
Chinese Journal OF Rice Science ›› 2022, Vol. 36 ›› Issue (2): 139-149.DOI: 10.16819/j.1001-7216.2022.210308
• Research Papers • Previous Articles Next Articles
WANG Yongxiang, YAN Hangang, XU Hancong, FU Yushuang, SHAN Zhuangzhuang, HU Xiaoqing, ZHANG Wenwei*( ), JIANG Ling
), JIANG Ling
Received:2021-03-19
Revised:2021-05-12
Online:2022-03-10
Published:2022-03-11
Contact:
ZHANG Wenwei
王永祥, 燕海刚, 徐含聪, 傅玉双, 单壮壮, 胡晓晴, 张文伟*( ), 江玲
), 江玲
通讯作者:
张文伟
基金资助:WANG Yongxiang, YAN Hangang, XU Hancong, FU Yushuang, SHAN Zhuangzhuang, HU Xiaoqing, ZHANG Wenwei, JIANG Ling. Effects of OsESV1 on Starch Synthesis in Rice[J]. Chinese Journal OF Rice Science, 2022, 36(2): 139-149.
王永祥, 燕海刚, 徐含聪, 傅玉双, 单壮壮, 胡晓晴, 张文伟, 江玲. OsESV1基因对水稻淀粉合成的影响[J]. 中国水稻科学, 2022, 36(2): 139-149.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2022.210308
| 引物 Primer | 正向引物序列 Forward primer sequence (5′-3′) | 反向引物序列 Reverse primer sequence (5′-3′) |
|---|---|---|
| Crispr-OsESV1 | GGCAAGCACCTGGTACAGGGAGAG | AAACCTCTCCCTGTACCAGGTGCT |
| OsESV1-TI | AGCTCCTCAATCCCGTGATA | CTCATACGAAAATGTGGT |
| PAN580-OsESV1-GFP | TCCGGAGCTAGCTCTAGAATGGCCGCGTGCTCCAGG | CGCCCTTGCTCACCATGGATCCTTCCAGAGGCGAAGGAGGCA |
| 1305-OsESV1-GFP | TCCGGAGCTAGCTCTAGAATGGCCGCGTGCTCCAGG | CGCCCTTGCTCACCATGGATCCTTCCAGAGGCGAAGGAGGCA |
| QRT-OsESV1 | GTGATACTCCGCCGAAGAGA | TCGTCTCCACTCTCCCTGTA |
| Actin | TGCTATGTACGTCGCCATCCAG | AATGAGTAACCACGCTCCGTCA |
| AD-OsESV1 | CATGGAGGCCGAATTCATGGCCGCGTGCTCCAGG | CGAGCTCGATGGATCCTCATTCCAGAGGCGAAGGAGG |
| BD-OsESV1 | CATGGAGGCCGAATTCATGGCCGCGTGCTCCAG | GCAGGTCGACGGATCCTCATTCCAGAGGCGAAGGAGG |
| AD-OsAGPS1 | GGAGGCCAGTGAATTCATGGCGATGATGGCGATG | CGAGCTCGATGGATCCTTATATGACTGTTCCGCTAG |
| BD-OsAGPS1 | CATGGAGGCCGAATTCATGGCGATGATGGCGATG | GCAGGTCGACGGATCCTTATATGACTGTTCCGCTAG |
| AD-OsAGPS2a | GGAGGCCAGTGAATTCATGGCGATGGCGGCAGCCAT | CGAGCTCGATGGATCCTCATATAACTGTTCCGCTAG |
| AD-OsAGPS2b | CATGGAGGCCGAATTCATGAATGTATTGGCATCTAAG | CGAGCTCGATGGATCCTCATATAACTGTTCCGCTAGG |
| AD-OsAGPL1 | CATGGAGGCCGAATTCATGCAGTTCAGCAGTGTGTTT | CGAGCTCGATGGATCCCTATATGACCTTCCCGTCC |
| AD-SSⅠ | CATGGAGGCCGAATTCATGGCGACGGCGGCGGGGAT | CGAGCTCGATGGATCCTTACATGACATATGGTTGATC |
| AD-SSⅡa | CATGGAGGCCGAATTCATGTCGTCGGCCGTCGTCGCGTC | CGAGCTCGATGGATCCTCACCATTGGTACTTGGCCTT |
| AD-SSⅡb | CATGGAGGCCGAATTCTTCACCTCCTCTTCGCCGCG | CGAGCTCGATGGATCCTCACCACTGGTACTTGGCCTT |
| AD-BEI | GGAGGCCAGTGAATTCATGCTGTGTCTCACC | CGAGCTCGATGGATCCCTCATTTGCAGTCTTC |
| AD-BEⅡb | CATGGAGGCCGAATTCATGGCGGCGCCGGCGTCTG | CGAGCTCGATGGATCCTCATTCCGCTGGAGCATA |
| p2YC-OsESV1 | ATTTACGAACGATAGTTAATTAAATGGCGATGGCCGCGTGCTCCAGG | CACTGCCACCTCCTCCACTAGTTTCCAGAGGCGAAGGAGG |
| p2YN-OsAGPS2a | ATTTACGAACGATAGTTAATTAAATGGCGATGGCGGCAGCCAT | CACTGCCACCTCCTCCACTAGTTATAACTGTTCCGCTAGGG |
| p2YN-OsAGPS1 | ATTTACGAACGATAGTTAATTAAATGGCGATGGCGATGATGGCGATG | CACTGCCACCTCCTCCACTAGTTATGACTGTTCCGCTA |
Table 1 Primers used in this study.
| 引物 Primer | 正向引物序列 Forward primer sequence (5′-3′) | 反向引物序列 Reverse primer sequence (5′-3′) |
|---|---|---|
| Crispr-OsESV1 | GGCAAGCACCTGGTACAGGGAGAG | AAACCTCTCCCTGTACCAGGTGCT |
| OsESV1-TI | AGCTCCTCAATCCCGTGATA | CTCATACGAAAATGTGGT |
| PAN580-OsESV1-GFP | TCCGGAGCTAGCTCTAGAATGGCCGCGTGCTCCAGG | CGCCCTTGCTCACCATGGATCCTTCCAGAGGCGAAGGAGGCA |
| 1305-OsESV1-GFP | TCCGGAGCTAGCTCTAGAATGGCCGCGTGCTCCAGG | CGCCCTTGCTCACCATGGATCCTTCCAGAGGCGAAGGAGGCA |
| QRT-OsESV1 | GTGATACTCCGCCGAAGAGA | TCGTCTCCACTCTCCCTGTA |
| Actin | TGCTATGTACGTCGCCATCCAG | AATGAGTAACCACGCTCCGTCA |
| AD-OsESV1 | CATGGAGGCCGAATTCATGGCCGCGTGCTCCAGG | CGAGCTCGATGGATCCTCATTCCAGAGGCGAAGGAGG |
| BD-OsESV1 | CATGGAGGCCGAATTCATGGCCGCGTGCTCCAG | GCAGGTCGACGGATCCTCATTCCAGAGGCGAAGGAGG |
| AD-OsAGPS1 | GGAGGCCAGTGAATTCATGGCGATGATGGCGATG | CGAGCTCGATGGATCCTTATATGACTGTTCCGCTAG |
| BD-OsAGPS1 | CATGGAGGCCGAATTCATGGCGATGATGGCGATG | GCAGGTCGACGGATCCTTATATGACTGTTCCGCTAG |
| AD-OsAGPS2a | GGAGGCCAGTGAATTCATGGCGATGGCGGCAGCCAT | CGAGCTCGATGGATCCTCATATAACTGTTCCGCTAG |
| AD-OsAGPS2b | CATGGAGGCCGAATTCATGAATGTATTGGCATCTAAG | CGAGCTCGATGGATCCTCATATAACTGTTCCGCTAGG |
| AD-OsAGPL1 | CATGGAGGCCGAATTCATGCAGTTCAGCAGTGTGTTT | CGAGCTCGATGGATCCCTATATGACCTTCCCGTCC |
| AD-SSⅠ | CATGGAGGCCGAATTCATGGCGACGGCGGCGGGGAT | CGAGCTCGATGGATCCTTACATGACATATGGTTGATC |
| AD-SSⅡa | CATGGAGGCCGAATTCATGTCGTCGGCCGTCGTCGCGTC | CGAGCTCGATGGATCCTCACCATTGGTACTTGGCCTT |
| AD-SSⅡb | CATGGAGGCCGAATTCTTCACCTCCTCTTCGCCGCG | CGAGCTCGATGGATCCTCACCACTGGTACTTGGCCTT |
| AD-BEI | GGAGGCCAGTGAATTCATGCTGTGTCTCACC | CGAGCTCGATGGATCCCTCATTTGCAGTCTTC |
| AD-BEⅡb | CATGGAGGCCGAATTCATGGCGGCGCCGGCGTCTG | CGAGCTCGATGGATCCTCATTCCGCTGGAGCATA |
| p2YC-OsESV1 | ATTTACGAACGATAGTTAATTAAATGGCGATGGCCGCGTGCTCCAGG | CACTGCCACCTCCTCCACTAGTTTCCAGAGGCGAAGGAGG |
| p2YN-OsAGPS2a | ATTTACGAACGATAGTTAATTAAATGGCGATGGCGGCAGCCAT | CACTGCCACCTCCTCCACTAGTTATAACTGTTCCGCTAGGG |
| p2YN-OsAGPS1 | ATTTACGAACGATAGTTAATTAAATGGCGATGGCGATGATGGCGATG | CACTGCCACCTCCTCCACTAGTTATGACTGTTCCGCTA |
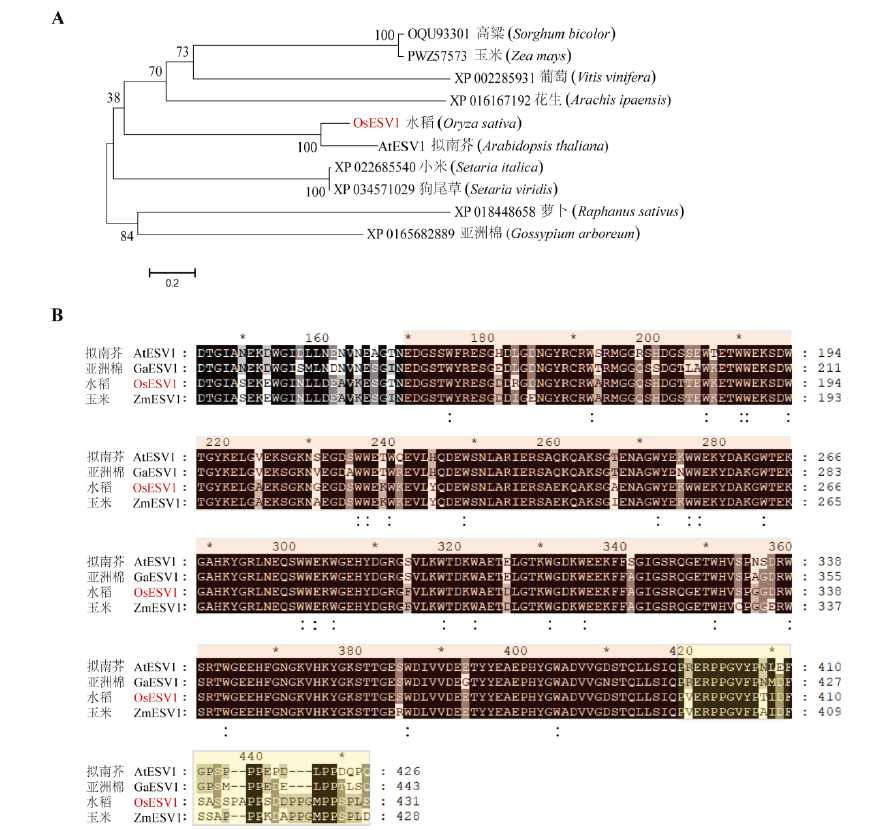
Fig. 1. The conservation analysis of OsESV1. A, Neighbor-joining tree of OsESV1 and its homologs. The proteins are named by GenBank protein accession numbers; B, Multiple sequence alignment of OsESV1 and its homologs. Asterisks represent the interval of twenty amino acids. The tryptophan (W)-rich region is marked by light red and those conserved tryptophan residues are indicated by colons. The proline (P)-rich region is marked by yellow.
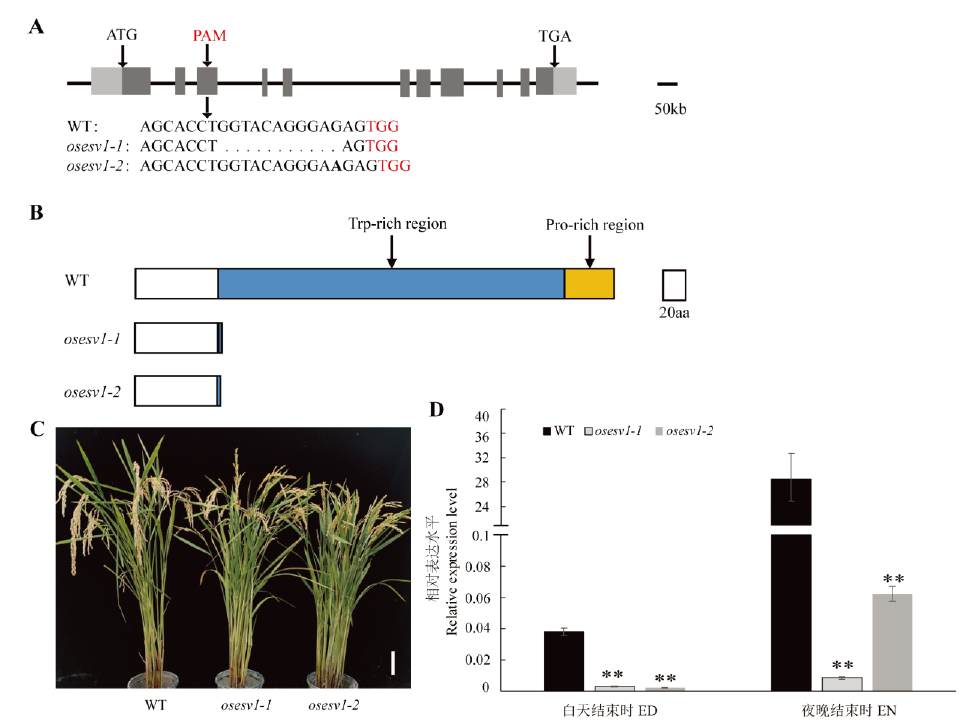
Fig. 2. Phenotype of the osesv1 mutants. A, Structure of the OsESV1 gene. The dark gray and light gray boxes represent the exon and UTR regions, respectively. The black lines connecting boxes are introns. The corresponding sequences of the wild type (WT) and osesv1 were compared below and a succession of points indicates the deleted sequence in osesv1. The inserted base is shown in bold. TGG (PAM) is marked by red; B, The protein structure of WT and osesv1; Arrows indicate the tryptophan-rich region and proline-rich region; C, WT and osesv1 plants. Bar=10cm; D, Expression level of OsESV1 in WT and osesv1 seedlings at end of day(ED) and end of night(EN). Values are mean ± SD (n=3). * and **indicate significant difference at 0.05 and 0.01 level, respectively (t-test). The same below.
| 材料 Materials | 株高 Plant height /cm | 穗长 Length of panicle /cm | 分蘖数 Tiller number | 每穗粒数 Grain number per panicle | 一次枝梗数 Primary rachis branch number | 千粒重 1000-grain weight /g |
|---|---|---|---|---|---|---|
| 野生型WT | 94.80±3.10 | 19.31±0.77 | 26.13±8.55 | 127.58±9.33 | 8.60±0.83 | 22.57±1.07 |
| osesv1-1 | 90.96±4.44** | 17.82±0.52** | 33.43±10.45* | 112.11±15.42* | 8.44±0.96 | 21.50±0.56 |
| osesv1-2 | 90.47±5.40** | 17.69±0.82** | 43.08±13.85** | 113.63±10.71** | 9.00±0.95 | 23.01±0.85 |
Table 2 Agronomic traits of the wild type (WT) and osesv1.
| 材料 Materials | 株高 Plant height /cm | 穗长 Length of panicle /cm | 分蘖数 Tiller number | 每穗粒数 Grain number per panicle | 一次枝梗数 Primary rachis branch number | 千粒重 1000-grain weight /g |
|---|---|---|---|---|---|---|
| 野生型WT | 94.80±3.10 | 19.31±0.77 | 26.13±8.55 | 127.58±9.33 | 8.60±0.83 | 22.57±1.07 |
| osesv1-1 | 90.96±4.44** | 17.82±0.52** | 33.43±10.45* | 112.11±15.42* | 8.44±0.96 | 21.50±0.56 |
| osesv1-2 | 90.47±5.40** | 17.69±0.82** | 43.08±13.85** | 113.63±10.71** | 9.00±0.95 | 23.01±0.85 |

Fig. 3. Starch contents in leaves of WT and osesv1. A and B show iodine staining and starch contents of WT and osesv1 leaves at end of day and end of night, respectively.
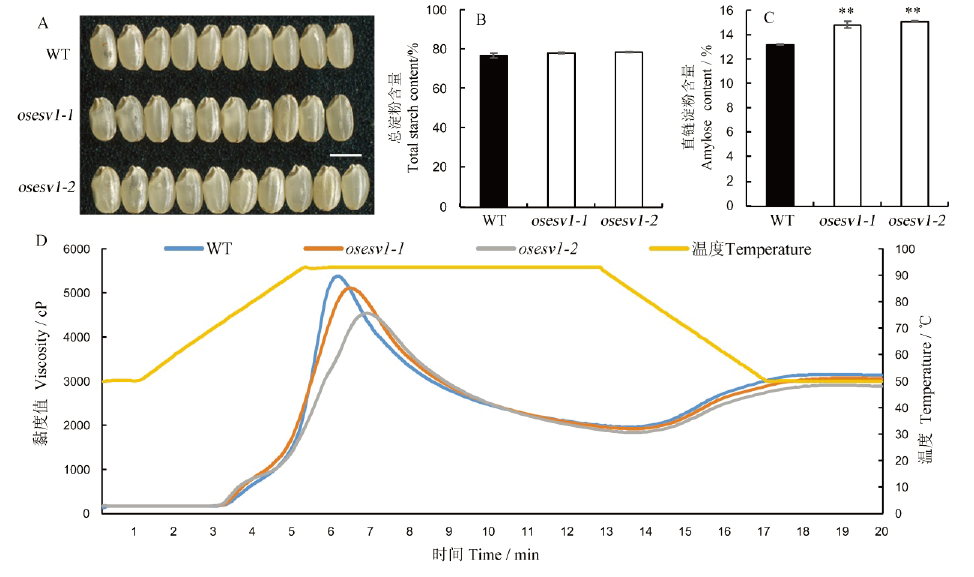
Fig. 4. Phenotype of osesv1 grains and physicochemical properties of endosperm starch. A, Phenotype of WT and osesv1 mature seeds. Bar=5 mm; B, Total starch contents of WT and osesv1 grains; C, Amylose content of WT and osesv1 grains; D, Viscosity profiles of endosperm starch of WT and osesv1.
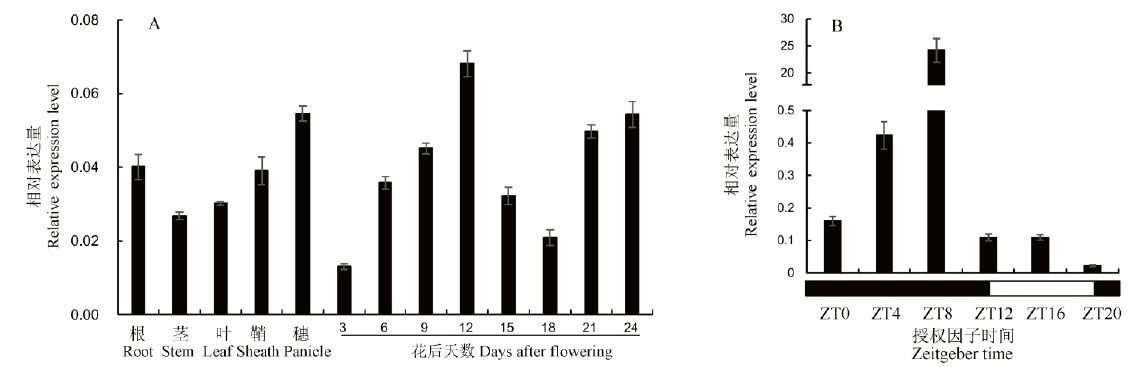
Fig. 5. Expression pattern of OsESV1. A, Expression level of OsESV1 in various tissues and endosperm at different developmental stages of WT; B, Diurnal expression pattern of OsESV1. The black and white boxes represent the timelines of night and day, respectively.
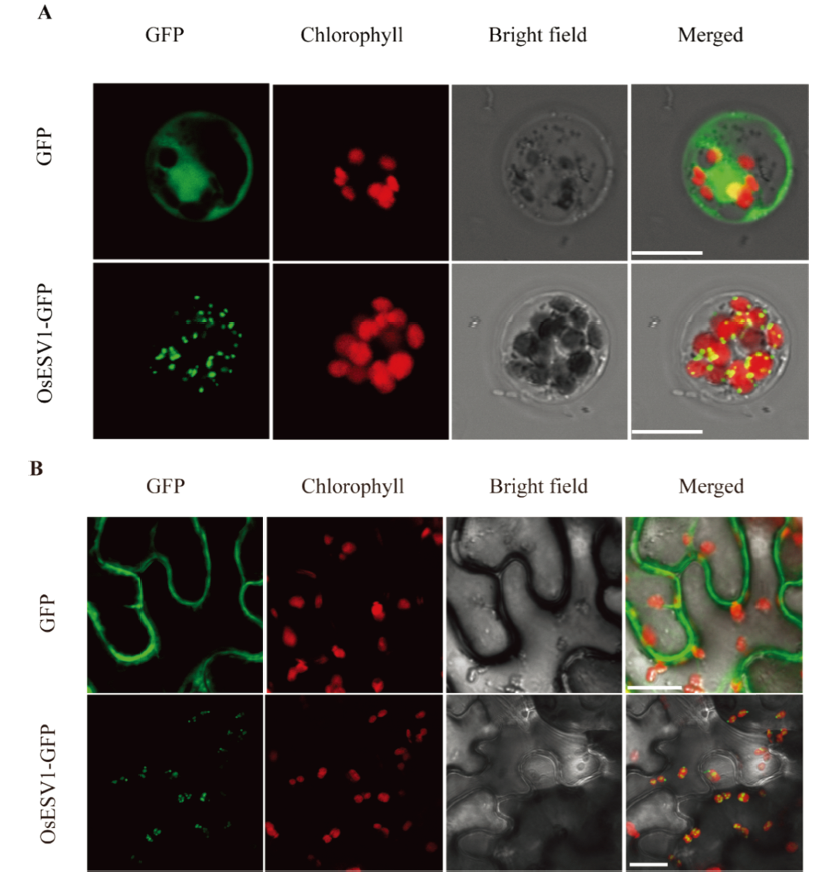
Fig. 6. Subcellular localization of OsESV1. A, Transient expression of OsESV1-GFP fusion protein in rice protoplasts. The empty vector pAN580-GFP was used as control. Bars=20 μm; B, Transient expression of OsESV1-GFP fusion protein in tobacco epidermal cells. The empty vector p1305-GFP was used as control. Bars=20 μm.
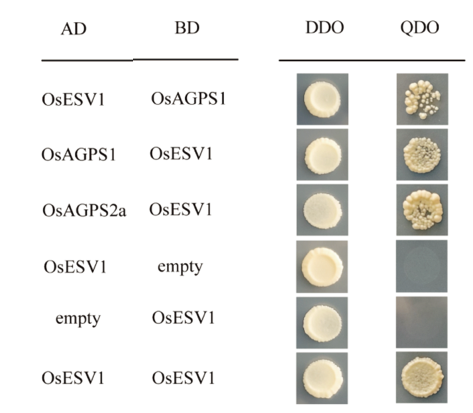
Fig. 7. Yeast two-hybrid assays show that OsESV1 can interact with OsAGPS2a and OsAGPS1. DDO(double dropout supplements), SD-Leu/-Trp; QDO(quadruple dropout supplements), SD-Leu/-Trp/ -His/-Ade.
| [1] | 侯立刚, 周广春, 严永峰, 全成哲, 马巍. 吉林省水稻产业发展现状与未来发展对策[J]. 北方水稻, 2015,45(2):73-75. |
| Hou L G, Zhou G C, Yan Y F, Quan C Z, Ma W. Analysis on the development status and the counter- measures of rice industry in Jilin[J]. North Rice, 2015,45(2):73-75. (in Chinese with English abstract) | |
| [2] | 方鹏飞, 李三峰, 焦桂爱, 谢黎虹, 胡培松, 魏祥进, 唐绍清. 水稻粉质胚乳突变体flo7的理化性质及基因定位[J]. 中国水稻科学, 2014,28(5):447-457. |
| Fang P F, Li S F, Jiao G A, Xie L H, Hu P S, Wei X J, Tang S Q. Physicochemical property analysis and gene mapping of a floury endosperm mutant flo7 in rice[J]. Chinese Journal of Rice Science, 2014,28(5):447-457. (in Chinese with English abstract) | |
| [3] | 蔡跃. 水稻心白胚乳突变体w59的基因克隆及功能分析[D]. 南京: 南京农业大学, 2018. |
| Cai Y. Gene cloning and functional analysis of rice white-core endosperm mutant w59[D]. Nanjing: Nanjing Agricultural University, 2018. (in Chinese with English abstract) | |
| [4] | Tetlow I J, Emes M J. Starch biosynjournal in the developing endosperms of grasses and cereals[J]. Agronomy, 2017,7(4):81. |
| [5] | Okita T W, Nakata P A, Anderson J M, Sowokinos J, Morell M, Preiss J. The subunit structure of potato tuber ADPglucose pyrophosphorylase[J]. Plant Physiology, 1990,93(2):785-790. |
| [6] | Geigenberger P. Regulation of starch biosynjournal in response to a fluctuating environment[J]. Plant Physiology, 2011,155(4):1566-1577. |
| [7] | Kaushik R P, Khush G S. Genetic analysis of endosperm mutants in rice Oryza sativa L.[J] Theoretical and Applied Genetics, 1991,83(2):146-152. |
| [8] | Wu Y, Pu C, Lin H, Huang H, Huang Y, Hong C, Chang M, Lin Y. Three novel alleles of FLOURY ENDOSPERM2 (FLO2) confer dull grains with low amylose content in rice[J]. Plant Science, 2015,233:44-52. |
| [9] | Nishio T, Iida S. Mutants having a low content of 16-kDa allergenic protein in rice (Oryza sativa L)[J]. Theoretical & Applied Genetics, 1993,86(2-3):317-321. |
| [10] | Kang H G, Park S, Matsuoka M, An G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate ortho- phosphate dikinase gene (OsPPDKB)[J]. The Plant Journal, 2010,42(6):901-911. |
| [11] | Ryoo N, Yu C, Park C S, Baik M Y, Baik M Y, Park I M, Cho M H, Bhoo S H, An G, Hahn T R, Jeon J S. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.)[J]. Plant Cell Reports, 2007,26(7):1083-1095. |
| [12] | Peng C, Wang Y, Liu F, Ren Y, Zhou K, Lü J, Zheng M, Zhao S, Zhang L, Wang C, Jiang L, Zhang X, Guo X, Bao Y, Wan J. FLOURY ENDOSPERM 6 encodes a CBM 48 domain-containing protein involved in compound granule formation and starch synjournal in rice endosperm[J]. The Plant Journal, 2014,77(6):917-930. |
| [13] | Zhang L, Ren Y, Lu B, Yang C, Feng Z, Liu Z, Chen J, Ma W, Wang Y, Yu X, Wang Y, Zhang W, Wang Y, Liu S, Wu F, Zhang X, Guo X, Bao Y, Jiang L, Wan J. FLOURY ENDOSPERM7 encodes a regulator of starch synjournal and amyloplast development essential for peripheral endosperm development in rice[J]. Journal of Experimental Botany, 2016(3):633-647. |
| [14] | Long W, Dong B, Wang Y, Pan P, Wang Y, Liu L, Chen X, Liu X, Liu S, Tian Y, Chen L, Wan J. FLOURY ENDOSPERM8, encoding the UDP-glucose pyrophos- phorylase 1, affects the synjournal and structure of starch in rice endosperm[J]. Journal of Plant Biology, 2017,60(5):513-522. |
| [15] | 刘艺, 朱小品, 刘喜, 田云录, 刘世家, 王云龙, 张文伟, 江玲, 王益华, 万建民. 水稻胚乳粉质突变体 flo9 的表型分析和基因定位[J]. 南京农业大学学报, 2018,41(4):616-624. |
| Liu Y, Zhu X P, Liu X, Tian Y L, Liu S J, Wang Y L, Zhang W W, Jiang L, Wang Y H, Wan J M. Phenotyping and gene-mapping of a floury endosperm mutant flo9 in rice[J]. Journal of Nanjing Agricultural University, 2018,41(4):616-624. (in Chinese with English abstract) | |
| [16] | Wu M, Ren Y, Cai M, Wang Y, Zhu S, Zhu J, Hao Y, Teng X, Zhu X, Jing R, Zhang H, Zhong M, Wang Y, Lei C, Zhang X, Guo X, Cheng Z, Lin Q, Wang J, Jiang L, Bao Y, Wang Y, Wan J. Rice FLOURY ENDOSPERM 10 encodes a pentatricopeptide repeat protein that is essential for the trans-splicing of mitochondrial nad1 intron 1 and endosperm development[J]. New Phytologist, 2019,223(2):736-750. |
| [17] | Zhu X, Teng X, Wang Y, Hao Y, Jing R, Wang Y, Liu Y, Zhu J, Wu M, Zhong M, Chen X, Zhang Y, Zhang W, Wang C, Wang Y, Wan J. FLOURY ENDOSPERM11 encoding a plastid heat shock protein 70 is essential for amyloplast development in rice[J]. Plant Science, 2018,277:89-99. |
| [18] | Zhong M, Liu X, Liu F, Ren Y, Wang Y, Zhu J, Teng X, Duan E, Wang F, Zhang H, Wu M, Hao Y, Zhu X, Jing R, Guo X, Jiang L, Wang Y, Wan J. FLOURY ENDOSPERM12 encoding alanine aminotransferase 1 regulates carbon and nitrogen metabolism in rice[J]. Journal of Plant Biology, 2019,62(1):61-73. |
| [19] | Hu T, Tian Y, Zhu J, Wang Y, Jing R, Lei J, Sun Y, Yu Y, Li J, Chen X, Zhu X, Hao Y, Liu L, Wang Y, Wan J. OsNDUFA9 encoding a mitochondrial complex I subunit is essential for embryo development and starch synjournal in rice[J]. Plant Cell Reports, 2018,37(12):1667-1679. |
| [20] | Xue M, Liu L, Yu Y, Zhu J, Gao H, Wang Y, Wan J. Lose-of-function of a rice nucleolus-localized pentatrico- peptide repeat protein is responsible for the floury endosperm14 mutant phenotypes[J]. Rice, 2019,12(1):1-15. |
| [21] | You X, Zhang W, Hu J, Jing R, Cai Y, Feng Z, Kong F, Zhang J, Yan H, Chen W, Chen X, Ma J, Tang X, Wang P, Zhu S, Liu L, Jiang L, Wan J. FLOURY ENDOSPERM15 encodes a glyoxalase I involved in compound granule formation and starch synjournal in rice endosperm[J]. Plant Cell Reports, 2019,38(3):345-359. |
| [22] | Teng X, Zhong M, Zhu X, Wang C, Ren Y, Wang Y, Zhang H, Jiang L, Wang D, Hao Y, Wu M, Zhu J, Zhang X, Guo X, Wang Y, Wan J. FLOURY ENDOSPERM16 encoding a NAD-dependent cytosolic malate dehydrogenase plays an important role in starch synjournal and seed development in rice[J]. Plant Biotechnology Journal, 2019,17(10):1914-1927. |
| [23] | Feike D, Seung D, Graf A, Bischof S, Ellick T, Coiro M, Soyk S, Eicke S, Mettler-Altmann T, Lu K J, Trick M, Zeeman S C, Smith A M. The starch granule-associated protein EARLY STARVATION1 is required for the control of starch degradation in Arabidopsis thaliana leaves[J]. The Plant Cell, 2016,28(6):1472-1489. |
| [24] | Malinova I, Mahto H, Brandt F, AL-Rawi S, Qasim H, Brust H, Hejazi M, Fettk J. EARLY STARVATION 1 specifically affects the phosphorylation action of starch- related dikinases[J]. The Plant Journal, 2018,95(1):126-137. |
| [25] | 汪秉琨, 张慧, 洪汝科, 张锦, 杨睿, 罗琼, 曾千春. CRISPR/Cas9系统编辑水稻Wx基因[J]. 中国水稻科学, 2018,32(1):35-42. |
| Wang B K, Zhang H, Hong R K, Zhang J W, Yang R, Luo Q, Zeng Q C. Wx gene editing via CRISPR/Cas9 system in rice[J]. Chinese Journal of Rice Science, 2018,32(1):35-42. (in Chinese with English abstract) | |
| [26] | 曾千春, 李旭刚, 马炳田, 陈松彪, 徐鸿林, 孟昆, 魏晓丽, 朱祯. 有效去除农杆菌和籼稻转化系统优化[J]. 分子植物育种, 2003,1(5):783-790. |
| Zeng Q C, Li X G, Ma B T, Chen S B, Xu H L, Meng K, Wei X L, Zhu Z. Efficient elimination of A. tumefaciens and optimization of Agrobacterium-mediated transformation of indica rice[J]. Molecular Plant Breeding, 2003,1(5):783-790. (in Chinese with English abstract) | |
| [27] | 王慧娜, 初志战, 马兴亮, 李日清, 刘耀光. 高通量的PCR 模板植物基因组 DNA 制备方法[J]. 作物学报, 2013,39(7):1200-1205. |
| Wang H N, Chu Z Z, Ma X L, Li R Q, Liu Y G. A high through-put protocol of plant genomic DNA preparation for PCR[J]. Acta Agronomica Sinica, 2013,39(7):1200-1205. (in Chinese with English abstract) | |
| [28] | Wang W, Wei X, Jiao G, Chen W, Wu Y, Sheng Z, Hu S, Xie L, Wang J, Tang S, Hu P. GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield[J]. Journal of Integrative Plant Biology, 2020,62(7):948-966. |
| [29] | Tong C, Chen Y, Tang F, Xu F, Huang Y, Chen H, Bao J. Genetic diversity of amylose content and RVA pasting parameters in 20 rice accessions grown in Hainan, China[J]. Food Chemistry, 2014,161:239-245. |
| [30] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method[J]. Methods, 2001,25(4):402-408. |
| [31] | Chen S, Tao L, Zeng L, Vega-Sanchez M E, Umemura K, Wang G. A highly efficient transient protoplast system for analyzing defence gene expression and protein- protein interactions in rice[J]. Molecular Plant Pathology, 2010,7(5):417-427. |
| [32] | Waadt R, Kudla J. In planta visualization of protein interactions using bimolecular fluorescence comple- mentation (BiFC)[J]. Cold Spring Harbor Protocols, 2008(4): pdb, prot4995. |
| [33] | Ohdan T, Francisco P B, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. Expression profiling of genes involved in starch synjournal in sink and source organs of rice[J]. Journal of Experimental Botany, 2005,56(422):3229-3244. |
| [34] | Lee S K, Hwang S K, Han M, Eom J S, Kang H G, Han Y, Choi S B, Cho M H, Bhoo S H, An G, Hahn T R, Okita T W, Jeon J S. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synjournal in the leaf and seed endosperm of rice (Oryza sativa L.)[J]. Plant Molecular Biology, 2007,65(4):531-546. |
| [35] | Seung D, Soyk S, Coiro M, Maier B A, Eicke S, Zeeman S C. PROTEIN TARGETING TO STARCH is required for localizing GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synjournal in Arabidopsis[J]. PLOS Biology, 2015,13(2):e1002080. |
| [36] | Abt M R, Zeeman S C. Evolutionary innovations in starch metabolism[J]. Current Opinion in Plant Biology, 2020,55:109-117. |
| [37] | Sakulsingharoj C, Choi S B, Hwang S K, Edwards G E, Bork J, Meyer C R, Preiss J, Okita T W. Engineering starch biosynjournal for increasing rice seed weight: the role of the cytoplasmic ADP-glucose pyropho- sphorylase[J]. Plant Science, 2004,167(6):1323-1333. |
| [38] | Cook F R, Fahy B, Trafford K. A rice mutant lacking a large subunit of ADP-glucose pyrophosphorylase has drastically reduced starch content in the culm but normal plant morphology and yield[J]. Functional Plant Biology, 2012,39:1068-1078. |
| [39] | Comparot-Moss S, Kötting O, Stettler M, Edner C, Graf A, Weise S E, Streb S, Lue W L, MacLean D, Mahlow S, Ritte G, Steup M, Chen J, Zeeman S C, Smith A M. A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves[J]. Plant Physiology, 2010,152(2):685-697. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||