
中国水稻科学 ›› 2025, Vol. 39 ›› Issue (1): 55-66.DOI: 10.16819/j.1001-7216.2025.231112
收稿日期:2023-11-15
修回日期:2023-12-12
出版日期:2025-01-10
发布日期:2025-01-14
通讯作者:
*email: liuchaolei@caas.cn;基金资助:
HU Fengyue, WANG Jian, WANG Chun, WANG Kejian*( ), LIU Chaolei*(
), LIU Chaolei*( )
)
Received:2023-11-15
Revised:2023-12-12
Online:2025-01-10
Published:2025-01-14
Contact:
*email: liuchaolei@caas.cn;摘要:
【目的】探究水稻DMP家族基因的单倍体诱导能力,为单倍体育种提供新的基因资源。【方法】筛选与ZmDMP基因高同源且在花粉中高表达的水稻DMP家族基因作为候选基因;利用CRISPR/Cas9多基因编辑技术,在籼粳杂交稻“春优84”中对筛选的DMP家族基因和单倍体诱导基因OsMTL创制单基因敲除以及多基因组合敲除突变体;对突变体进行形态学观察和花粉育性鉴定;调查统计转基因T0材料的结实率和单倍体诱导率。【结果】在水稻RAP-DB数据库中共检索到13个玉米单倍体诱导基因ZmDMP的同源基因。OsDMP1和OsDMP2与ZmDMP同源性最高,相似度分别为41.24% 和37.32%;OsDMP1和OsDMP3在花药中表达量最高。因此,选择OsDMP1、OsDMP2、OsDMP3基因为候选基因。通过CRISPR/Cas9基因编辑技术创制了OsDMP1、OsDMP2、OsDMP3的单基因敲除和组合敲除突变体(osdmp1、osdmp2、osdmp3、osdmp1-osdmp2、osdmp1-osdmp3、osdmp1-osdmp2-osdmp3),OsMTL单基因敲除突变体(osmtl),以及OsDMP1、OsDMP2、OsDMP3与OsMTL基因的组合敲除突变体(osmtl-osdmp1、osmtl-osdmp2、osmtl-osdmp3、osmtl-osdmp1-osdmp2、osmtl-osdmp1-osdmp3、osmtl- osdmp1-osdmp2-osdmp3)。表型考察发现,相比较野生型,所有突变体的植株形态和花粉育性均未发生显著变化,而仅在包含osmtl的单基因敲除和多基因组合敲除突变体中结实率发生显著下降。单倍体鉴定结果显示,OsDMP1、OsDMP2、OsDMP3与OsMTL组合突变的单倍体诱导效率分别为0.4% ± 0.6%、2.6% ± 2.8%、1.4% ± 0.6%、1.5% ±1.3%、2.1% ± 2.4%、2.2% ± 0.6%,与osmtl突变体(1.5% ± 0.5%)无显著差异。然而,当OsDMP家族基因单个突变或多个组合突变时均无单倍体产生。【结论】本研究利用CRISPR/Cas9基因编辑技术成功创制了水稻OsDMP1、OsDMP2、OsDMP3、OsMTL单基因和多基因组合敲除突变体,发现了OsDMP1、OsDMP2和OsDMP3均无独立单倍体诱导能力,也不能提升OsMTL基因的单倍体诱导效率。本研究促进了对水稻中OsDMP同源基因的了解,为后续单倍体诱导基因研究提供参考。
胡风越, 王健, 王春, 王克剑, 刘朝雷. 水稻DMP1、DMP2、DMP3基因突变体的创制及其单倍体诱导能力鉴定[J]. 中国水稻科学, 2025, 39(1): 55-66.
HU Fengyue, WANG Jian, WANG Chun, WANG Kejian, LIU Chaolei. Generation of Rice DMP1, DMP2 and DMP3 Mutants and Identification of Their Haploid Induction Ability[J]. Chinese Journal OF Rice Science, 2025, 39(1): 55-66.
| 重命名Rename | 基因登录号Accession number | 描述 Description | 长度Length/aa | 相似性Identity | E值 E value |
|---|---|---|---|---|---|
| OsDMP1 | LOC_Os05g48840 | Protein of unknown function DUF679 family protein | 239 | 137 | 3e−33 |
| OsDMP2 | LOC_Os08g01530 | Protein of unknown function DUF679 family protein | 225 | 132 | 1e−31 |
| OsDMP3 | LOC_Os01g29240 | Protein of unknown function DUF679 family protein | 226 | 120 | 4e−28 |
| OsDMP4 | LOC_Os07g22510 | Protein of unknown function DUF679 family protein | 263 | 119 | 9e−28 |
| OsDMP5 | LOC_Os03g25440 | Protein of unknown function DUF679 family protein | 194 | 112 | 2e−25 |
| OsDMP6 | LOC_Os06g24490 | Protein of unknown function DUF679 family protein | 254 | 110 | 5e−25 |
| OsDMP7 | LOC_Os01g65992 | Protein of unknown function DUF679 domain containing protein | 240 | 107 | 4e−24 |
| OsDMP8 | LOC_Os01g27100 | Protein of unknown function DUF679 domain containing protein | 319 | 107 | 7e−24 |
| OsDMP9 | LOC_Os01g29330 | Protein of unknown function DUF679 family protein | 241 | 100 | 8e−22 |
| OsDMP10 | LOC_Os01g29280 | Protein of unknown function DUF679 family protein | 265 | 96.3 | 1e−20 |
| OsDMP11 | LOC_Os07g45080 | Protein of unknown function DUF679 family protein | 189 | 90.9 | 5e−19 |
| OsDMP12 | LOC_Os01g27120 | Protein of unknown function DUF679 domain containing protein | 226 | 83.2 | 9e−17 |
| OsDMP13 | LOC_Os12g23310 | Protein of unknown function DUF679 domain containing protein | 111 | 53.1 | 1e−7 |
表1 水稻DMP同源家族基因
Table 1. The DMP homologous genes in rice
| 重命名Rename | 基因登录号Accession number | 描述 Description | 长度Length/aa | 相似性Identity | E值 E value |
|---|---|---|---|---|---|
| OsDMP1 | LOC_Os05g48840 | Protein of unknown function DUF679 family protein | 239 | 137 | 3e−33 |
| OsDMP2 | LOC_Os08g01530 | Protein of unknown function DUF679 family protein | 225 | 132 | 1e−31 |
| OsDMP3 | LOC_Os01g29240 | Protein of unknown function DUF679 family protein | 226 | 120 | 4e−28 |
| OsDMP4 | LOC_Os07g22510 | Protein of unknown function DUF679 family protein | 263 | 119 | 9e−28 |
| OsDMP5 | LOC_Os03g25440 | Protein of unknown function DUF679 family protein | 194 | 112 | 2e−25 |
| OsDMP6 | LOC_Os06g24490 | Protein of unknown function DUF679 family protein | 254 | 110 | 5e−25 |
| OsDMP7 | LOC_Os01g65992 | Protein of unknown function DUF679 domain containing protein | 240 | 107 | 4e−24 |
| OsDMP8 | LOC_Os01g27100 | Protein of unknown function DUF679 domain containing protein | 319 | 107 | 7e−24 |
| OsDMP9 | LOC_Os01g29330 | Protein of unknown function DUF679 family protein | 241 | 100 | 8e−22 |
| OsDMP10 | LOC_Os01g29280 | Protein of unknown function DUF679 family protein | 265 | 96.3 | 1e−20 |
| OsDMP11 | LOC_Os07g45080 | Protein of unknown function DUF679 family protein | 189 | 90.9 | 5e−19 |
| OsDMP12 | LOC_Os01g27120 | Protein of unknown function DUF679 domain containing protein | 226 | 83.2 | 9e−17 |
| OsDMP13 | LOC_Os12g23310 | Protein of unknown function DUF679 domain containing protein | 111 | 53.1 | 1e−7 |
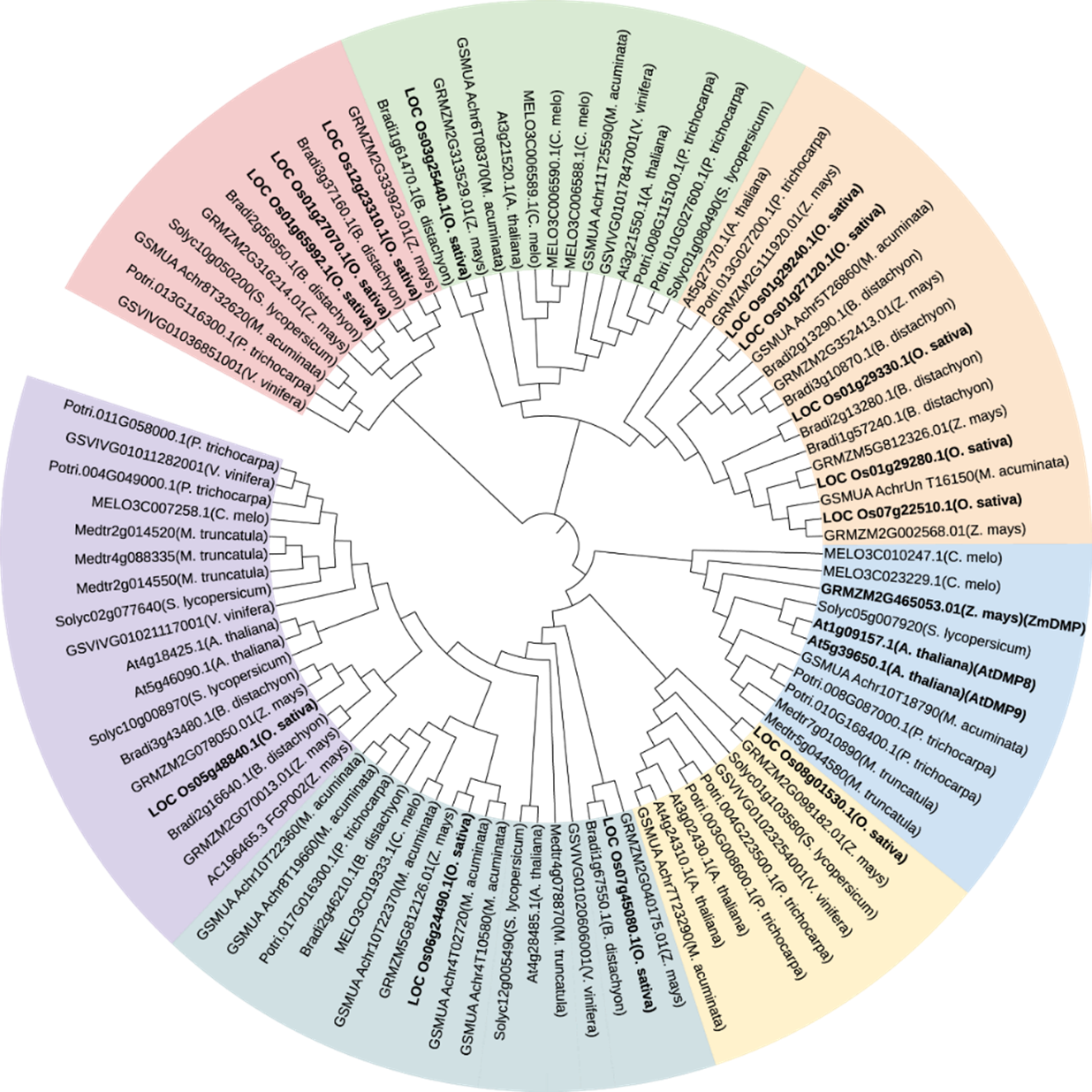
图1 不同物种DMP蛋白系统进化分析 A. thaliana: 拟南芥;B. distachyon: 二穗短柄草;C. melo: 甜瓜;M. acuminata: 小果野芭蕉;M. truncatula: 苜蓿;P. trichocarpa: 三角叶杨;S. lycopersicum: 番茄; V. vinifera: 葡萄;Z. mays: 玉米。采用MEGA7软件中的最大似然法构建,Bootstrap为1000。ZmDMP、AtDMP8、AtDMP以及水稻中DMP同源蛋白加粗显示。
Fig. 1. Phylogenetic analysis of DMP proteins in various species A. thaliana, Arabidopsis thaliana; B. distachyon, Brachypodium distachyon; C. melo, Cucumis melo; M. acuminate, Musa acuminata; M. truncatula, Medicago truncatul; P. trichocarpa, Populus trichocarpa; S. lycopersicum, Solanum lycopersicum; V. vinifera, Vitis vinifera; Z. mays, Zea mays. The maximum likelihood method in MEGA7 software was adopted. Bootstrap = 1000. ZmDMP, AtDMP8, AtDMP and rice DMP homologous proteins are shown in bold.
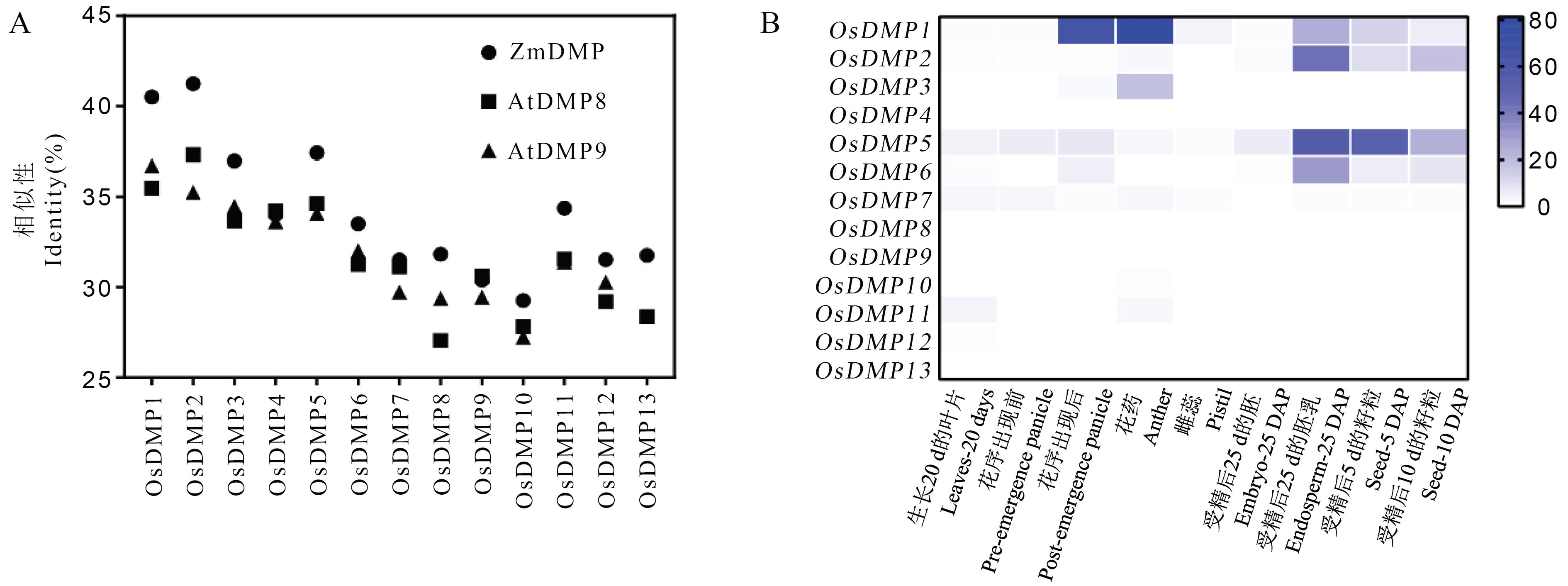
图2 同源比对与表达谱分析筛选水稻DMP家族候选基因 A: OsDMP家族蛋白与ZmDMP、AtDMP8、AtDMP9蛋白序列相似性比较;B: OsDMP家族基因的表达谱预测。
Fig. 2. Candidate genes of rice DMP family by homologous comparison and expression profile analysis A, Similarity analysis of OsDMP family proteins with ZmDMP, AtDMP8 and AtDMP9 proteins; B, Online expression analysis of OsDMP family genes.
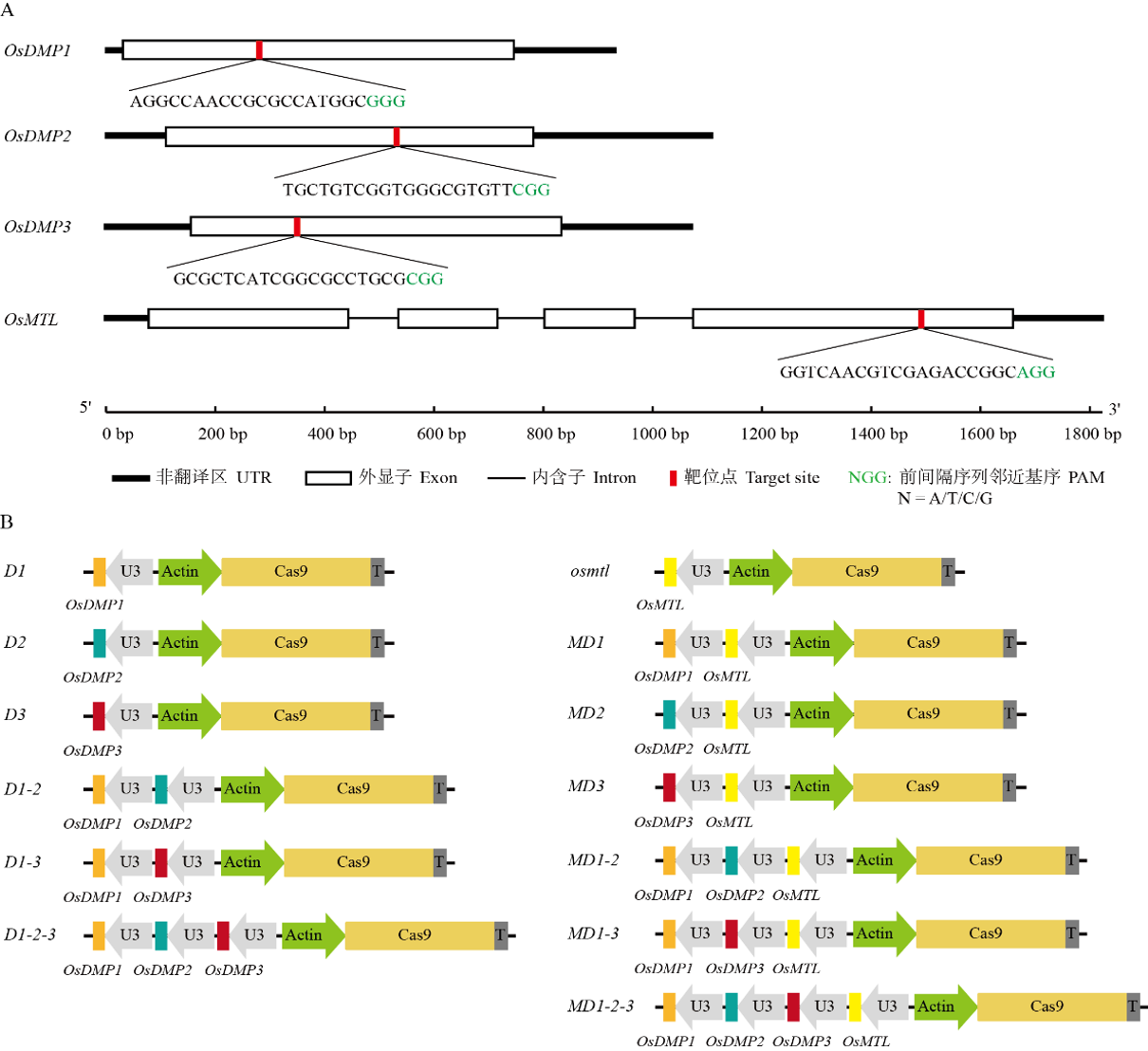
图3 OsDMP1、OsDMP2、OsDMP3和OsMTL的CRISPR/Cas9多基因敲除载体结构示意图 靶基因gRNA的启动子为U3,Cas9蛋白的启动子为Actin。
Fig. 3. Structure schematic diagram of CRISPR/Cas9 multi-gene knockout vectors for OsDMP1, OsDMP2, OsDMP3 and OsMTL genes The promoter of the target gene gRNA is U3, and the promoter of Cas9 protein is Actin.
| 编号 Code | 转化株数 No. of transgenic lines | 突变株数 No. of mutant lines | 突变株占比 (突变株数/转化株数) Proportion of mutant lines (No. of mutant lines / No. of transgenic lines) (%) | 双等位突变株数 No. of biallelic mutant lines | 双等位突变株占比 (双等位突变株数/转化株数) Proportion of biallelic mutant plants (No. of biallelic mutant lines/No. of transgenic lines) (%) |
|---|---|---|---|---|---|
| D1 | 24 | 21 | 87.5 | 18 | 75.0 |
| D2 | 21 | 17 | 81.0 | 14 | 66.7 |
| D3 | 39 | 38 | 97.4 | 36 | 92.3 |
| D1-2 | 24 | 24 | 100.0 | 19 | 79.2 |
| D1-3 | 24 | 23 | 95.8 | 11 | 45.8 |
| D1-2-3 | 37 | 35 | 94.6 | 24 | 64.9 |
| osmtl | 22 | 15 | 68.2 | 12 | 54.5 |
| MD1 | 23 | 21 | 91.3 | 15 | 65.2 |
| MD2 | 21 | 13 | 61.9 | 12 | 57.1 |
| MD3 | 24 | 19 | 79.2 | 12 | 50.0 |
| MD1-2 | 24 | 15 | 62.5 | 9 | 37.5 |
| MD1-3 | 24 | 19 | 79.2 | 14 | 58.3 |
| MD1-2-3 | 40 | 33 | 82.5 | 14 | 35.0 |
表2 转基因T0植株突变比例
Table 2. Proportion of mutations in T0 transgenic plants
| 编号 Code | 转化株数 No. of transgenic lines | 突变株数 No. of mutant lines | 突变株占比 (突变株数/转化株数) Proportion of mutant lines (No. of mutant lines / No. of transgenic lines) (%) | 双等位突变株数 No. of biallelic mutant lines | 双等位突变株占比 (双等位突变株数/转化株数) Proportion of biallelic mutant plants (No. of biallelic mutant lines/No. of transgenic lines) (%) |
|---|---|---|---|---|---|
| D1 | 24 | 21 | 87.5 | 18 | 75.0 |
| D2 | 21 | 17 | 81.0 | 14 | 66.7 |
| D3 | 39 | 38 | 97.4 | 36 | 92.3 |
| D1-2 | 24 | 24 | 100.0 | 19 | 79.2 |
| D1-3 | 24 | 23 | 95.8 | 11 | 45.8 |
| D1-2-3 | 37 | 35 | 94.6 | 24 | 64.9 |
| osmtl | 22 | 15 | 68.2 | 12 | 54.5 |
| MD1 | 23 | 21 | 91.3 | 15 | 65.2 |
| MD2 | 21 | 13 | 61.9 | 12 | 57.1 |
| MD3 | 24 | 19 | 79.2 | 12 | 50.0 |
| MD1-2 | 24 | 15 | 62.5 | 9 | 37.5 |
| MD1-3 | 24 | 19 | 79.2 | 14 | 58.3 |
| MD1-2-3 | 40 | 33 | 82.5 | 14 | 35.0 |
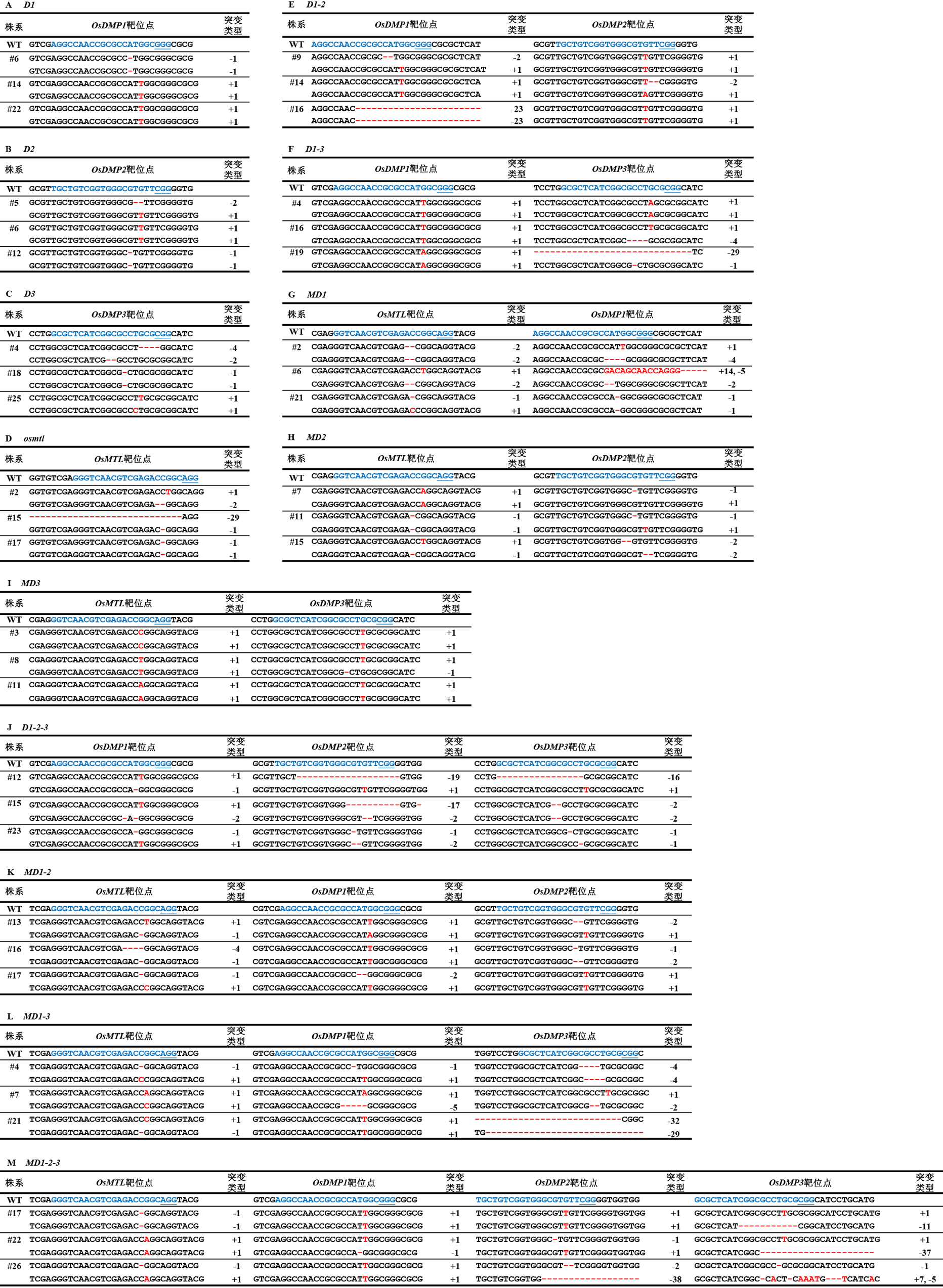
图4 转基因T0植株突变类型 蓝色字母表示靶位点序列;蓝色下划线表示原间隔相邻基序(Protospacer adjacent motif, PAM);红色字母表示“插入”突变的碱基;红色中划线表示“缺失”突变。
Fig. 4. Mutations of transgenic T0 plants Blue letters indicate the target site sequences; blue underscores indicate the protospacer adjacent motif (PAM); red letters indicate ‘insertion’ mutant bases; and red hyphens indicate ‘deletion’ mutations.
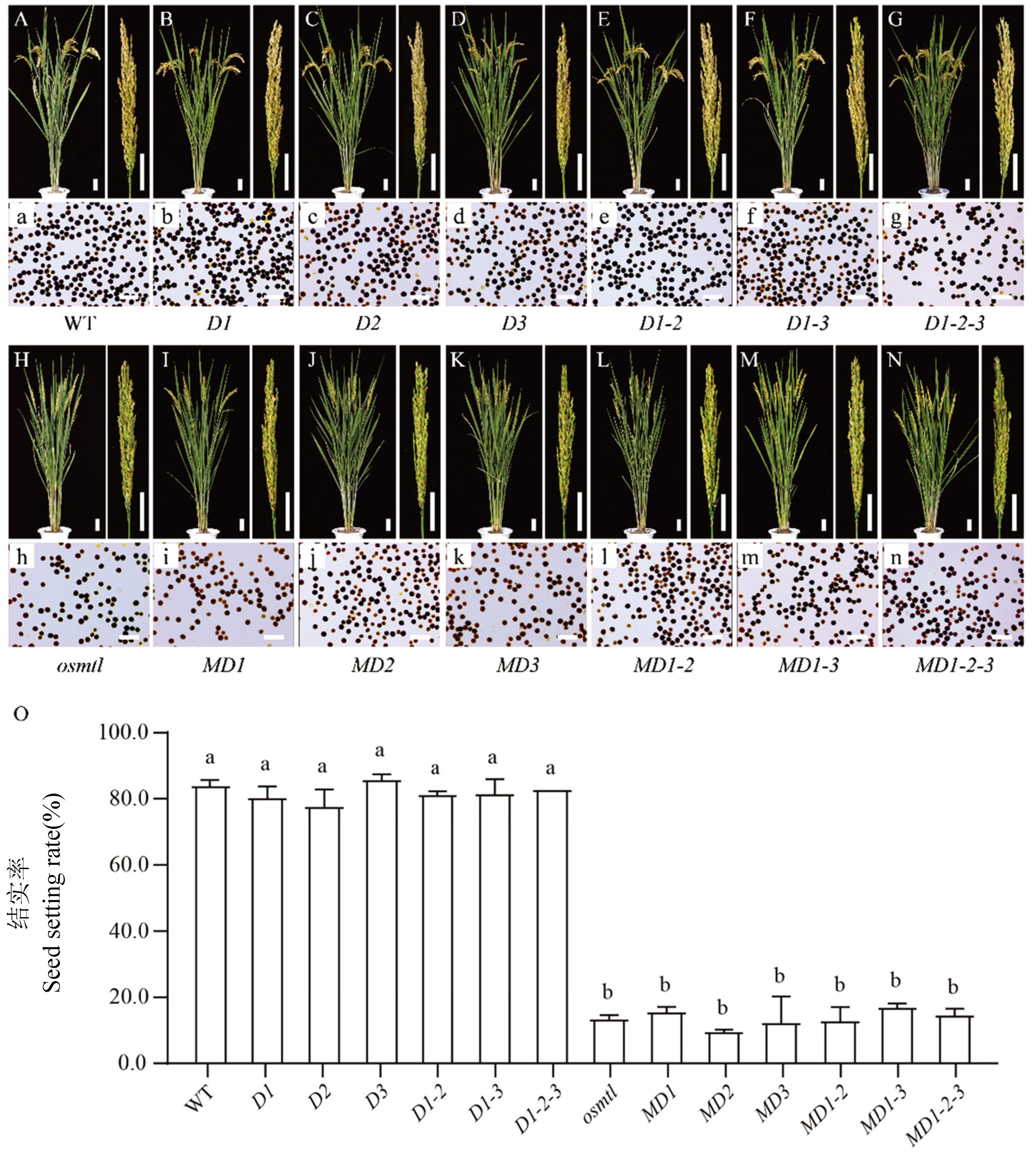
图5 野生型和突变体形态学观察、花粉育性分析和结实率统计 A~N: 成熟期植株的株型、穗型,标尺为5 cm;a~n: 花粉碘染结果,标尺为100 μm;O: 野生型和突变体结实率比较和分析,采用邓肯新复极差法 (Duncan’s multiple-range test) 进行均值差异显著性分析,字母a、b表示在α = 0.05水平差异显著。
Fig. 5. Morphologic observation, pollen fertility analysis and seed setting rate of wild-type and mutants A-N, The morphology of mature plant and panicle, bar = 5 cm; a-n, Results of pollen iodine staining, bar = 100 μm; O, Comparison and analysis of seed setting rate in wild-type and mutants. Duncan's Multiple Range test was used to analyze the significance of mean difference, and letters a and b indicate significant difference at α = 0.05 level.
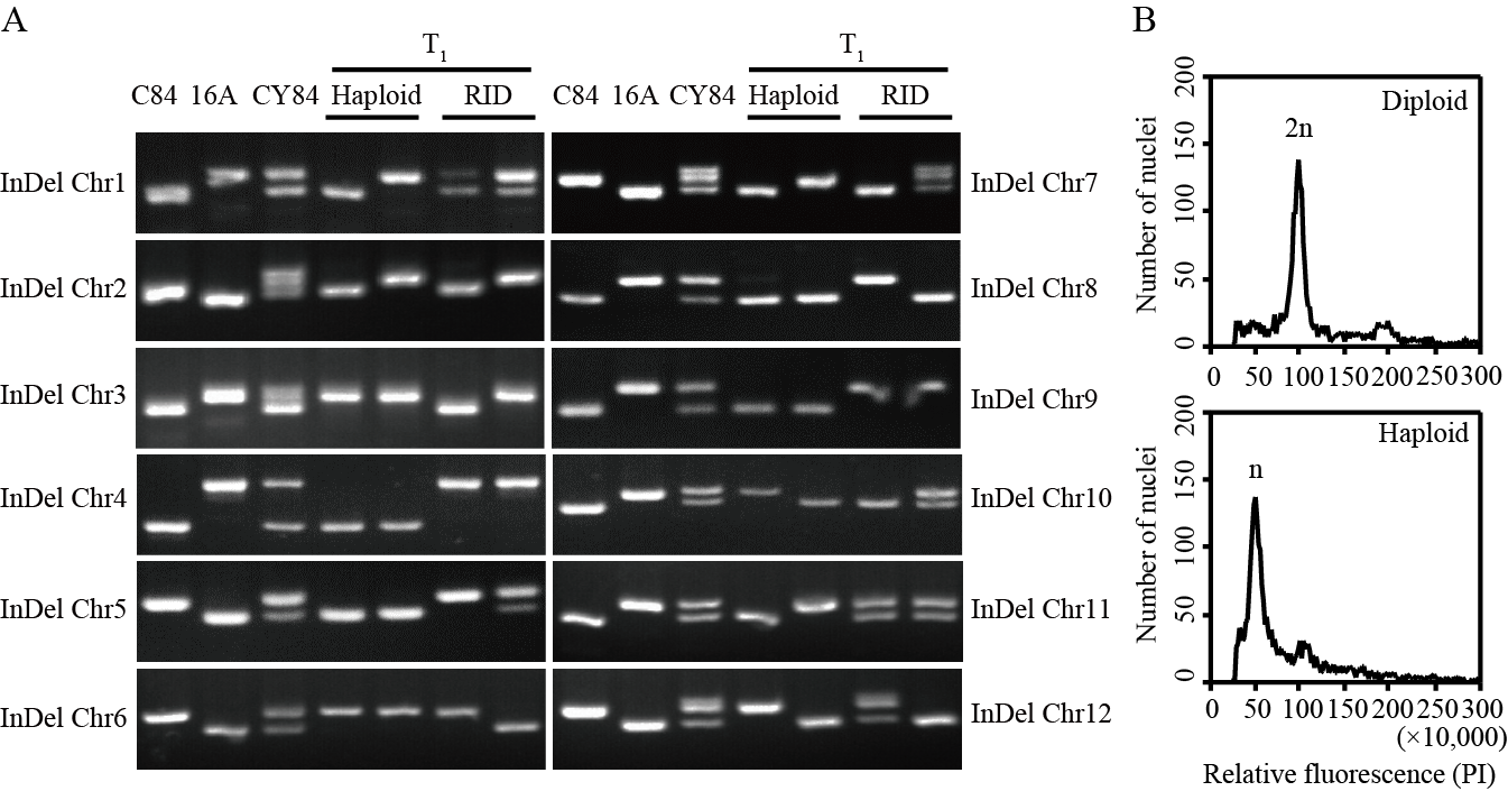
图6 分子标记与流式细胞术鉴定植株倍性 A: 水稻12条染色体上的InDel标记在不同材料中的凝胶电泳带型; chr: 染色体(chromosome)的简写;C84: 春优84的父本;16A: 春优84的母本春江16A;Haploid: 单倍体;RID: 重组自交二倍体。B: 二倍体和单倍体的流式细胞术分析;PI: 碘化丙啶(propidium iodide)。
Fig. 6. Ploidy identification with InDel markers and flow cytometry A, Gel electrophoresis band pattern of InDel markers on 12 chromosomes of rice in different materials; chr, Abbreviation of chromosome; C84, Male parent of CY84; 16A, Female parent of CY84; RID, Recombinant inbred diploid. B, Ploidy analysis of diploid and haploid by flow cytometry; PI, Propidium iodide.
| 编号 Number | 基因型 Genotype | 单倍体诱导效率%(单倍体数/检测数) Haploid induction rate % (Haploid/Tested number) | 均值±标准差Mean±SD(%) | ||
|---|---|---|---|---|---|
| 株系1 Line 1 | 株系2 Line 2 | 株系3 Line 3 | |||
| WT | WT | 0.0%(0/92) | 0.0%(0/93) | 0.0%(0/93) | 0.0 |
| D1 | osdmp1 | 0.0%(0/179) | 0.0%(0/186) | 0.0%(0/172) | 0.0 |
| D2 | osdmp2 | 0.0%(0/182) | 0.0%(0/179) | 0.0%(0/186) | 0.0 |
| D3 | osdmp3 | 0.0%(0/186) | 0.0%(0/183) | 0.0%(0/185) | 0.0 |
| D1-2 | osdmp1-osdmp2 | 0.0%(0/978) | 0.0%(0/165) | 0.0%(0/172) | 0.0 |
| D1-3 | osdmp1-osdmp3 | 0.0%(0/1662) | 0.0%(0/176) | 0.0%(0/175) | 0.0 |
| D1-2-3 | osdmp1-osdmp2-osdmp3 | 0.0%(0/1393) | 0.0%(0/181) | 0.0%(0/169) | 0.0 |
| osmtl | osmtl | 1.1%(1/93) | 1.3%(1/78) | 2.2%(2/93) | 1.5±0.5 |
| MD1 | osmtl-osdmp1 | 0.0%(0/102) | 0.0%(0/104) | 1.2%(1/86) | 0.4±0.6 ns |
| MD2 | osmtl-osdmp2 | 1.3%(1/77) | 6.5%(4/62) | 0.0%(0/58) | 2.6±2.8 ns |
| MD3 | osmtl-osdmp3 | 2.2%(2/90) | 0.9%(1/107) | 1.2%(1/82) | 1.4±0.6 ns |
| MD1-2 | osmtl-osdmp1-osdmp2 | 3.2%(3/94) | 0.0%(0/84) | 1.4%(1/72) | 1.5±1.3 ns |
| MD1-3 | osmtl-osdmp1-osdmp3 | 5.4%(5/93) | 0.9%(0/108) | 0.0%(0/64) | 2.1±2.4 ns |
| MD1-2-3 | osmtl-osdmp1-osdmp2-osdmp3 | 1.6%(2/129) | 1.9%(2/108) | 3.1%(3/96) | 2.2±0.6 ns |
表3 不同类型突变体单倍体诱导效率
Table 3. Haploid induction rate of different type mutants
| 编号 Number | 基因型 Genotype | 单倍体诱导效率%(单倍体数/检测数) Haploid induction rate % (Haploid/Tested number) | 均值±标准差Mean±SD(%) | ||
|---|---|---|---|---|---|
| 株系1 Line 1 | 株系2 Line 2 | 株系3 Line 3 | |||
| WT | WT | 0.0%(0/92) | 0.0%(0/93) | 0.0%(0/93) | 0.0 |
| D1 | osdmp1 | 0.0%(0/179) | 0.0%(0/186) | 0.0%(0/172) | 0.0 |
| D2 | osdmp2 | 0.0%(0/182) | 0.0%(0/179) | 0.0%(0/186) | 0.0 |
| D3 | osdmp3 | 0.0%(0/186) | 0.0%(0/183) | 0.0%(0/185) | 0.0 |
| D1-2 | osdmp1-osdmp2 | 0.0%(0/978) | 0.0%(0/165) | 0.0%(0/172) | 0.0 |
| D1-3 | osdmp1-osdmp3 | 0.0%(0/1662) | 0.0%(0/176) | 0.0%(0/175) | 0.0 |
| D1-2-3 | osdmp1-osdmp2-osdmp3 | 0.0%(0/1393) | 0.0%(0/181) | 0.0%(0/169) | 0.0 |
| osmtl | osmtl | 1.1%(1/93) | 1.3%(1/78) | 2.2%(2/93) | 1.5±0.5 |
| MD1 | osmtl-osdmp1 | 0.0%(0/102) | 0.0%(0/104) | 1.2%(1/86) | 0.4±0.6 ns |
| MD2 | osmtl-osdmp2 | 1.3%(1/77) | 6.5%(4/62) | 0.0%(0/58) | 2.6±2.8 ns |
| MD3 | osmtl-osdmp3 | 2.2%(2/90) | 0.9%(1/107) | 1.2%(1/82) | 1.4±0.6 ns |
| MD1-2 | osmtl-osdmp1-osdmp2 | 3.2%(3/94) | 0.0%(0/84) | 1.4%(1/72) | 1.5±1.3 ns |
| MD1-3 | osmtl-osdmp1-osdmp3 | 5.4%(5/93) | 0.9%(0/108) | 0.0%(0/64) | 2.1±2.4 ns |
| MD1-2-3 | osmtl-osdmp1-osdmp2-osdmp3 | 1.6%(2/129) | 1.9%(2/108) | 3.1%(3/96) | 2.2±0.6 ns |
| [1] | 陈海强, 刘会云, 王轲, 张双喜, 叶兴国. 植物单倍体诱导技术发展与创新[J]. 遗传, 2020, 42(5): 466-482. |
| Chen H Q, Liu H Y, Wang K, Zhang S X, Ye X G. Development and innovation of haploid induction technologies in plants[J]. Hereditas (Beijing), 2020, 42(5): 466-482. (in Chinese with English abstract) | |
| [2] | Dunwell J M. Haploids in flowering plants: origins and exploitation[J]. Plant Biotechnology Journal, 2010, 8(4): 377-424. |
| [3] | 相志国, 海燕, 康明辉, 赵永英. 单倍体的产生途径及其在作物遗传育种中的应用[J]. 河南农业科学, 2011, 40(11): 17-21. |
| Xiang Z G, Hai Y, Kang M H, Zhao Y Y. Generation ways of haploid and its application in crop genetics and breeding[J]. Journal of Henan Agricultural Sciences, 2011, 40(11): 17-21. | |
| [4] | Coe E H. A line of maize with high haploid frequency[J]. American Naturalist, 1959, 93(873): 381-382. |
| [5] | Prigge V, Xu X W, Li L, Babu R, Chen S J, Atlin G N, Melchinger A E. New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize[J]. Genetics, 2012, 190(2): 781-793. |
| [6] | Kelliher T, Starr D, Richbourg L, Chintamanani S, Delzer B, Nuccio M L, Green J, Chen Z, McCuiston J, Wang W, Liebler T, Bullock P, Martin B. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction[J]. Nature, 2017, 542(7639): 105-109. |
| [7] | Gilles L M, Khaled A, Laffaire J B, Chaignon S, Gendrot G, Laplaige J, Bergès H, Beydon G, Bayle V, Barret P, Comadran J, Martinant J P, Rogowsky P M, Widiez T. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize[J]. The EMBO Journal, 2017, 36(6): 707-717. |
| [8] | Liu C X, Li X, Meng D X, Zhong Y, Chen C, Dong X, Xu X W, Chen B J, Li W, Li L, Tian X L, Zhao H M, Song W B, Luo H S, Zhang Q H, Lai J S, Jin W W, Yan J B, Chen S J. A 4-bp insertion at ZmPLA1 encoding a putative phospholipase a generates haploid induction in maize[J]. Molecular Plant, 2017, 10(3): 520-522. |
| [9] | Yao L, Zhang Y, Liu C, Liu Y, Wang Y, Liang D, Liu J, Sahoo G, Kelliher T. OsMATL mutation induces haploid seed formation in indica rice[J]. Nature Plants, 2018, 4(8): 530-533. |
| [10] | Liu C X, Zhong Y, Oi X L, Chen M, Liu Z K, Chen C, Tian X L, Li I L, Jiao Y Y, Wang D, Wang Y W, Li M R, Xin M M, Liu W X, Jin W W, Chen S J. Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat[J]. Plant Biotechnology Journal, 2020, 18(2): 316-318. |
| [11] | Cheng Z X, Sun Y, Yang S H, Zhi H, Yin T, Ma X J, Zhang H S, Diao X M, Guo Y, Li X H, Wu C Y, Sui Y. Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica)[J]. Plant Biotechnology Journal, 2021, 19(6): 1089-1091. |
| [12] | Zhong Y, Liu C X, Qi X L, Jiao Y Y, Wang D, Wang Y W, Liu Z K, Chen C, Chen B J, Tian X L, Li J L, Chen M, Dong X, Xu X W, Li L, Li W, Liu W X, Jin W W, Lai J S, Chen S J. Mutation of ZmDMP enhances haploid induction in maize[J]. Nature Plants, 2019, 5(6): 575-580. |
| [13] | Zhong Y, Chen B J, Li M R, Wang D, Jiao Y Y, Qi X L, Wang M, Liu Z K, Chen C, Wang Y W, Chen M, Li J L, Xiao Z J, Cheng D H, Liu W X, Boutilier K, Liu C X, Chen S J. A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis[J]. Nature Plants, 2020, 6(5): 466-472. |
| [14] | Wang N, Xia X Z, Jiang T, Li L L, Zhang P C, Niu L F, Cheng H M, Wang K J, Lin H. In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula[J]. Plant Biotechnology Journal, 2022, 20(1): 22-24. |
| [15] | Li Y F, Li D, Xiao Q, Wang H D, Wen J, Tu J X, Shen J X, Fu T D, Yi B. An in planta haploid induction system in Brassica napus[J]. Journal of Integrative Plant Biology, 2022, 64(6): 1140-1144. |
| [16] | Zhong Y, Wang Y W, Chen B J, Liu J C, Wang D, Li M R, Qi X L, Liu C X, Boutilier K, Chen S J. Establishment of a dmp based maternal haploid induction system for polyploid Brassica napus and Nicotiana tabacum[J]. Journal of Integrative Plant Biology, 2022, 64(6): 1281-1294. |
| [17] | Wang C, Shen L, Fu Y P, Yan C J, Wang K J. A simple CRISPR/Cas9 system for multiplex genome editing in rice[J]. Journal of Genetics and Genomics, 2015, 42(12): 703-706. |
| [18] | Wang C, Liu Q, Shen Y, Hua Y F, Wang J J, Lin J R, Wu M G, Sun T T, Cheng Z K, Mercier R, Wang K J. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes[J]. Nature Biotechnology, 2019, 37(3): 283-286. |
| [19] | Liu Q, Wang C, Jiao X Z, Zhang H W, Song L L, Li Y X, Gao C X, Wang K J. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems[J]. Science China: Life Sciences, 2019, 62(1): 1-7. |
| [20] | Ma X L, Chen L T, Zhu Q L, Chen Y L, Liu Y G. Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products[J]. Molecular Plant, 2015, 8(8): 1285-1287. |
| [21] | 曹跃炫, 严绘景, 王克剑, 刘朝雷. 苗期快速分选水稻人工无融合生殖克隆种子[J]. 中国水稻科学, 2022, 36(6): 656-662. |
| Cao Y X, Yan H J, Wang K J, Liu C L. Rapid identification of rice clonal seeds generated by synthetic apomixis at seedling stage[J]. Chinese Journal of Rice Science, 2022, 36(6): 656-662. (in Chinese with English abstract) | |
| [22] | Liu C L, He Z X, Zhang Y, Hu F Y, Li M Q, Liu Q, Huang Y, Wang J, Zhang W, Wang C, Wang K J. Synthetic apomixis enables stable transgenerational transmission of heterotic phenotypes in hybrid rice[J]. Plant Communications, 2023, 4(2): 100470. |
| [23] | Liu C L, Wang J, Lu H W, Huang Y, Yan H J, Liang H, Wang C, Wang K J. Engineering synthetic apomixis in different hybrid rice varieties using the Fix strategy[J]. New Crops, 2024(1): 100003. |
| [24] | Zhao X, Xu X W, Xie H X, Chen S J, Jin W W. Fertilization and uniparental chromosome elimination during crosses with maize haploid inducers[J]. Plant Physiology, 2013, 163(2): 721-731. |
| [25] | Qiu F Z, Liang Y L, Li Y, Liu Y Z, Wang L M, Zheng Y L. Morphological, cellular and molecular evidences of chromosome random elimination in vivo upon haploid induction in maize[J]. Current Plant Biology, 2014(1): 83-90. |
| [26] | Karimi-Ashtiyani R, Ishii T, Niessen M, Stein N, Heckmann S, Gurushidze M, Banaei-Moghaddam A M, Fuchs J, Schubert V, Koch K, Weiss O, Demidov D, Schmidt K, Kumlehn J, Houben A. Point mutation impairs centromeric CENH3 loading and induces haploid plants[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(36): 11211-11216. |
| [27] | Kelliher T, Starr D, Wang W, Mccuiston J, Zhong H, Nuccio M L, Martin B. Maternal haploids are preferentially induced by CENH3-tailswap transgenic complementation in maize[J]. Frontiers in Plant Science, 2016, 7: 414. |
| [28] | Lü J, Yu K, Wei J, Gui H P, Liu C X, Liang D W, Wang Y L, Zhou H J, Carlin R, Rich R, Lu T C, Que Q D, Wang W C, Zhang X P, Kelliher T. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3[J]. Nature Biotechnology, 2020, 38(12): 1397-1401. |
| [29] | Jiang C L, Sun J, Li R, Yan S J, Chen W, Guo L, Qin G C, Wang P C, Luo C, Huang W J, Zhang Q H, Fernie A R, Jackson D, Li X, Yan J B. A reactive oxygen species burst causes haploid induction in maize[J]. Molecular Plant, 2022, 15(6): 943-955. |
| [30] | Sarkar K R, Coe E H. A genetic analysis of the origin of maternal haploids in maize[J]. Genetics, 1966, 54(2): 453-464. |
| [31] | Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds[J]. Nature, 2019, 565(7737): 91-95. |
| [32] | Conner J A, Podio M, Ozias-Akins P. Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes[J]. Plant Reproduction, 2017, 30(1): 41-52. |
| [33] | Wei X, Liu C L, Chen X, Lu H W, Wang J, Yang S L, Wang K J. Synthetic apomixis with normal hybrid rice seed production[J]. Molecular Plant, 2023, 16(3): 489-492. |
| [34] | Underwood C J, Vijverberg K, Rigola D, Okamoto S, Oplaat C, Camp R, Radoeva T, Schauer S E, Fierens J, Jansen K, Mansveld S, Busscher M, Xiong W, Datema E, Nijbroek K, Blom E J, Bicknell R, Catanach A, Erasmuson S, Winefield C, van Tunen A J, Prins M, Schranz M E, van Dijk P J. A PARTHENOGENESIS allele from apomictic dandelion can induce egg cell division without fertilization in lettuce[J]. Nature Genetics, 2022, 54(1): 84-93. |
| [35] | Zhang X C, Shi C, Li S L, Zhang B, Luo P, Peng X B, Zhao P, Dresselhaus T, Sun M X. A female in vivo haploid-induction system via mutagenesis of egg cell-specific peptidases[J]. Molecular Plant, 2023, 16(2): 471-480. |
| [36] | Cyprys P, Lindemeier M, Sprunck S. Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins[J]. Nature Plants, 2019, 5(3): 253-257. |
| [1] | 吴金水, 唐江英, 谭立, 过志强, 杨娟, 张鑫臻, 陈桂芳, 王建龙, 施婉菊. 水稻对砷的吸收与转运机理及农艺阻控策略 [J]. 中国水稻科学, 2025, 39(2): 143-155. |
| [2] | 马唯一, 朱济邹, 朱旺, 耿孝宇, 张翔, 刁刘云, 汪璐璐, 孟天瑶, 高平磊, 陈英龙, 戴其根, 韦还和. 盐害和干旱对稻米品质形成的影响及生理机制研究进展 [J]. 中国水稻科学, 2025, 39(2): 156-170. |
| [3] | 张来桐, 杨乐, 刘洪, 赵学明, 程涛, 徐振江. 水稻香味物质的研究进展 [J]. 中国水稻科学, 2025, 39(2): 171-186. |
| [4] | 冯涛, 张朝阳, 黄新妮, 王月, 钟旭志, 冯志明, 刘欣, 左示敏, 欧阳寿强. Osa-miR166i-3p介导活性氧积累途径正调控水稻纹枯病抗性 [J]. 中国水稻科学, 2025, 39(2): 187-196. |
| [5] | 龚蒙萌, 宋书锋, 邱牡丹, 董皓, 张龙辉, 李磊, 李斌, 谌伟军, 李懿星, 王天抗, 雷东阳, 李莉. 水稻叶色基因OsClpP6的功能研究 [J]. 中国水稻科学, 2025, 39(2): 197-208. |
| [6] | 闫影, 王凯, 张丽霞, 胡泽军, 叶俊华, 杨航, 顾春军, 吴书俊. 利用分子聚合育种培育优质多抗粳稻新品种沪香粳216 [J]. 中国水稻科学, 2025, 39(2): 209-219. |
| [7] | 徐月梅, 彭诗燕, 孙志伟, 王志琴, 朱宽宇, 杨建昌. 不同耐低磷水稻品种的内源激素水平差异及其与产量和磷利用率的关系 [J]. 中国水稻科学, 2025, 39(2): 231-244. |
| [8] | 随晶晶, 赵桂龙, 金欣, 卜庆云, 唐佳琦. 水稻孕穗期耐冷调控的分子及生理机制研究进展[J]. 中国水稻科学, 2025, 39(1): 1-10. |
| [9] | 任宁宁, 孙永建, 申聪聪, 朱双兵, 李慧菊, 张志远, 陈凯. 水稻中胚轴研究进展[J]. 中国水稻科学, 2025, 39(1): 11-23. |
| [10] | 张丰勇, 应晓平, 张健, 杨隆维, 应杰政. 半矮秆基因sd1调控水稻重要农艺性状的研究进展[J]. 中国水稻科学, 2025, 39(1): 24-32. |
| [11] | 陈智慧, 陶亚军, 范方军, 许扬, 王芳权, 李文奇, 古丽娜尔·巴合提别克, 蒋彦婕, 朱建平, 李霞, 杨杰. 水稻抽穗期调控基因Hd6功能标记的开发及应用[J]. 中国水稻科学, 2025, 39(1): 47-54. |
| [12] | 陈书融, 朱练峰, 秦碧蓉, 王婕, 朱旭华, 田文昊, 朱春权, 曹小闯, 孔亚丽, 张均华, 金千瑜. 增氧灌溉下配施硝化抑制剂对水稻生长、产量和氮肥利用的影响[J]. 中国水稻科学, 2025, 39(1): 92-100. |
| [13] | 吴猛, 倪川, 康钰莹, 毛雨欣, 叶苗, 张祖建. 水稻分蘖早发特性的品种间差异及其氮素响应[J]. 中国水稻科学, 2025, 39(1): 101-114. |
| [14] | 王晓茜, 蔡创, 宋练, 周伟, 杨雄, 顾歆悦, 朱春梧. 开放式大气CO2浓度升高和温度升高对扬稻6号稻米品质的影响[J]. 中国水稻科学, 2025, 39(1): 115-127. |
| [15] | 江敏, 王广伦, 李明璐, 苗波, 李明煊, 石春林. 基于模型的水稻高温热害风险评估与动态预警[J]. 中国水稻科学, 2025, 39(1): 128-142. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||