
Chinese Journal OF Rice Science ›› 2022, Vol. 36 ›› Issue (3): 237-247.DOI: 10.16819/j.1001-7216.2022.210117
• Research Papers • Previous Articles Next Articles
LI Zhaowei1, SUN Congying1, LING Donglan1, ZENG Huiling1, ZHANG Xiaomei1, FAN Kai2, LIN Wenxiong1,*( )
)
Received:2021-01-17
Revised:2021-04-22
Online:2022-05-10
Published:2022-05-11
Contact:
LIN Wenxiong
李兆伟1, 孙聪颖1, 零东兰1, 曾慧玲1, 张晓妹1, 范凯2, 林文雄1,*( )
)
通讯作者:
林文雄
基金资助:LI Zhaowei, SUN Congying, LING Donglan, ZENG Huiling, ZHANG Xiaomei, FAN Kai, LIN Wenxiong. Construction of osarf7 Mutants in Rice Based on CRISPR/Cas9 Technology and Investigation on Their Agronomic Traits[J]. Chinese Journal OF Rice Science, 2022, 36(3): 237-247.
李兆伟, 孙聪颖, 零东兰, 曾慧玲, 张晓妹, 范凯, 林文雄. 利用CRISPR/Cas9创建osarf7突变体及其农艺性状调查[J]. 中国水稻科学, 2022, 36(3): 237-247.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2022.210117
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) | 用途Usage |
|---|---|---|
| OsARF7-T1-fwd | gccgACACGAACCTGTGCCACCGC | 靶序列1构建 |
| OsARF7-T1-rev | aaacGCGGTGGCACAGGTTCGTGT | Construction of the target sequence 1 |
| OsARF7-T2-fwd | gttGAGCTATGGCATGCTTGCGC | 靶序列2构建 |
| OsARF7-T2-rev | aaacGCGCAAGCATGCCATAGCT | Construction of the target sequence 2 |
| Hyp-F | ACGGTGTCGTCCATCACAGTTTGCC | 阳性转基因植株鉴定 |
| Hyp-R | TTCCGGAAGTGCTTGACATTGGGGA | Identification of the positive transgenic plant |
| OsARF7-T1-F | ACCACCACCTCCTCCTCAC | 靶点1测序检测 |
| OsARF7-T1-R | TCGCACGAATCAGCACAAC | Sequencing for target 1 |
| OsARF7-T2-F | TTACGGATCTGAGAACCTG | 靶点2测序检测 |
| OsARF7-T2-R | TAACTACATTGCCGAAGAA | Sequencing for target 2 |
Table 1. Sequences of gRNA targets and other primers used in this study.
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) | 用途Usage |
|---|---|---|
| OsARF7-T1-fwd | gccgACACGAACCTGTGCCACCGC | 靶序列1构建 |
| OsARF7-T1-rev | aaacGCGGTGGCACAGGTTCGTGT | Construction of the target sequence 1 |
| OsARF7-T2-fwd | gttGAGCTATGGCATGCTTGCGC | 靶序列2构建 |
| OsARF7-T2-rev | aaacGCGCAAGCATGCCATAGCT | Construction of the target sequence 2 |
| Hyp-F | ACGGTGTCGTCCATCACAGTTTGCC | 阳性转基因植株鉴定 |
| Hyp-R | TTCCGGAAGTGCTTGACATTGGGGA | Identification of the positive transgenic plant |
| OsARF7-T1-F | ACCACCACCTCCTCCTCAC | 靶点1测序检测 |
| OsARF7-T1-R | TCGCACGAATCAGCACAAC | Sequencing for target 1 |
| OsARF7-T2-F | TTACGGATCTGAGAACCTG | 靶点2测序检测 |
| OsARF7-T2-R | TAACTACATTGCCGAAGAA | Sequencing for target 2 |
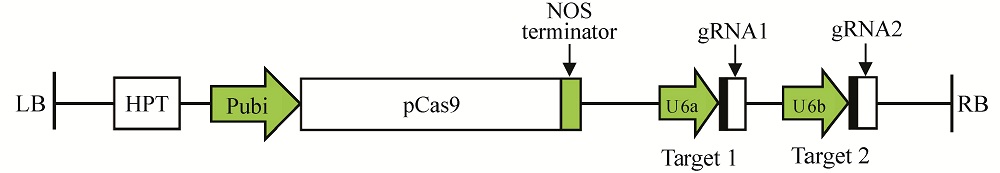
Fig. 2. Diagrammatic sketch of pYLCRISPR/Cas9-ARF7-T12 recombinant vector. Cloning of two gRNA cassettes into the pYLCRISPR/Cas9-MT vector. LB, Left border; RB, Right border.
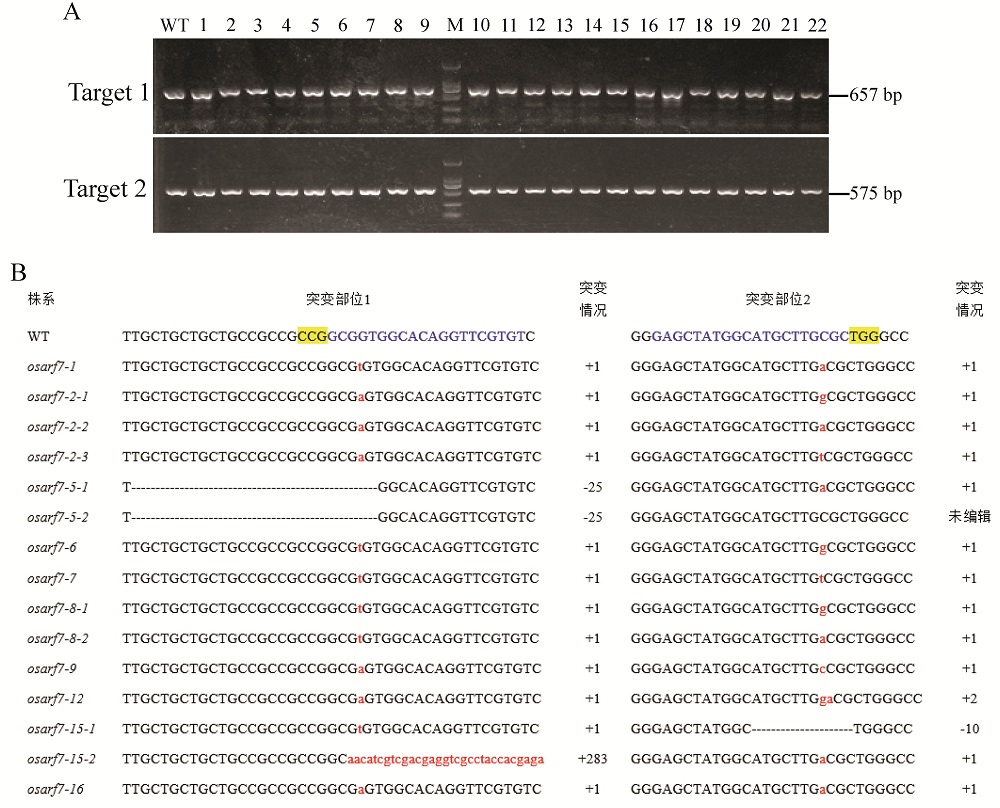
Fig. 5. PCR identification and sequence alignment of osarf7 mutants compared to the WT line. A, PCR amplification results of the DNA fragments near the OsARF7 locus for partial osarf7 T2 mutants, the fragment length of target 1 and 2 is 657 bp and 575 bp, respectively, and WT is ZH11; B, Sequence alignment of the edited locus for the osarf7 mutants and WT line. The blue letters are the target genome sequence; The yellow highlighted letters denote PAM; Dashes strikethrough indicate the deleted bases; Insertion nucleotides are shown in red lowercase letters; –, Deletion; +, Insertion; WT, Wild type.
| 编号 Number | 潜在脱靶位置 Putative off-target locus | 潜在脱靶序列 Sequence of the putative off-target site (5′-3′) | 错配碱基数 No. of mismatched bases | 测试株数 No. of plants tested | 突变株数 No. of plants with mutations |
|---|---|---|---|---|---|
| Off-target 1-1 | Chr08: 12898391-12898413 | ACCCTAACCTCTCCCACCGC | 4 | 9 | 0 |
| Off-target 1-2 | Chr10: 15856432-15856454 | AAACGGACGTGTGCCGCCGC | 4 | 9 | 0 |
| Off-target 1-3 | Chr02: 27855035-27855057 | AGACGGGCCTGTGCCAACGC | 4 | 9 | 0 |
| Off-target 1-4 | Chr12: 12787156-12787178 | CCAAGAACCTGCGCCGCCGC | 4 | 9 | 0 |
| Off-target 1-5 | Chr07: 2126716-2126738 | ACAGCAGCCGGTGCCACCGC | 4 | 9 | 0 |
| Off-target 2-1 | Chr12: 26007034-26007056 | GAGCTGTGGCATGCATGCGC | 2 | 10 | 0 |
| Off-target 2-2 | Chr02: 3488312-3488334 | GAGTTGTGGCATGCATGCGC | 3 | 9 | 0 |
| Off-target 2-3 | Chr04: 34285188-34285210 | GAGCTATGGCACGCCTGCGC | 2 | 8 | 0 |
| Off-target 2-4 | Chr06: 4926812-4926834 | GAGCTATGGCATGCTTGTGC | 1 | 10 | 0 |
| Off-target 2-5 | Chr04: 22007551-22007573 | GAGCTATGGCATGCTTGTGC | 1 | 10 | 0 |
| Off-target 2-6 | Chr01: 40696054-40696076 | GAGCTGTGGCACGCCTGCGC | 3 | 10 | 0 |
Table 2. Mutation detections in the putative CRISPR/Cas9-OsARF7-T12 off-target sites.
| 编号 Number | 潜在脱靶位置 Putative off-target locus | 潜在脱靶序列 Sequence of the putative off-target site (5′-3′) | 错配碱基数 No. of mismatched bases | 测试株数 No. of plants tested | 突变株数 No. of plants with mutations |
|---|---|---|---|---|---|
| Off-target 1-1 | Chr08: 12898391-12898413 | ACCCTAACCTCTCCCACCGC | 4 | 9 | 0 |
| Off-target 1-2 | Chr10: 15856432-15856454 | AAACGGACGTGTGCCGCCGC | 4 | 9 | 0 |
| Off-target 1-3 | Chr02: 27855035-27855057 | AGACGGGCCTGTGCCAACGC | 4 | 9 | 0 |
| Off-target 1-4 | Chr12: 12787156-12787178 | CCAAGAACCTGCGCCGCCGC | 4 | 9 | 0 |
| Off-target 1-5 | Chr07: 2126716-2126738 | ACAGCAGCCGGTGCCACCGC | 4 | 9 | 0 |
| Off-target 2-1 | Chr12: 26007034-26007056 | GAGCTGTGGCATGCATGCGC | 2 | 10 | 0 |
| Off-target 2-2 | Chr02: 3488312-3488334 | GAGTTGTGGCATGCATGCGC | 3 | 9 | 0 |
| Off-target 2-3 | Chr04: 34285188-34285210 | GAGCTATGGCACGCCTGCGC | 2 | 8 | 0 |
| Off-target 2-4 | Chr06: 4926812-4926834 | GAGCTATGGCATGCTTGTGC | 1 | 10 | 0 |
| Off-target 2-5 | Chr04: 22007551-22007573 | GAGCTATGGCATGCTTGTGC | 1 | 10 | 0 |
| Off-target 2-6 | Chr01: 40696054-40696076 | GAGCTGTGGCACGCCTGCGC | 3 | 10 | 0 |
| 株系 Line | 株高 Plant height / cm | 总分蘖数 Tiller number per plant | 有效穗数 Effective panicle per plant | 穗长 Length of main panicle /cm | 穗粒数 Grain number per panicle | 结实率 Seed setting rate / % | 千粒重 1000-grain weight / g | 单株产量 Grain yield per plant / g |
|---|---|---|---|---|---|---|---|---|
| osarf7-1 | 73.5±1.6 ab | 11.2±1.3 a | 24.6±2.1 a | 119.3±30.8 ab | 63.4±19.0 bc | 23.3±2.9 e | 8.6±4.6 ab | |
| osarf7-2-1 | 72.2±3.4 ab | 20.6±3.3 a | 7.4±1.5 abc | 22.8±1.5 ab | 135.9±34.9 ab | 74.5±14.7 abc | 25.5±0.2 bcde | 12.9±6.3 ab |
| osarf7-2-2 | 67.3±4.9 b | 11.0±2.6 e | 5.0±1.0 c | 23.5±2.5 ab | 131.8±35.0 ab | 82.5±8.8 ab | 23.5±0.3 de | 9.3±4.2 ab |
| osarf7-2-3 | 72.7±1.5 ab | 20.0±1.0 a | 7.3±3.5 abc | 23.7±1.2 ab | 167.5±13.8 a | 78.4±5.8 abc | 22.6±0.6 e | 9.0±3.3 ab |
| osarf7-5-1 | 67.3±3.2 b | 12.7±1.2 de | 5.0±0.9 c | 22.6±1.2 ab | 119.4±12.1 ab | 81.8±5.2 ab | 25.5±0.6 bcde | 10.4±0.5 ab |
| osarf7-5-2 | 70.8±3.2 ab | 12.8±1.5 de | 6.2±0.8 bc | 22.8±2.0 ab | 112.8±29.5 b | 75.9±9.5 abc | 25.2±1.3 cde | 9.7±1.2 ab |
| osarf7-6 | 68.5±3.6 b | 14.5±1.1 bcd | 7.6±1.8 bc | 23.0±1.5 ab | 126.2±23.0 ab | 69.9±11.9 abc | 24.4±1.2 de | 11.3±3.0 ab |
| osarf7-7 | 73.0±3.7 ab | 20.2±1.8 a | 10.2±1.9 ab | 23.2±1.4 ab | 135.9±21.2 ab | 73.7±14.6 abc | 25.0±1.5 cde | 14.5±4.2 ab |
| osarf7-8-1 | 74.8±2.2 a | 13.8±0.5 bcde | 6.5±2.1 bc | 23.7±2.0 ab | 144.8±34.6 ab | 76.5±10.0 abc | 25.4±1.2 bcde | 14.1±3.0 ab |
| osarf7-8-2 | 75.4±2.4 a | 16.0±0.7 bc | 7.0±2.6 bc | 23.1±1.8 ab | 140.5±32.2 ab | 83.21±9.3 a | 24.7±0.7 cde | 12.0±3.3 ab |
| osarf7-9 | 72.3±0.8 ab | 15.5±1.0 bc | 6.8±1.9 bc | 22.3±1.6 ab | 119.9±23.1 ab | 86.5±8.3 a | 26.7±1.6 bcd | 12.5±2.4 ab |
| osarf7-12 | 75.0±2.0 a | 17.3±0.6 ab | 6.3±1.5 bc | 24.0±1.0 ab | 139.8±36.0 ab | 90.8±3.3 a | 23.6±0.8 cde | 17.4±9.8 a |
| osarf7-15-1 | 75.8±2.4 a | 13.7±1.2 de | 5.1±1.7 c | 21.9±1.3 b | 117.2±22.5 b | 83.0±5.9 a | 28.1±1.1 ab | 10.0±3.2 ab |
| osarf7-15-2 | 74.7±1.4 a | 11.9±1.2 de | 4.9±2.3 c | 21.8±1.9 b | 122.0±24.5 ab | 79.1±15.0 ab | 27.4±1.3 abc | 9.4±3.1 ab |
| osarf7-16 | 72.0±2.3ab | 15.4±1.6 bc | 5.6±0.9 c | 21.8±1.3 b | 103.0±26.7 b | 57.2±26.4 c | 30.1±2.2 a | 13.0±3.9 b |
| WT | 71.2±1.9 ab | 11.3±0.6 de | 10.2±1.8 ab | 20.8±1.6 b | 111.8±15.0 b | 88.4±4.4 a | 24.3±0.7 de | 14.2±4.8 ab |
Table 3. Agronomic traits of different types of osarf7 mutants and its corresponding wild type grown in paddy field.
| 株系 Line | 株高 Plant height / cm | 总分蘖数 Tiller number per plant | 有效穗数 Effective panicle per plant | 穗长 Length of main panicle /cm | 穗粒数 Grain number per panicle | 结实率 Seed setting rate / % | 千粒重 1000-grain weight / g | 单株产量 Grain yield per plant / g |
|---|---|---|---|---|---|---|---|---|
| osarf7-1 | 73.5±1.6 ab | 11.2±1.3 a | 24.6±2.1 a | 119.3±30.8 ab | 63.4±19.0 bc | 23.3±2.9 e | 8.6±4.6 ab | |
| osarf7-2-1 | 72.2±3.4 ab | 20.6±3.3 a | 7.4±1.5 abc | 22.8±1.5 ab | 135.9±34.9 ab | 74.5±14.7 abc | 25.5±0.2 bcde | 12.9±6.3 ab |
| osarf7-2-2 | 67.3±4.9 b | 11.0±2.6 e | 5.0±1.0 c | 23.5±2.5 ab | 131.8±35.0 ab | 82.5±8.8 ab | 23.5±0.3 de | 9.3±4.2 ab |
| osarf7-2-3 | 72.7±1.5 ab | 20.0±1.0 a | 7.3±3.5 abc | 23.7±1.2 ab | 167.5±13.8 a | 78.4±5.8 abc | 22.6±0.6 e | 9.0±3.3 ab |
| osarf7-5-1 | 67.3±3.2 b | 12.7±1.2 de | 5.0±0.9 c | 22.6±1.2 ab | 119.4±12.1 ab | 81.8±5.2 ab | 25.5±0.6 bcde | 10.4±0.5 ab |
| osarf7-5-2 | 70.8±3.2 ab | 12.8±1.5 de | 6.2±0.8 bc | 22.8±2.0 ab | 112.8±29.5 b | 75.9±9.5 abc | 25.2±1.3 cde | 9.7±1.2 ab |
| osarf7-6 | 68.5±3.6 b | 14.5±1.1 bcd | 7.6±1.8 bc | 23.0±1.5 ab | 126.2±23.0 ab | 69.9±11.9 abc | 24.4±1.2 de | 11.3±3.0 ab |
| osarf7-7 | 73.0±3.7 ab | 20.2±1.8 a | 10.2±1.9 ab | 23.2±1.4 ab | 135.9±21.2 ab | 73.7±14.6 abc | 25.0±1.5 cde | 14.5±4.2 ab |
| osarf7-8-1 | 74.8±2.2 a | 13.8±0.5 bcde | 6.5±2.1 bc | 23.7±2.0 ab | 144.8±34.6 ab | 76.5±10.0 abc | 25.4±1.2 bcde | 14.1±3.0 ab |
| osarf7-8-2 | 75.4±2.4 a | 16.0±0.7 bc | 7.0±2.6 bc | 23.1±1.8 ab | 140.5±32.2 ab | 83.21±9.3 a | 24.7±0.7 cde | 12.0±3.3 ab |
| osarf7-9 | 72.3±0.8 ab | 15.5±1.0 bc | 6.8±1.9 bc | 22.3±1.6 ab | 119.9±23.1 ab | 86.5±8.3 a | 26.7±1.6 bcd | 12.5±2.4 ab |
| osarf7-12 | 75.0±2.0 a | 17.3±0.6 ab | 6.3±1.5 bc | 24.0±1.0 ab | 139.8±36.0 ab | 90.8±3.3 a | 23.6±0.8 cde | 17.4±9.8 a |
| osarf7-15-1 | 75.8±2.4 a | 13.7±1.2 de | 5.1±1.7 c | 21.9±1.3 b | 117.2±22.5 b | 83.0±5.9 a | 28.1±1.1 ab | 10.0±3.2 ab |
| osarf7-15-2 | 74.7±1.4 a | 11.9±1.2 de | 4.9±2.3 c | 21.8±1.9 b | 122.0±24.5 ab | 79.1±15.0 ab | 27.4±1.3 abc | 9.4±3.1 ab |
| osarf7-16 | 72.0±2.3ab | 15.4±1.6 bc | 5.6±0.9 c | 21.8±1.3 b | 103.0±26.7 b | 57.2±26.4 c | 30.1±2.2 a | 13.0±3.9 b |
| WT | 71.2±1.9 ab | 11.3±0.6 de | 10.2±1.8 ab | 20.8±1.6 b | 111.8±15.0 b | 88.4±4.4 a | 24.3±0.7 de | 14.2±4.8 ab |
| [1] | 郭韬, 余泓, 邱杰, 李家洋, 韩斌, 林鸿宣. 中国水稻遗传学研究进展与分子设计育种[J]. 中国科学: 生命科学, 2019, 49: 1-28. |
| Guo T, Yu H, Qiu J, Li J Y, Han B, Lin H X. Advances in rice genetics and breeding by molecular design in China[J]. Scientia Sinica Vitae, 2019, 49: 1-28. (in Chinese with English abstract) | |
| [2] | Woodward A W, Bartel B. Auxin: Regulation, action, and interaction[J]. Annals of Botany, 2005, 95(5): 707-735. |
| [3] | Guilfoyle T J, Hagen G. Auxin response factors[J]. Current Opinion in Plant Biology, 2007, 10: 453-460. |
| [4] | Simonini S, Deb J, Moubayidin L, Stephenson P, Valluru M, Freire-Rios A, Sorefan K, Weijers D, Friml J, Østergaard L. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis[J]. Genes & Development, 2016, 30: 2286-2296. |
| [5] | Guilfoyle T, Hagen G, Ulmasov T, Murfett J. How does auxin turn on genes[J]. Plant Physiology, 1998, 118: 341-347. |
| [6] | Okushima Y, Overvoorde P J, Arima K, Alonso J M, Chan A, Chang C, Ecker J R, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19[J]. Plant Cell, 2005, 17(2): 444-463. |
| [7] | Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa)[J]. Gene, 2007, 394(1): 13-24. |
| [8] | Xing H, Pudake R N, Guo G, Xing G, Hu Z, Zhang Y, Sun Q, Ni Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize[J]. BMC Genomics, 2011, 12(1): 178. |
| [9] | Ha C V, Le D T, Nishiyama R, Watanabe Y, Sulieman S, Tran U T, Mochida K, Dong N V, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. The auxin response factor transcription factor family in soybean: genome-wide identification and expression analyses during development and water stress[J]. DNA Research, 2013, 20(5): 511-524. |
| [10] | Wu J, Wang F, Cheng L, Kong F, Peng Z, Liu S, Yu X, Lu G. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum[J]. Plant Cell Reports, 2011, 30(11): 2059-2073. |
| [11] | Liu S Q, Hu L F. Genome-wide analysis of the auxin response factor gene family in cucumber[J]. Genetics and Molecular Research, 2013, 12(4): 4317-4331. |
| [12] | 刘瑞娥, 胡长贵, 孙玉强. 植物生长素反应因子研究进展[J]. 植物生理学报, 2011, 47(7): 669-679. |
| Liu R E, Hu C G, Sun Y Q. Advances in plant auxin response factors[J]. Journal of Plant Physiology, 2011, 47(7): 669-679. (in Chinese with English abstract) | |
| [13] | 张赛娜. OsARF19调控水稻叶夹角的分子机制[D]. 杭州: 浙江大学, 2014: 32-33. |
| Zhang S N. Molecular mechanism of OsARF19 controlling leaf angle in rice (Oryza sativa)[D]. Hangzhou: Zhejiang University, 2014: 32-33. (in Chinese with English abstract) | |
| [14] | Ulmasov T, Hagen G, Guilfoyle T J. Activation and repression of transcription by auxin response factors[J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96: 5844-5849. |
| [15] | Waller F, Furuya M, Nick P. OsARF1, an auxin response factor from rice, is auxin-regulated and classifies as a primary auxin responsive gene[J]. Plant Molecular Biology, 2002, 50(3): 415-425. |
| [16] | 汪高航. 利用点突变研究水稻OsARF的功能[D]. 杭州: 浙江大学, 2010: 22-24. |
| Wang G H. Preliminary study of the functions of OsARFs by mutation[D]. Hangzhou: Zhejiang University, 2010: 22-24. (in Chinese with English abstract) | |
| [17] | 淳俊, 王文国, 王胜华, 陈放. 水稻(Oryza sativa L.)愈伤组织发育过程Auxin-miR167-ARF8信号通路的研究[J]. 四川大学学报, 2013, 50(4): 863-868. |
| Chun J, Wang W G, Wang S H, Chen F. The study on Auxin-miR167-ARF8 signal pathway during the growth and development process of rice callus[J]. Journal of Sichuan University, 2013, 50(4): 863-868. (in Chinese with English abstract) | |
| [18] | Shen C, Yue R, Yang Y, Zhang L, Sun T, Tie S, Wang H. OsARF16 is involved in cytokinin-mediated inhibition of phosphate transport and phosphate signaling in rice (Oryza sativa L.)[J]. PLoS One, 2014, 9(11): e112906. |
| [19] | Shen C, Yue R, Sun T, Zhang L, Yang Y, Wang H. OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.)[J]. Plant Science, 2015, 231: 148-158. |
| [20] | Sato Y, Nishimura A, Ito M, Ashikari M, Hirano H Y, Matsuoka M. Auxin response factor family in rice[J]. Genes & Genetic Systems, 2001, 76: 373-380. |
| [21] | Ellis C M, Nagpal P, Young J C, Hagen G, Guilfoyle T J, Reed J W. Auxin response factor1 and Auxin response factor1 regulate senescence and floral organ abscission in Arabidopsis thaliana[J]. Development, 2005, 132(20): 4563-4574. |
| [22] | Doudna J A, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096. |
| [23] | Belhaj K, Chaparro-Garcia A, Kamoun S, Patron N J, Nekrasov V. Editing plant genomes with CRISPR/ Cas9[J]. Current Opinion in Biotechnology, 2015, 32: 76-84. |
| [24] | 王加峰, 郑才敏, 刘维, 罗文龙, 王慧, 陈志强, 郭涛. 基于CRISPR/Cas9技术的水稻千粒重基因tgw6突变体的创建[J]. 作物学报, 2016, 42(8): 1160-1167. |
| Wang J F, Zheng C M, Liu W, Luo W L, Wang H, Chen Z Q, Guo T. Construction of tgw6 mutants in rice based on CRISPR/Cas9 technology[J]. Acta Agronomy Sinica, 2016, 42(8): 1160-1167. (in Chinese with English abstract) | |
| [25] | 周延彪, 赵新辉, 唐晓丹, 周在为, 庄楚雄, 杨远柱. 基于CRISPR/Cas9技术的水稻反光敏不育基因csa突变体的获得[J]. 杂交水稻, 2018, 33(6): 68-74. |
| Zhou Y B, Zhao X H, Tang X D, Zhou Z W, Zhuang C X, Yang Y Z. Acquisition of mutants of the reverse photoperiod-sensitive genic male sterility gene csa in rice based on CRISPR/Cas9 technology[J]. Hybrid Rice, 2018, 33(6): 68-74. (in Chinese with English abstract) | |
| [26] | 黄忠明, 周延彪, 唐晓丹, 赵新辉, 周在为, 符星学, 王凯, 史江伟, 李艳锋, 符辰建, 杨远柱. 基于CRISPR/Cas9技术的水稻温敏不育基因tms5突变体的构建[J]. 作物学报, 2018, 44(6): 844-851. |
| Huang Z M, Zhou Y B, Tang X D, Zhao X H, Zhou Z W, Fu X X, Wang K, Shi J W, Li Y F, Fu C J, Yang Y Z. Construction of tms5 mutants in rice based on CRISPR/Cas9 technology[J]. Acta Agronomy Sinica, 2018, 44(6): 844-851. (in Chinese with English abstract) | |
| [27] | Ma X L, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu Y G. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicotplants[J]. Molecular Plant, 2015, 8: 1274-1284. |
| [28] | Qi Y H, Wang S K, Shen C J, Zhang S N, Chen Y, Xu Y X, Liu Y, Wu Y R, Jiang D A. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa)[J]. New Phytologist, 2012, 193: 109-120. |
| [29] | Bibikova M, Beumer K, Trautman J K, Carroll D. Enhancing gene targeting with designed zinc finger nucleases[J]. Science, 2003, 300: 764. |
| [30] | Huang P, Xiao A, Zhou M G, Zhu Z Y, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs[J]. Nature Biotechnology, 2011, 29: 699-700. |
| [31] | 季新, 李飞, 晏云, 孙红正, 张静, 李俊周, 彭廷, 杜彦修, 赵全志. 基于CRISPR/Cas9系统的水稻光敏色素互作因子OsPIL15基因编辑[J]. 中国农业科学, 2017, 50(15): 2861-2871. |
| Ji X, Li F, Yan Y, Sun H Z, Zhang J, Li J Z, Peng T, Du Y X, Zhao Q Z. CRISPR/Cas9 system-based editing of phytochrome-interacting factor OsPIL15[J]. Scientia Agricultura Sinica, 2017, 50(15): 2861-2871. (in Chinese with English abstract) | |
| [32] | 唐丽, 李曜魁, 张丹, 毛毕刚, 吕启明, 胡远艺, 韶也, 彭彦, 赵炳然, 夏石头. 基于基因组编辑技术的水稻靶向突变特征及遗传分析[J]. 遗传, 2016, 38(8): 746-755. |
| Tang L, Li Y K, Zhang D, Mao B G, Lü Q M, Hu Y Y, Shao Y, Peng Y, Zhao B R, Xia S T. Characteristic and inheritance analysis of targeted mutagenesis mediated by genome editing in rice[J]. Hereditas, 2016, 38(8): 746-755. (in Chinese with English abstract) | |
| [33] | Xu M, Zhu L, Shou H, Wu P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice[J]. Plant Cell Physiology, 2005, 46(10): 1674-168. |
| [34] | Chen Y, Fan X, Song W, Zhang Y, Xu G. Over- expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1[J]. Plant Biotechnology Journal, 2012, 10(2): 139-149. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||