
Chinese Journal OF Rice Science ›› 2021, Vol. 35 ›› Issue (3): 259-268.DOI: 10.16819/j.1001-7216.2021.01204
• Research Papers • Previous Articles Next Articles
Xuemei DENG1,#, Peng HU1,#, Yueying WANG1, Yi WEN1, Yiqing TAN1, Hao WU2, Kaixiong WU1, Junge WANG1, Linlin HOU1, Lixin ZHU1, Li ZHU1, Guang CHEN1, Dali ZENG1, Guangheng ZHANG1, Longbiao GUO1, Zhenyu GAO1, Deyong REN1, Qian QIAN1,*( ), Jiang HU1,*(
), Jiang HU1,*( )
)
Received:2020-06-10
Revised:2020-08-06
Online:2021-05-10
Published:2021-05-10
Contact:
Qian QIAN, Jiang HU
About author:#These authors contributed equally to this work;
邓雪梅1,#, 胡鹏1,#, 王月影1, 文艺1, 谭义青1, 伍豪2, 吴凯雄1, 王俊格1, 侯琳琳1, 朱黎欣1, 朱丽1, 陈光1, 曾大力1, 张光恒1, 郭龙彪1, 高振宇1, 任德勇1, 钱前1,*( ), 胡江1,*(
), 胡江1,*( )
)
通讯作者:
钱前,胡江
作者简介:#共同第一作者;
基金资助:Xuemei DENG, Peng HU, Yueying WANG, Yi WEN, Yiqing TAN, Hao WU, Kaixiong WU, Junge WANG, Linlin HOU, Lixin ZHU, Li ZHU, Guang CHEN, Dali ZENG, Guangheng ZHANG, Longbiao GUO, Zhenyu GAO, Deyong REN, Qian QIAN, Jiang HU. Identification and Fine Mapping of a Grain Width Mutant gw4 in Rice[J]. Chinese Journal OF Rice Science, 2021, 35(3): 259-268.
邓雪梅, 胡鹏, 王月影, 文艺, 谭义青, 伍豪, 吴凯雄, 王俊格, 侯琳琳, 朱黎欣, 朱丽, 陈光, 曾大力, 张光恒, 郭龙彪, 高振宇, 任德勇, 钱前, 胡江. 水稻粒宽突变体gw4的鉴定与基因定位[J]. 中国水稻科学, 2021, 35(3): 259-268.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2021.01204
| 分子标记 | 正向引物序列(5'-3') | 反向引物序列(5'-3') | 实验目的 | |
|---|---|---|---|---|
| Marker | Forward primer sequence (5'-3') | Reverse primer sequence (5'-3') | Purpose | |
| BS6 | AGTACACATGGGGAGTAGTTGG | GATGGCAATTGAAAAGTGACC | 定位Mapping | |
| BS11 | TGCAGGTACACACACATACCC | GGAAGTAGCATCCATGCAACTA | 定位Mapping | |
| BS15 | TTTGCAGCTGGCTAGCTTA | ACTTCCATAATTACGGGTCCT | 定位Mapping | |
| BS16 | GGCAAATAATTAATAGCATGGTCC | TGGATGATTTCTGTTAGTGGTTCA | 定位Mapping | |
| BS18 | TGGTTACAACGACACAACTGC | TCACTCATACCAACCTTGCG | 定位Mapping | |
| EX49 | GTGCCTCCTTTTCTTGGAGAT | CACAGTCATCTCACTTGCAATC | 定位Mapping | |
| BS19 | ACGTGTCTTGGGGCTGTGTC | TCCCTACACTTATTGCCCCTCT | 测序Sequencing | |
| BS20 | TCCTCATTACATGTCCCCATCC | CCCTATCTGAACGCAAGTGCT | 测序Sequencing | |
| BS21 | GTGGTAGCATGTGCTCTCCCT | AGGGTTGGCAGAGTTGTCAGA | 测序Sequencing | |
| BS22 | TACCATGTTGCACCTGAATACCT | CAACCCTCCTGGTGATTCCC | 测序Sequencing | |
| BS23 | CCAGTTGTTAGGGCTGTGTCTG | AGATTAGGGAGCCAATGCGA | 测序Sequencing | |
| BS24 | GATGCTCTGTTTGGACAGCCT | TGTCGTCAGCATGTTGCCTG | 测序Sequencing | |
| BS25 | ACCAACCAACCCCTTGAGTCT | TGGAAATCGGAGAGATTATACGTAC | 测序Sequencing | |
| Histon | GGTCAACTTGTTGATTCCCCTCT | AACCGCAAAATCCAAAGAACG | RT-PCR | |
| RT-GW2 | ATCGTGGACAAGGGCACCTGC | CCGTCCCGCTCGCGAGGCATG | RT-PCR | |
| RT-GW5 | AATCCAGACGGCATTCAGAG | CTTCACCAGCGCCTTGAG | RT-PCR | |
| RT-GW7 | CCCTAGCATCGACACCAAGT | TCGAGGACAGAGATGGGACT | RT-PCR | |
| RT-GW8 | ACAGCCAGATCCCATGAACT | GCGTGTAGTATGGGCTCTCC | RT-PCR | |
| RT-GS2 | TGCGTCCCTTCTTTGATGAGT | ACAGTTGGGTGCCTGAGAATG | RT-PCR | |
| RT-GS3 | CGGAAGAACTCCTGATCCATTC | CACTTGCTCTGCACAAACAGC | RT-PCR | |
| RT-GS5 | GTTCTCGGTACTGCGTGGAAG | ACTCCACAAACCTCCCAGCA | RT-PCR | |
| RT-GL6 | TCCAAACCAGATGTTGGTGA | GCCGTCGACTACATCTCGTT | RT-PCR | |
| RT-LOC_Os04g01590 | TTGGTGATGTCCCCATACAA | ACCAAATCTCTTCCCCTGCT | RT-PCR | |
Table 1. Markers developed for mapping of gw4 and quantitative real-time PCR (qRT- PCR) analysis.
| 分子标记 | 正向引物序列(5'-3') | 反向引物序列(5'-3') | 实验目的 | |
|---|---|---|---|---|
| Marker | Forward primer sequence (5'-3') | Reverse primer sequence (5'-3') | Purpose | |
| BS6 | AGTACACATGGGGAGTAGTTGG | GATGGCAATTGAAAAGTGACC | 定位Mapping | |
| BS11 | TGCAGGTACACACACATACCC | GGAAGTAGCATCCATGCAACTA | 定位Mapping | |
| BS15 | TTTGCAGCTGGCTAGCTTA | ACTTCCATAATTACGGGTCCT | 定位Mapping | |
| BS16 | GGCAAATAATTAATAGCATGGTCC | TGGATGATTTCTGTTAGTGGTTCA | 定位Mapping | |
| BS18 | TGGTTACAACGACACAACTGC | TCACTCATACCAACCTTGCG | 定位Mapping | |
| EX49 | GTGCCTCCTTTTCTTGGAGAT | CACAGTCATCTCACTTGCAATC | 定位Mapping | |
| BS19 | ACGTGTCTTGGGGCTGTGTC | TCCCTACACTTATTGCCCCTCT | 测序Sequencing | |
| BS20 | TCCTCATTACATGTCCCCATCC | CCCTATCTGAACGCAAGTGCT | 测序Sequencing | |
| BS21 | GTGGTAGCATGTGCTCTCCCT | AGGGTTGGCAGAGTTGTCAGA | 测序Sequencing | |
| BS22 | TACCATGTTGCACCTGAATACCT | CAACCCTCCTGGTGATTCCC | 测序Sequencing | |
| BS23 | CCAGTTGTTAGGGCTGTGTCTG | AGATTAGGGAGCCAATGCGA | 测序Sequencing | |
| BS24 | GATGCTCTGTTTGGACAGCCT | TGTCGTCAGCATGTTGCCTG | 测序Sequencing | |
| BS25 | ACCAACCAACCCCTTGAGTCT | TGGAAATCGGAGAGATTATACGTAC | 测序Sequencing | |
| Histon | GGTCAACTTGTTGATTCCCCTCT | AACCGCAAAATCCAAAGAACG | RT-PCR | |
| RT-GW2 | ATCGTGGACAAGGGCACCTGC | CCGTCCCGCTCGCGAGGCATG | RT-PCR | |
| RT-GW5 | AATCCAGACGGCATTCAGAG | CTTCACCAGCGCCTTGAG | RT-PCR | |
| RT-GW7 | CCCTAGCATCGACACCAAGT | TCGAGGACAGAGATGGGACT | RT-PCR | |
| RT-GW8 | ACAGCCAGATCCCATGAACT | GCGTGTAGTATGGGCTCTCC | RT-PCR | |
| RT-GS2 | TGCGTCCCTTCTTTGATGAGT | ACAGTTGGGTGCCTGAGAATG | RT-PCR | |
| RT-GS3 | CGGAAGAACTCCTGATCCATTC | CACTTGCTCTGCACAAACAGC | RT-PCR | |
| RT-GS5 | GTTCTCGGTACTGCGTGGAAG | ACTCCACAAACCTCCCAGCA | RT-PCR | |
| RT-GL6 | TCCAAACCAGATGTTGGTGA | GCCGTCGACTACATCTCGTT | RT-PCR | |
| RT-LOC_Os04g01590 | TTGGTGATGTCCCCATACAA | ACCAAATCTCTTCCCCTGCT | RT-PCR | |
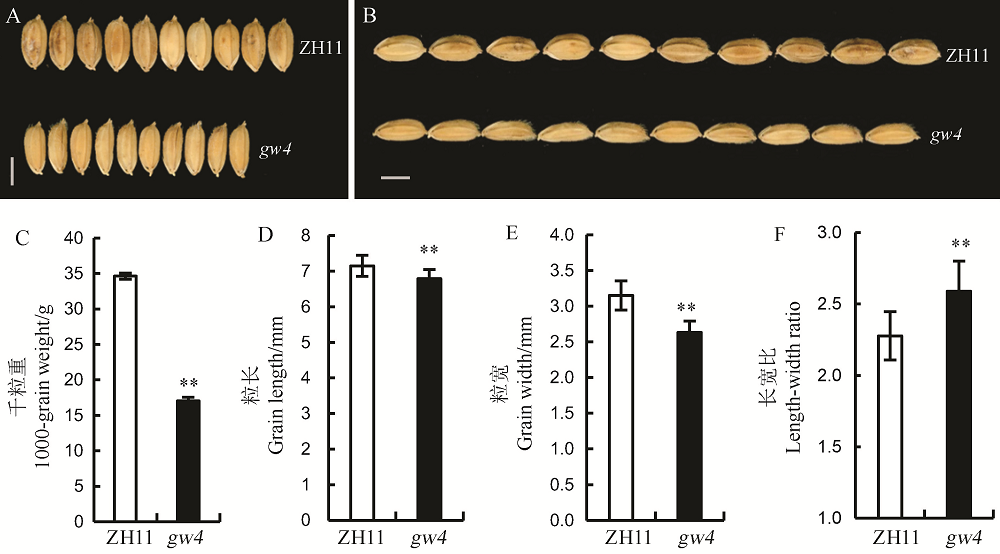
Fig. 1. Grains size of the wild type Zhonghua 11(ZH11) and its mutant gw4. A and B, Mature grain of WT and gw4, bar= 0.25 cm; C-F, Bars represent standard error(n=13). **indicate significant difference between WT and gw4 by t-test(P<0.01). ZH11, Zhonghua 11.
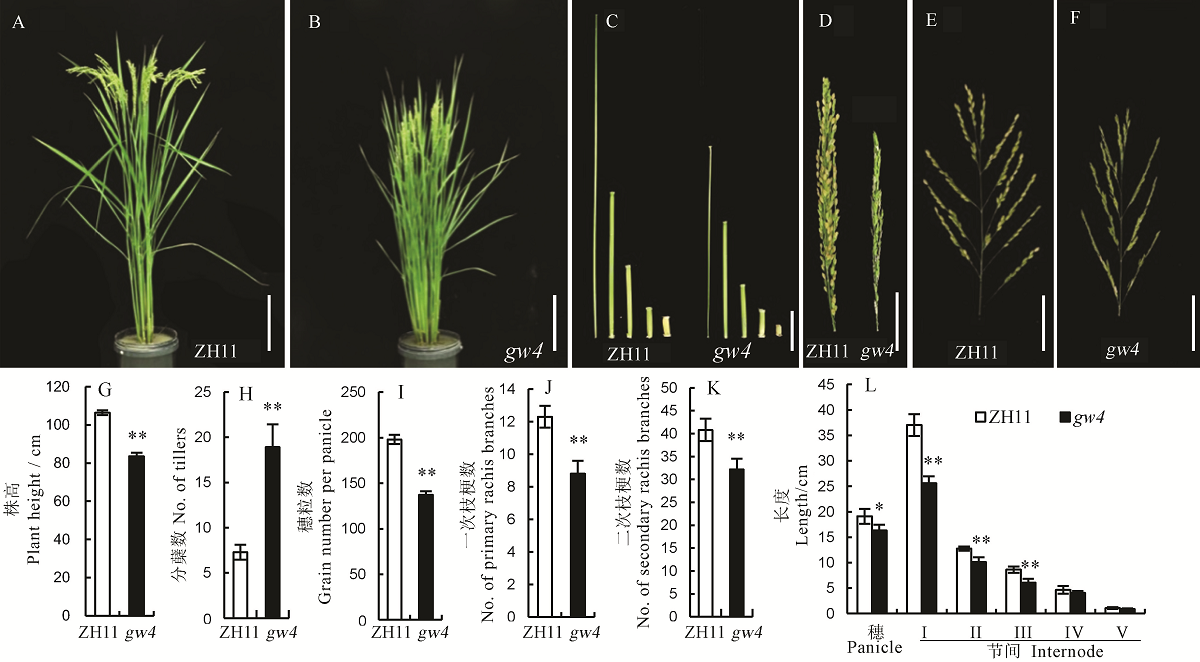
Fig. 2. Phenotype of the wild type Zhonghua 11(ZH11) and its mutant gw4. A and B, Phenotype of Zhonghua 11(ZH11) and gw4, bar=20 cm; C, Internodes, bar=5 cm; D, E and F, Panicles, bar=5 cm; G-L, Bars represent standard error (n=10). ** and * indicate significant difference between WT and gw4 by t-test(P<0.01, P<0.05), respectively.
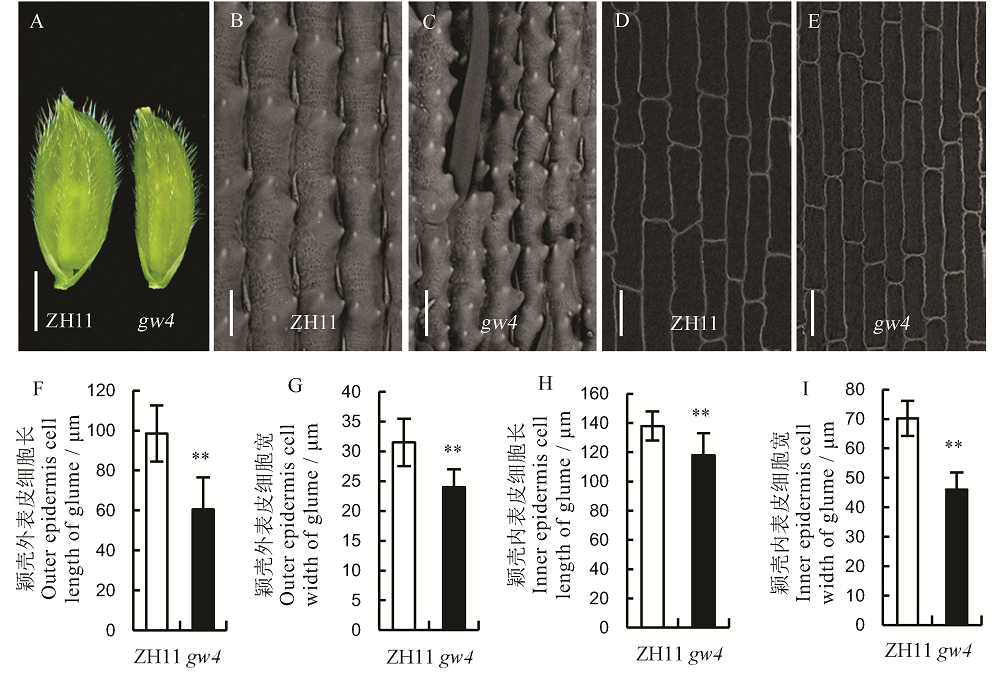
Fig. 3. Glumes of the wild type(ZH11) and its mutant gw4. A, Glume, bar=0.25 cm; B and C, Outer epidermis of glume, bar=50 μm; D and E, Inner epidermis of glume; bar=50 μm; Bars represent standard error (n=10). **indicate significant difference between WT and gw4 by t-test(P<0.01).
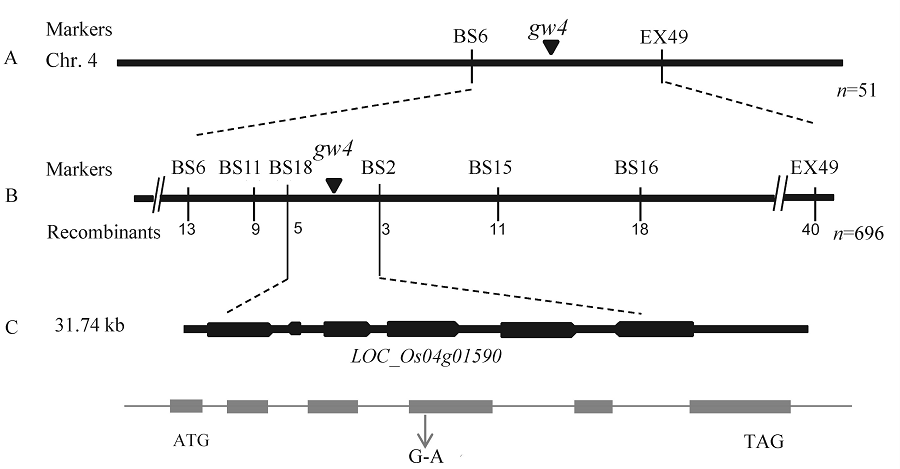
Fig. 4. Fine mapping of gw4. A, gw4 was primarily mapped on chromosome 4; B, gw4 was narrowed to a 31.74 kb genomic region; C, Total six genes was predicted in this region.
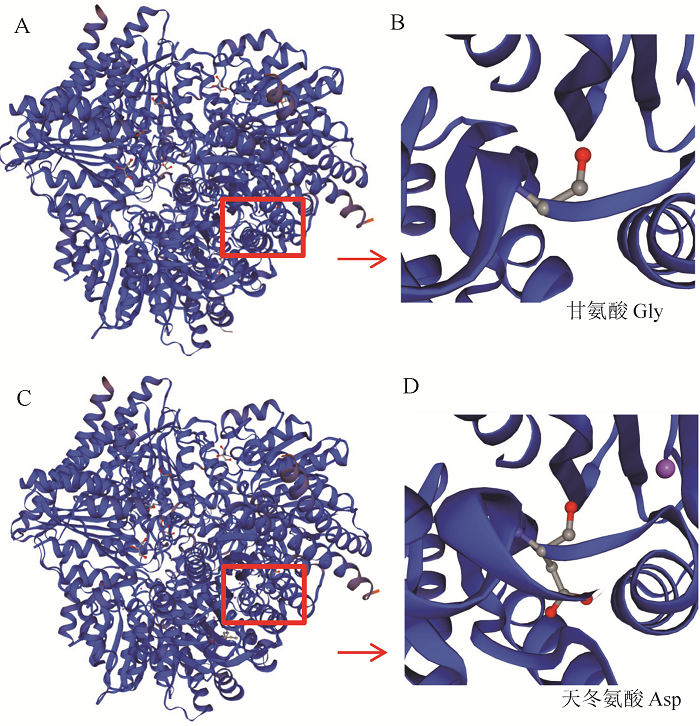
Fig. 5. Three-dimensional structures of proteins GW4 and gw4. A and B, Protein domain of GW4; C and D, Protein domain of gw4; B and D, Enlarged size of A and C; The red box indicate the differences in protein folding.
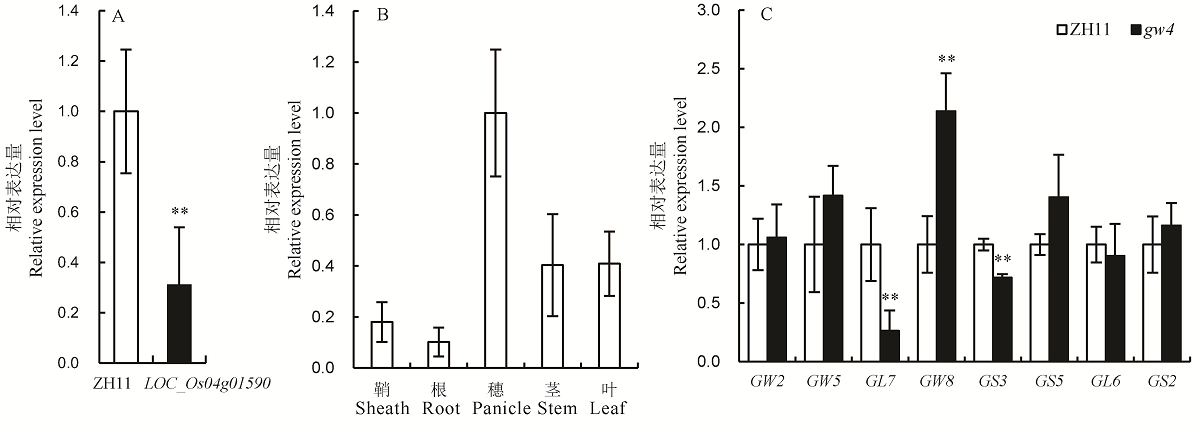
Fig. 6. Expression analysis of LOC_Os04g01590 and grain shape genes. A, Expression analysis of LOC_Os04g01590 in Zhonghua 11(ZH11) and gw4; B, Relative transcript levels of LOC_Os04g01590 in various organs; C, Expression analysis of grain shape genes in the ZH11 and gw4. Set the transcription level in the panicle of ZH11 to 1.0. Bars represent standard deviation (n=3). ** indicates significant difference between ZH11 and gw4 by t-test (P<0.01).
| 基因 Gene | 功能注释 Function annotation |
|---|---|
| LOC_Os04g01560 | 表达蛋白Expressed protein |
| LOC_Os04g01570 | 蔗糖酶/果胶甲基酯酶抑制剂家族蛋白Invertase/Pectin methylesterase inhibitor family protein |
| LOC_Os04g01580 | 表达蛋白Expressed protein |
| LOC_Os04g01590 | 精氨酸酶Arginase |
| LOC_Os04g01600 | 赤藓酸-4-磷酸脱氢酶结构域蛋白Erythroate-4-phosphate dehydrogenase domain protein |
| LOC_Os04g01610 | 表达蛋白Expressed protein |
Table 2 Predicted genes in fine-mapping region.
| 基因 Gene | 功能注释 Function annotation |
|---|---|
| LOC_Os04g01560 | 表达蛋白Expressed protein |
| LOC_Os04g01570 | 蔗糖酶/果胶甲基酯酶抑制剂家族蛋白Invertase/Pectin methylesterase inhibitor family protein |
| LOC_Os04g01580 | 表达蛋白Expressed protein |
| LOC_Os04g01590 | 精氨酸酶Arginase |
| LOC_Os04g01600 | 赤藓酸-4-磷酸脱氢酶结构域蛋白Erythroate-4-phosphate dehydrogenase domain protein |
| LOC_Os04g01610 | 表达蛋白Expressed protein |
| [1] | Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Xu J, Ye W, Meng X, Liu R, Chen H, Jing Y, Wang Y, Zhu X, Li J, Qian Q.Copy number variation at the GL7 locus contributes to grain size diversity in rice[J]. Nature Genetics, 2015, 47(8): 944-948. |
| [2] | 尉鑫, 曾智锋, 杨维丰, 韩婧, 柯善文. 水稻粒形遗传调控研究进展[J]. 安徽农业科学, 2019, 47(5): 21-28. |
| Wei X, Zeng Z F, Yang Wei F, Han J, Ke S W.Research progress on genetic regulation of rice grain shape[J]. Journal of Anhui Agricultural Sciences, 2019, 47(5): 21-28. (in Chinese with English abstract) | |
| [3] | Yan S, Zou G, Li S, Wang H, Liu H, Zhai G, Guo P, Song H, Yan C, Tao Y.Seed size is determined by the combinations of the genes controlling different seed characteristics in rice[J]. Theoretical and Applied Genetics, 2011, 123: 1173-1181. |
| [4] | Huang K, Wang D, Duan P, Zhang B, Xu R, Li N, Li Y.WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice[J]. Plant Journal, 2017, 91(5): 849-860. |
| [5] | Shi C, Ren Y, Liu L, Wang F, Zhang H, Tian P, Pan T, Wang Y, Jing R, Liu T, Wu F, Lin Q, Lei C, Zhang X, Zhu S, Guo X, Wang J, Zhao Z, Wang J, Zhai H, Cheng Z, Wan J.Ubiquitin Specific Protease 15 has an important role in regulating grain width and size in rice[J]. Plant Physiology, 2019, 180(1): 381-391. |
| [6] | Chen Y, Xu Y, Luo W, Li W, Chen N, Zhang D, Chong K.The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice[J]. Plant Physiology, 2013, 163(4): 1673-1685. |
| [7] | Hu X, Qian Q, Xu T, Zhang Y, Dong G, Gao T, Xie Q, Xue Y.The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric Gα subunit to regulate Brassinosteroid-mediated growth in rice[J]. PLoS Genetics, 2013, 9(3): e1003391. |
| [8] | Liu Q, Han R, Wu K, Zhang J, Ye Y, Wang S, Chen J, Pan Y, Li Q, Xu X, Zhou J, Tao D, Wu Y, Fu X.G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice[J]. Nature Communications, 2018, 9(1): 852. |
| [9] | Zhang D P, Zhou Y, Yin J F, Yan X J, Lin S, Xu W F, Baluška F, Wang Y P, Xia Y J, Liang G H, Liang J S. Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation[J]. Journal of Experimental Botany, 2015(20): 6371. |
| [10] | Swain D M, Sahoo R K, Srivastava V K, Tripathy B C, Tuteja R, Tuteja N.Function of heterotrimeric G-protein γ subunit RGG1 in providing salinity stress tolerance in rice by elevating detoxification of ROS[J]. Planta, 2017, 245(2): 367-383. |
| [11] | Yadav D K, Islam S M, Tuteja N.Rice heterotrimeric G-protein gamma subunits (RGG1 and RGG2) are differentially regulated under abiotic stress[J]. Plant Signaling & Behavior, 2012, 7(7): 733-740. |
| [12] | Xu R, Duan P, Yu H, Zhou Z, Zhang B, Wang R, Li J, Zhang G, Zhuang S, Lü J, Li N, Chai T, Tian Z, Yao S, Li Y.Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice[J]. Molecular Plant, 2018, 11(6): 860-873. |
| [13] | Guo T, Chen K, Dong N Q, Shi C L, Ye W W, Gao J P, Shan J X, Lin H X.GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice[J]. Plant Cell, 2018, 30(4): 871-888. |
| [14] | Yi J, Lee Y S, Lee D Y, Cho M H, Jeon J S, An G. OsMPK6 plays a critical role in cell differentiation during early embryogenesis in Oryza sativa[J]. Journal of Experimental Botany, 2016(8): 2425-2437. |
| [15] | Xia D, Zhou H, Liu R, Dan W, Li P, Wu B, Chen J, Wang L, Gao G, Zhang Q, He Y.GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice[J]. Molecular Plant, 2018, 11(5): 754-756. |
| [16] | 梁文化, 赵春芳, 张善磊, 张亚东, 朱镇, 赵庆勇, 陈涛, 王才林. 水稻粒型基因GLW7的功能标记开发与鉴定//江苏省遗传学会. 2017年学术研讨会—“技术创新与遗传学发展”论文摘要集[C]. 南京: 江苏省遗传学会, 2017: 1. |
| Liang W H, Zhao C F, Zhang S L, Zhang Y D, Zhu Z, Zhao Q Y, Chen T, Wang C L.Development and identification of functional markers of rice grain type gene GLW7//Jiangsu Society of Genetics. Academic Seminar of Jiangsu Society of Genetics in 2017 Meeting—“Technology Innovation and Genetics Development” Abstracts Collection[C]. Nanjing: Jiangsu Genetics Society, 2017: 1. (in Chinese) | |
| [17] | Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, Zhu L, Dong G, Guo L, Zeng D, Zhang G, Xie L, Xiong G, Li J, Qian Q.A Rare allele of GS2 enhances grain size and grain yield in rice[J]. Molecular Plant, 2015: 1455-1465. |
| [18] | Wang S, Li S, Liu Q, Wu K, Zhang J, Wang S, Wang Y, Chen X, Zhang Y, Gao C, Wang F, Huang H, Fu X.The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality[J]. Nature Genetics, 2015, 47(8): 949. |
| [19] | 徐乾坤, 余海平, 夏赛赛, 崔元江, 俞晓琦, 刘贺,曾大力, 胡江, 张强, 高振宇, 张光恒, 朱丽, 沈兰, 郭龙彪, 饶玉春, 钱前, 任德勇. C2H2锌指蛋白LRG1调控水稻小穗的发育[J]. 科学通报, 2020, 65(9): 753-764. |
| Xu Q K, Yu H P, Xia S S, Cui Y J, Yu X Q, Liu H, Zeng D L, Hu J, Zhang Q, Gao Z Y, Zhang G H, Zhu L, Shen L, Guo L B, Rao Y C, Qian Q, Ren D Y.C2H2 zinc finger protein LRG1 regulates the development of rice spikelets[J]. Science Bulletin, 2020, 65(9): 753-764. (in Chinese with English abstract) | |
| [20] | Mori M, Tomita C, Sugimoto K, Hasegawa M, Hayashi N, Dubouzet JG, Ochiai H, Sekimoto H, Hirochika H, Kikuchi S.Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice[J]. Plant Molecular Biology, 2007, 63(6): 847-860. |
| [21] | Yan S, Zou G, Li S, Wang H, Liu H, Zhai G, Guo P, Song H, Yan C, Tao Y.Seed size is determined by the combinations of the genes controlling different seed characteristics in rice[J]. Theoretical and Applied Genetics, 2011, 123(7): 1173-1181. |
| [22] | Jia S, Xiong Y, Xiao P, Wang X, Yao J.OsNF-YC10, a seed preferentially expressed gene regulates grain width by affecting cell proliferation in rice[J]. Plant Science, 2019, 280: 219-227. |
| [23] | Prasad K, Parameswaran S, Vijayraghavan U.OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs[J]. Plant Journal, 2010, 43(6): 915-928. |
| [24] | Zhao M, Liu B, Wu K, Ye Y, Huang S, Wang S, Wang Y, Han R, Liu Q, Fu X, Wu Y.Regulation of OsmiR156h through alternative polyadenylation improves grain yield in rice[J]. PLoS One, 2015, 10(5): e0126154. |
| [25] | Miao C, Wang D, He R, Liu S, Zhu J K.Mutations in MIR396e and MIR396f increase grain size and modulate shoot architecture in rice[J]. Plant Biotechnology Journal, 2020, 18(2): 491-501. |
| [26] | Zhang Y C, Yu Y, Wang C Y, Li Z Y, Liu Q, Xu J, Liao J Y, Wang X J, Qu L H, Chen F, Xin P, Yan C, Chu J, Li H Q, Chen Y Q.Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching[J]. Nature Biotechnology, 2013, 31(9): 848-852. |
| [27] | Sun W, Xu X H, Li Y, Xie L, He Y, Li W, Lu X, Sun H, Xie X.OsmiR530 acts downstream of OsPIL15 to regulate grain yield in rice[J]. New Phytology, 2019, 226(3): 823-837. |
| [28] | Chen J, Gao H, Zheng X M, Jin M, Weng J F, Ma J, Ren Y, Zhou K, Wang Q, Wang J, Wang J L, Zhang X, Cheng Z, Wu C, Wang H, Wan J M.An evolutionarily conserved gene, FUWA, plays a role in determining panicle architecture, grain shape and grain weight in rice[J]. Plant Journal, 2015, 83(3): 427-438. |
| [29] | Abe Y, Mieda K, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y.The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice[J]. Genes & Genetic Systems, 2011, 85(5): 327-339. |
| [30] | Wu T, Shen Y, Zheng M, Yang C, Chen Y, Feng Z, Liu X, Liu S, Chen Z, Lei C, Wang J, Jiang L, Wan J.Gene SGL, encoding a kinesin-like protein with transactivation activity, is involved in grain length and plant height in rice[J]. Plant Cell Reports, 2014, 33(2): 235-244. |
| [31] | Segami S, Kono I, Ando T, Yano M, Kitano H, Miura K, Iwasaki Y.Small and round seed 5 gene encodes alpha-tubulin regulating seed cell elongation in rice[J]. Rice, 2011, 5(1): 4. |
| [32] | Wu L, Ren D, Hu S, Li G, Dong G, Jiang L, Hu X, Ye W, Cui Y, Zhu L, Hu J, Zhang G, Gao Z, Zeng D, Qian Q, Guo L.Down-regulation of a nicotinate phosphoribosyltransferase gene, OsNaPRT1, leads to withered leaf tips[J]. Plant Physiology, 2016, 171(2): 1085. |
| [33] | Hu Z, Lu S J, Wang M J, He H, Sun L, Wang H, Liu X H, Jiang L, Sun J L, Xin X, Kong W, Chu C, Xue H W, Yang J, Luo X, Liu J X. A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice[J]. Molecular Plant, 2018(5): 736-749. |
| [34] | Xiong H, Yu J, Miao J, Li J, Zhang H, Wang X, Liu P, Zhao Y, Jiang C, Yin Z, Li Y, Guo Y, Fu B, Wang W, Li Z, Ali J, Li Z,. Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging[J]. Plant Physiology, 2018, 178(1): 451-467. |
| [35] | Gao X, Zhang J Q, Zhang X, Zhou J, Jiang Z, Huang P, Tang Z, Bao Y, Cheng J, Tang H, Zhang W, Zhang H, Huang J.Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling[J]. Plant Cell, 2019, 31(5): 1077-1093. |
| [36] | Zhao D S, Li Q F, Zhang C Q, Zhang C, Yang Q Q, Pan L X, Ren X Y, Lu J, Gu M H, Liu Q Q.GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality[J]. Nature Communications, 2018, 9(1): 1240. |
| [37] | Xu C, Liu Y, Li Y, Xu X, Xu C, Li X, Xiao J, Zhang Q.Differential expression of GS5 regulates grain size in rice[J]. Journal of Experimental Botany, 2015, 66(9): 2611-2623. |
| [38] | Weng J, Gu S, Wan X, Gao H, Guo T, Su N, Lei C, Zhang X, Cheng Z, Guo X, Wang J, Jiang L, Zhai H, Wan J.Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight[J]. Cell Research, 2008, 18(12): 1199-1209. |
| [39] | Ma X, Cheng Z, Qin R, Qiu Y, Heng Y, Yang H, Ren Y, Wang X, Bi J, Ma X, Zhang X, Wang J, Lei C, Guo X, Wang J, Wu F, Jiang L, Wang H, Wan J.OsARG encodes an arginase that plays critical roles in panicle development and grain production in rice[J]. Plant Journal, 2013, 73(2): 190-200. |
| [40] | Hang L?, Yu H, Ma B, Liu G, Wang J, Wang J, Gao R, Li J, Liu J, Xu J, Zhang Y, Li Q, Huang X, Xu J, Li J, Qian Q, Han B, He Z, Li J,. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice[J]. Nature Communications, 2017, 8: 14789. |
| [41] | Wang Y, Shang L, Yu H, Zeng L, Hu J, Ni S, Rao Y, Li S, Chu J, Meng X, Wang L, Hu P, Yan J, Kang S, Qu M, Lin H, Wang T, Wang Q, Hu X, Chen H, Wang B, Gao Z, Guo L, Zeng D, Zhu X, Xiong G, Li J, Qian Q.A strigolactone biosynthesis gene contributed to the green revolution in rice[J]. Molecular Plant, 2020, 13(6): 923-932. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||