25 | 75 | 27 | 43 |
| 浙江Zhejiang | 119.95 | 30.07 | 29 | 35 | 8 |
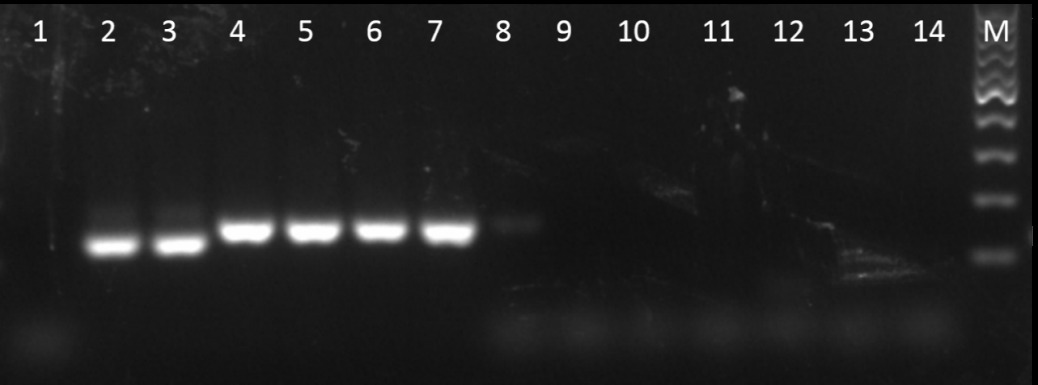
图2 褐飞虱属通用引物ITS1-F/R的特异性验证 1―水;2~3―褐飞虱;4~5―伪褐飞虱;6~7―拟褐飞虱;8―白背飞虱;9―灰飞虱;10―烟翅白背飞虱;11―稗飞虱;12―廖飞虱;13―短头飞虱;14―黑边梅塔飞虱;M―100 bp DNA标准品
Fig. 2. Specificity analysis of the inter-speies general primes ITS1-F/R. 1, H2O; 2-3, N. lugens; 4-5, N. muiri; 6-7, N. bakeri; 8, S. furcifera; 9, L. striatellus; 10, S. kolophon; 11, S. longifurcifera; 12, H. gayasana; 13, E. nawaii; 14, M. propinqua; M, 100 bp DNA marker.
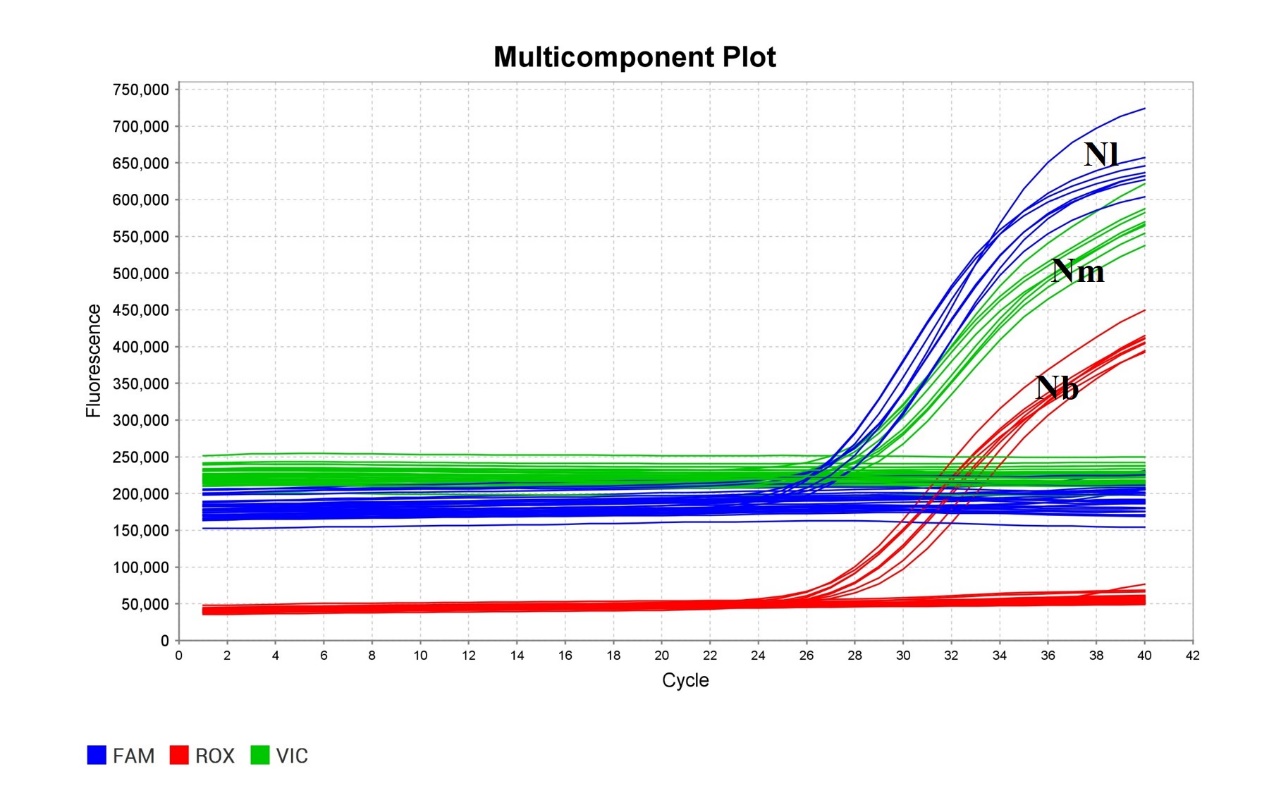
图3 直接多重TaqMan qPCR体系的特异性试验 Nl―褐飞虱;Nm―伪褐飞虱;Nb―拟褐飞虱。
Fig. 3. Specificity analysis of the direct multiplex TaqMan qPCR assay. Nl, N. lugens; Nm, N.muiri; Nb, N. bakeri.
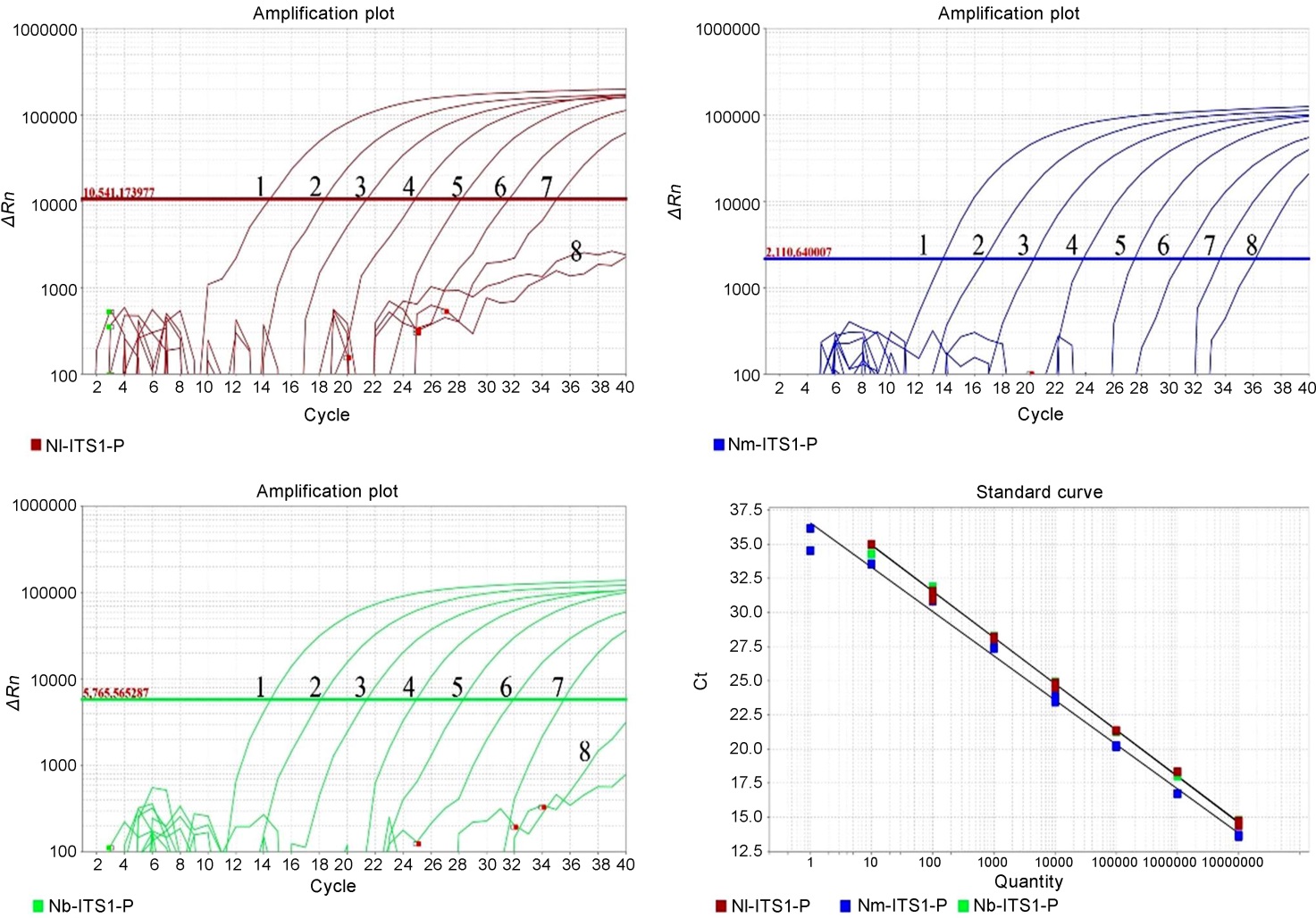
图4 直接多重TaqMan qPCR灵敏性测试试验 1~8分别代表1.0×107、1.0×106、1.0×105、1.0×104、1.0×103、1.0×102、1.0×101、1.0×100 拷贝/μL。
Fig. 4. Sensitivity analysis of the direct multiplex TaqMan qPCR assay. 1-8 represent 1.0×107, 1.0×106, 1.0×105, 1.0×104, 1.0×103, 1.0×102, 1.0×101, 1.0×100 copies /μL, respectively.
| 样品编号 Sample No. | 形态鉴定 Morphological identification | Ct值Cycle of threshold | ||
|---|---|---|---|---|
| Nl 探针 Nl probe | Nm 探针 Nm probe | Nb 探针 Nb probe | ||
| 1 | Nl | 24.91 | Un | Un |
| 2 | Nm | Un | 26.72 | Un |
| 3 | Nl | 25.50 | Un | Un |
| 4 | Nb | Un | Un | 24.68 |
| 5 | Nm | Un | 26.75 | Un |
| 6 | Nm | Un | 27.78 | Un |
| 7 | Nl | 26.33 | Un | Un |
| 8 | Nb | Un | Un | 24.28 |
| 9 | Nm | Un | 27.34 | Un |
| 10 | Nl | 25.46 | Un | Un |
| 11 | Pc | 21.80 | 21.75 | 22.12 |
| 12 | Nc | Un | Un | Un |
表3 直接多重TaqMan qPCR检测体系对382个样本检测的部分结果
Table 3. Partial dmTqPCR assay results for the 382 Nilaparvata species individuals.
| 样品编号 Sample No. | 形态鉴定 Morphological identification | Ct值Cycle of threshold | ||
|---|---|---|---|---|
| Nl 探针 Nl probe | Nm 探针 Nm probe | Nb 探针 Nb probe | ||
| 1 | Nl | 24.91 | Un | Un |
| 2 | Nm | Un | 26.72 | Un |
| 3 | Nl | 25.50 | Un | Un |
| 4 | Nb | Un | Un | 24.68 |
| 5 | Nm | Un | 26.75 | Un |
| 6 | Nm | Un | 27.78 | Un |
| 7 | Nl | 26.33 | Un | Un |
| 8 | Nb | Un | Un | 24.28 |
| 9 | Nm | Un | 27.34 | Un |
| 10 | Nl | 25.46 | Un | Un |
| 11 | Pc | 21.80 | 21.75 | 22.12 |
| 12 | Nc | Un | Un | Un |
| [1] | 丁锦华. 褐飞虱属分类及其种类的研究[J]. 南京农业大学学报, 1981(1): 78-85. |
| Ding J H. Taxonomic study of Nilaparvata (Hemiptera: Delphacidae)[J]. Journal of Nanjing Agricultural College, 1981(1): 78-85. (in Chinese) | |
| [2] | 程遐年, 吴进才, 马飞. 褐飞虱研究与防治[M]. 北京: 中国农业出版社, 2002. |
| Cheng X N, Wu J C, Ma F. Research and Management of Nilaparvata lugens[M]. Beijing: China Agricultural Press, 2002. (in Chinese) | |
| [3] | Guo L Z, Liang A P, Jiang G M. Four new species and a new record of Delphacidae (Hemiptera) from China[J]. Orient Insects, 2005, 39: 161-174. |
| [4] | 崔亚丽, 何佳春, 罗举, 赖凤香, 傅强. 褐飞虱近似种伪褐飞虱和拟褐飞虱的寄主植物[J]. 中国水稻科学, 2013, 27(1): 105-110. |
| Cui Y L, He J C, Luo J, Lai F X, Fu Q. Host plants of Nilaparvala muiri China and N. bakeri(Muir), two sibling species of N. lugens (Stål)[J]. Chinese Journal of Rice Science, 2013, 27(1): 105-110. (in Chinese with English abstract) | |
| [5] | 罗举, 傅强, 陆志坚, 吴彩谦, 李一波, 段德康, 刘玉坤, 张志涛. 测报灯下褐飞虱及其两种近似种的数量动态[J]. 中国水稻科学, 2010, 24(3): 315-319. |
| Luo J, Fu Q, Lu Z J, Wu C Q, Li Y B, Duan D K, Liu Y K, Zhang Z T. Population dynamics of Nilaparvata lugens and its two sibling species under black light trap[J]. Chinese Journal of Rice Science, 2010, 24(3): 315-319. (in Chinese) | |
| [6] | 蒋天梅, 虞根聪, 赖朝晖, 蒋晔. 象山县测报灯下褐飞虱及其近似种的数量动态[J]. 中国植保导刊, 2018, 38(7): 43-45. |
| Jiang T M, Yu G C, Lai C H, Jiang Y. Population dynamics of Nilaparvata lugens and its two sibling species under light trap in Xiangshan[J]. China Plant Protection, 2018, 38(7): 43-45. (in Chinese) | |
| [7] | 丁锦华, 胡春林, 傅强, 何佳春, 谢茂成. 中国稻区常见飞虱原色图鉴[M]. 杭州: 浙江科学技术出版社, 2012. |
| Ding J H, Hu C L, Fu Q, He J C, Xie M C. Primary Color Atlas of Common Planthoppers in Rice Areas of China[M]. Hangzhou: Zhejiang Science and Technology Press, 2012. (in Chinese) | |
| [8] | 芮金富, 周建平, 石磊. 褐飞虱与拟褐飞虱和伪褐飞虱的识别[J]. 现代农业科技, 2011(5): 191. |
| Rui J F, Zhou J P, Shi L. Morphological identification of Nilaparvata lugens from its two sibling species, Nilaparvata muiri and Nilaparvata bakeri[J]. Modern Agriculture Technology, 2011(5): 191. (in Chinese) | |
| [9] | 崔亚丽. 褐飞虱及其两近似种的生物学及分子标记的研究[D]. 北京: 中国农业科学院, 2012: 26-39. |
| Cui Y L. Studies on the biological characteristics and molecular markers for Nilaparvata lugens (Stål) and its two sibling species N. muiri China and N. bakeri (Muir)[D]. Beijing: Chinese Academy of Agricultural Sciences, 2012: 26-39. (in Chinese) | |
| [10] | Liu S H, Luo J, Liu R, Zhang C G, Duan D K, Chen H M, Bei W Y, Tang J. Identification of Nilaparvata lugens and its two sibling species (N. bakeri and N. muiri) by direct multiplex PCR[J]. Journal of Economic Entomology, 2018, 111(6): 2869-2875. |
| [11] | 罗举, 唐健, 王爱英, 杨保军, 刘淑华. 基于重组酶介导扩增-侧流层析试纸条的褐飞虱快速鉴定方法[J]. 中国水稻科学, 2022, 36(3): 96-104. |
| Luo J, Tang J, Wang A Y, Yang B J, Liu S H. A rapid detection assay of Nilaparvata lugens based on recombinase aided amplification-lateral flow dipstick technology[J]. Chinese Journal of Rice Science, 2022, 36(1): 96-104. (in Chinese with English abstract) | |
| [12] | 中华人民共和国国家质量监督检验检疫总局/中国国家标准化管理委员会: 稻飞虱测报调查规范. GB/T 15794—2009[S]. 北京: 中国标准出版社, 2009. |
| General Administration of Quality Supervision, Inspection and Quarantine/ Standardization Adminis- tration of the People's Republic of China: Standard for Survey of Rice Planthopper: GB/T 15794—2009[S]. Beijing: China Standard Press, 2009. (in Chinese) | |
| [13] | 黄丽莉, 阙海勇, 车飞. 茶园茶黄蓟马及其近似种的DNA条形码鉴定[J]. 植物检疫, 2014, 28(6): 68-72. |
| Huang L L, Que H Y, Che F. Molecular identification of Scirtothrips dorsalis and related species through DNA barcoding in tea garden[J]. Plant Quarantin, 2014, 28(6): 68-72. (in Chinese) | |
| [14] | 张磊, 靳明辉, 张丹丹, 姜玉英, 刘杰, 吴孔明, 萧玉涛. 入侵云南草地贪夜蛾的分子鉴定[J]. 植物保护, 2019, 45(2): 19-24. |
| Zhang L, Jin M H, Zhang D D, Jiang Y Y, Liu J, Wu K M, Xiao Y T. Molecular identification of invasive fall armyworm Spodoptera frugiperda in Yunnan Province[J]. Plant Protection, 2019, 45(2): 19-24. (in Chinese) | |
| [15] | 吴霞, 张桂芬, 万方浩. 基于TaqMan实时荧光定量PCR技术的西花蓟马快速检测[J]. 应用昆虫学报, 2011, 48(3): 497-503. |
| Wu X, Zhang G F, Wan F H. TaqMan real-time fluorescent quantitative PCR for identification of Frankliniella occidentalis[J]. Chinese Journal of Applied Entomology, 2011, 48(3): 497-503. (in Chinese with English abstract) | |
| [16] | 李茵, 刘英, 双巧云, 彭徽, 黄丽莉. TaqMan实时荧光定量PCR检测茶黄蓟马[J]. 植物检疫, 2021, 35(6): 43-47. |
| Li Y, Liu Y, Shuang Q Y, Peng H, Huang L L. TaqMan real-time fluorescent quantitative PCR for identification of Scirtothrips dorsalis Hoods[J]. Plant Quarantin, 2021, 35(6): 43-47. (in Chinese) | |
| [17] | 唐慧骥, 党志浩, 郭长宁, 王晓晶, 苗小星, 郭保生, 徐超. 应用重组酶介导核酸扩增技术快速鉴定新菠萝灰粉蚧[J]. 植物检疫, 2019, 33(2): 37-42. |
| Tang H J, Dang Z H, Guo C N, Wang X J, Miao X X, Guo B S, Xu C. Rapid detection for Dysmicoccus neobrevipes by recombinase-aid amplification[J]. Plant Quarantin, 2019, 33(2): 37-42. (in Chinese) | |
| [18] | Agarwal A, Cunningham J P, Valenzuela I, Blacket M J. A diagnostic LAMP assay for the destructive grapevine insect pest, phylloxera (Daktulosphaira vitifoliae)[J]. Scientific Reports, 2020, 10: 21229. |
| [19] | Agarwal A, Rako L, Schutze M K, Starkie M L, Tay W T, Rodoni B C, Blacket M J. A diagnostic LAMP assay for rapid identification of an invasive plant pest, fall armyworm Spodoptera frugiperda(Lepidoptera: Noctuidae)[J]. Scientific Reports, 2022, 12: 1116 |
| [20] | Lu C C, Dai T T, Zhang H F, Zeng D D, Wang Y C, Yang W F, Zheng X B. A novel LAMP assay using hot water in vacuum insulated bottle for rapid detection of the soybean red crown rot pathogen Calonectria ilicicola[J]. Australasian Plant Pathology, 2022, 51: 251-259. |
| [21] | Ponting S, Tomkies V, Stainton K. Rapid identification of the invasive Small hive beetle (Aethina tumida) using LAMP[J]. Pest Management Science, 2021, 77(3): 1476-1481. |
| [22] | Nam H Y, Kwon M, Kim H J, Kim J. Development of a species diagnostic molecular tool for an invasive pest, Mythimna loreyi, Using LAMP[J]. Insects, 2020, 11(11): 817. |
| [23] | 徐浪, 林伟, 黄蓬英, 张伟锋, 卢小雨, 汪莹, 郑耘, 焦懿, 娄定风, 余道坚. 应用TaqMan MGB探针快速检测新菠萝灰粉蚧和菠萝灰粉蚧[J]. 植物检疫, 2016, 30(4): 38-41. |
| Xu L, Lin W, Huang P Y, Zhang W F, Lu X Y, Zhen Y, Jiao Y, Lou D F, Yu D J. Rapid detection of Dysmicoccus neobrevipes and D. brevipes (Hemiptera: Diaspididae) using TaqMan MGB probe[J]. Plant Quarantin, 2016, 30(4): 38-41. (in Chinese) | |
| [24] | Aguirre C, Sanchez E, Olivares N, Hinrichsen P. Multiplex TaqMan real-time PCR assay for sensitive detection of two weevil species(Coleoptera: Curculionidae)[J]. Journal of Economic Entomology, 2021, 114(1): 90-99. |
| [25] | Huang Q Y, Chen D M, Du C, Liu Q Q, Lin S, Liang L L, Xu Y, Liao Y Q, Li Q G. Highly multiplex PCR assays by coupling the 5'-flap endonuclease activity of Taq DNA polymerase and molecular beacon reporters[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(9): 1-11. |
| [26] | 姚晶, 郭晓军, 王甦, 王昱超, 罗晨, 张帆, 李绍清. 利用TaqMan等位基因技术鉴定烟粉虱MEAM1和MED隐种[J]. 昆虫学报. 2013, 56(1): 98-103. |
| Yao J, Guo X J, Wang S, Wang Y C, Luo C, Zhang F, Li S Q. Discrimination of cryptic species MEAM1 and MED of Bemisia tabaci (Hemiptera: Aleyrodidae) by using TaqMan allele-selective PCR[J]. Acta Entomologica Sinica, 2013, 56 (1): 98-103. (in Chinese with English abstract) |
| [1] | 杨奇欣, 赖凤香, 何佳春, 魏琪, 王渭霞, 万品俊, 傅强. 不同抗感水稻品种对褐飞虱胁迫的高光谱响应特征[J]. 中国水稻科学, 2024, 38(1): 81-90. |
| [2] | 程玲, 黄福钢, 邱一埔, 王心怡, 舒宛, 邱永福, 李发活. 籼稻材料570011抗褐飞虱基因的遗传分析及鉴定[J]. 中国水稻科学, 2023, 37(3): 244-252. |
| [3] | 何佳春, 何雨婷, 万品俊, 魏琪, 赖凤香, 陈祥盛, 傅强. 温度对褐飞虱天敌黄腿双距螯蜂生物学特性的影响[J]. 中国水稻科学, 2022, 36(3): 318-326. |
| [4] | 罗举, 唐健, 王爱英, 杨保军, 刘淑华. 基于重组酶介导扩增-侧流层析试纸条的褐飞虱快速鉴定方法[J]. 中国水稻科学, 2022, 36(1): 96-104. |
| [5] | 马银花, 李萍芳, 董文静, 易松望, 杨芳, 杜波, 金晨钟. 水稻抗性蛋白OsRRK1抗褐飞虱机理分析[J]. 中国水稻科学, 2020, 34(6): 512-519. |
| [6] | 潘磊, 王利华, 朱凤, 韩阳春, 王培, 方继朝. 褐飞虱小分子量热激蛋白基因表达特性和功能[J]. 中国水稻科学, 2020, 34(1): 37-45. |
| [7] | 朱永生, 白建林, 谢鸿光, 吴方喜, 罗曦, 姜身飞, 何炜, 陈丽萍, 蔡秋华, 谢华安, 张建福. 聚合白背飞虱和褐飞虱抗性基因创制杂交水稻恢复系[J]. 中国水稻科学, 2019, 33(5): 421-428. |
| [8] | 何佳春, 李波, 谢茂成, 赖凤香, 胡国文, 傅强. 新烟碱类及其他稻田杀虫剂对褐飞虱的室内药效评价[J]. 中国水稻科学, 2019, 33(5): 467-478. |
| [9] | 降好宇, 曾盖, 郝明, 黄湘桂, 肖应辉. 广谱抗稻瘟病种质75-1-127的褐飞虱抗性基因鉴定及分子标记辅助选择育种[J]. 中国水稻科学, 2019, 33(3): 227-234. |
| [10] | 张珏锋, 李芳, 钟海英, 陈建明. 制霉菌素对褐飞虱若虫解毒酶、尿酸酶含量的影响[J]. 中国水稻科学, 2019, 33(2): 186-190. |
| [11] | 朱欢欢, 陈洋, 万品俊, 王渭霞, 赖凤香, 傅强. 共生菌Arsenophonus、水稻品种和温度对褐飞虱黄绿绿僵菌发病率的影响[J]. 中国水稻科学, 2017, 31(6): 643-651. |
| [12] | 单丹, 王利华, 张月亮, 韩阳春, 牛洪涛, 潘磊, 方继朝. 褐飞虱热激蛋白70在不同温度胁迫下的差异表达特性研究[J]. 中国水稻科学, 2017, 31(5): 533-541. |
| [13] | 赵晨星, 俞叶微, 许益鹏, 俞晓平. 褐飞虱两个dynamin-1-like基因的克隆、多克隆抗体制备及 表达定位[J]. 中国水稻科学, 2017, 31(4): 345-354. |
| [14] | 陈龙飞, 万品俊, 王渭霞, 傅强, 朱廷恒. 褐飞虱NlTgo基因的克隆及功能研究[J]. 中国水稻科学, 2016, 30(6): 653-660. |
| [15] | 郑瑜, 何佳春, 万品俊, 赖凤香, 孙燕群, 林晶晶, 傅强. 褐飞虱IR56种群的致害特征[J]. 中国水稻科学, 2016, 30(5): 552-558. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||