
中国水稻科学 ›› 2021, Vol. 35 ›› Issue (6): 535-542.DOI: 10.16819/j.1001-7216.2021. 210205
吴先美1,#, 李三峰1,#, 胡萍1, 何瑞1, 焦然2, 毛一剑1, 鲁草林1, 胡娟2, 林晗2, 吴荣梁1, 朱旭东1, 饶玉春2,*( ), 王跃星1,*(
), 王跃星1,*( )
)
收稿日期:2021-02-16
修回日期:2021-03-23
出版日期:2021-11-10
发布日期:2021-11-10
通讯作者:
饶玉春,王跃星
作者简介:#共同第一作者
基金资助:
Xianmei WU1,#, Sanfeng LI1,#, Ping HU1, Rui HE1, Ran JIAO2, Yijian MAO1, Caolin LU1, Juan HU2, Han LIN2, Rongliang WU1, Xudong ZHU1, Yuchun RAO2,*( ), Yuexing WANG1,*(
), Yuexing WANG1,*( )
)
Received:2021-02-16
Revised:2021-03-23
Online:2021-11-10
Published:2021-11-10
Contact:
Yuchun RAO, Yuexing WANG
About author:#These authors contributed equally to this work
摘要:
【目的】克隆水稻分蘖相关基因,为构建理想株型水稻,提高粮食产量提供理论基础及有利基因资源。【方法】在常规大田种植条件下分别比较突变体htd3(high-tillering dwarf 3)与野生型在幼苗期、抽穗期和成熟期表型及主要农艺性状差异,利用图位克隆方法克隆候选基因,利用荧光定量PCR方法分析HTD3及独脚金内酯和脱落酸相关基因的表达水平,测序比对分析HTD3在147份种质资源中的自然变异情况。【结果】与野生型相比,突变体htd3的分蘖芽生长较快,分蘖数和有效穗数显著增多,株高、一次枝梗数和每穗粒数显著降低,结实率和千粒重没有显著变化。遗传分析表明,htd3多分蘖的性状受一对隐性核基因控制。图位克隆将HTD3基因定位在第12染色体CM8和CM10之间约63.5 kb的物理区间内,互补转基因实验证明该区间内LOC_Os12g21710为控制突变体多分蘖表型的基因。HTD3在野生型和突变体中呈组成型表达,该基因突变会引起部分独脚金内酯和脱落酸相关基因的表达水平上调。水稻品种中HTD3编码区G2674A自然变异使得分蘖数显著增多。【结论】HTD3是最近报道的T20/MIT1基因的一个新的等位基因,HTD3突变导致水稻出现分蘖适度增加,株高略矮的表型,在培育理想株型水稻和高产育种上具有较大的应用潜力。
吴先美, 李三峰, 胡萍, 何瑞, 焦然, 毛一剑, 鲁草林, 胡娟, 林晗, 吴荣梁, 朱旭东, 饶玉春, 王跃星. 水稻分蘖调控基因HTD3的克隆与功能研究[J]. 中国水稻科学, 2021, 35(6): 535-542.
Xianmei WU, Sanfeng LI, Ping HU, Rui HE, Ran JIAO, Yijian MAO, Caolin LU, Juan HU, Han LIN, Rongliang WU, Xudong ZHU, Yuchun RAO, Yuexing WANG. Cloning and Functional Analysis of Rice Tillering Regulatory Gene HTD3[J]. Chinese Journal OF Rice Science, 2021, 35(6): 535-542.
| 标记 Marker | 正向引物 Forward primer (5′→3′) | 反向引物 Reverse primer (5′→3′) |
|---|---|---|
| B12-6 | TGAAGCGGTTTGACTTTGACC | GGGGTGAAAACTGGTAGGGT |
| B12-9 | TCCTTCTCGTTTATGAACTTATGG | TAGAGCAAAGCAGAGCCCAG |
| M9 | TAATCCCCTGCACTCCATCC | GTGAACAACCAGCCGAGAAT |
| M17 | GACTTTCTAGCATTGCCCACA | GCATTAACTGGGGCCATTGT |
| M25 | GAGACGGCCAGCTTAGGTAG | CAGTGCTACAGAAACAGGGC |
| M29 | CCGAACTCCAGTTTGTGAGG | CGGAAATCTGACGCTGGTAT |
| M35 | AAAAGAACAACACAGCCCCT | ACAAGGAGCAATCTGGACCA |
| CM5 | TTGTGAACAAGAGCCAACGG | CGCTGTTGGGCATTCTTTAAAG |
| CM8 | GTGTGATCCATGGGTAGCCT | CAGAGCCCTATTAGTCTATTGCT |
| CM10 | TCACATGATACCTCGCGAGT | CAGACGATTCTACACAACAGGA |
| CM13 | AGCAGCCAAGATTAAGGAGGA | CGTCAGAGTGATTAGCAAAAGGA |
表1 HTD3精细定位所用分子标记
Table 1 Sequences of markers for HTD3 mapping.
| 标记 Marker | 正向引物 Forward primer (5′→3′) | 反向引物 Reverse primer (5′→3′) |
|---|---|---|
| B12-6 | TGAAGCGGTTTGACTTTGACC | GGGGTGAAAACTGGTAGGGT |
| B12-9 | TCCTTCTCGTTTATGAACTTATGG | TAGAGCAAAGCAGAGCCCAG |
| M9 | TAATCCCCTGCACTCCATCC | GTGAACAACCAGCCGAGAAT |
| M17 | GACTTTCTAGCATTGCCCACA | GCATTAACTGGGGCCATTGT |
| M25 | GAGACGGCCAGCTTAGGTAG | CAGTGCTACAGAAACAGGGC |
| M29 | CCGAACTCCAGTTTGTGAGG | CGGAAATCTGACGCTGGTAT |
| M35 | AAAAGAACAACACAGCCCCT | ACAAGGAGCAATCTGGACCA |
| CM5 | TTGTGAACAAGAGCCAACGG | CGCTGTTGGGCATTCTTTAAAG |
| CM8 | GTGTGATCCATGGGTAGCCT | CAGAGCCCTATTAGTCTATTGCT |
| CM10 | TCACATGATACCTCGCGAGT | CAGACGATTCTACACAACAGGA |
| CM13 | AGCAGCCAAGATTAAGGAGGA | CGTCAGAGTGATTAGCAAAAGGA |
| 引物名称 Primer name | 引物序列 Primer sequences (5′→3′) |
|---|---|
| HTD3-CPT-EcoRⅠ-F | ACGAATTCGAGCTCGGTACCTGGTTCTGTGACTAAAGCGC |
| HTD3-CPT-HindⅢ-R | GGCCAGTGCCAAGCTTTCTCCGGGGCCCTGAATATTCCTCT |
表2 扩增HTD3完整表达单元的引物
Table 2 Primers for amplifying the complete expression unit of HTD3.
| 引物名称 Primer name | 引物序列 Primer sequences (5′→3′) |
|---|---|
| HTD3-CPT-EcoRⅠ-F | ACGAATTCGAGCTCGGTACCTGGTTCTGTGACTAAAGCGC |
| HTD3-CPT-HindⅢ-R | GGCCAGTGCCAAGCTTTCTCCGGGGCCCTGAATATTCCTCT |
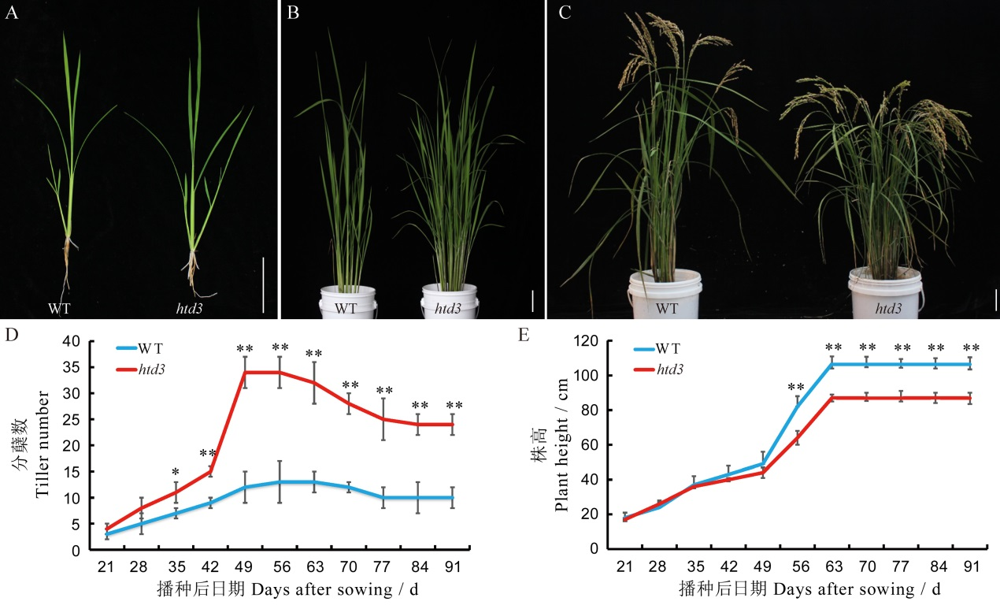
图1 野生型和htd3的形态性状比较 A~C-野生型和htd3在苗期(A),分蘖期(B)和成熟期(C)的植株表型,标尺为6 cm;D,E-野生型和htd3的分蘖数(D)和株高(E)随播种后日期的动态变化。数据为平均值±标准差(n=15);采用t测验,**表示在0.01水平上差异显著,*表示在0.05水平上差异显著。
Fig. 1. Morphological characterization of WT and htd3. A~C, Plant phenotypes of wild-type (WT) and htd3 in the seedling stage (A), tillering stage (B) , and mature stage (C) , bar= 6 cm; D~E, Tiller number (D) and plant height (E) of WT and htd3 after sowing. Values are means ± SD (n=15), t-test, *, ** indicate significant difference at P<0.05 and P<0.01, respectively.
| 农艺性状Agronomic trait | 野生型WT | 突变体htd3 |
|---|---|---|
| 株高Plant height /cm | 97.08±2.78 | 83.76±1.54* |
| 有效穗数Effective panicles | 11.0±1.0 | 28.0±2.0** |
| 穗长Panicle length/cm | 21.52±0.98 | 20.82±0.62 |
| 一次枝梗数 Primary branches | 12.0±2.0 | 9.0±1.0* |
| 二次枝梗数Secondary branches | 26.0±3.0 | 23.0±4.0 |
| 每穗粒数Grains per panicle | 121.0±11.0 | 104.0±16.0* |
| 每株总粒数Total grains per plant | 1331±28 | 2018±36** |
| 千粒重1000-grain weight/g | 23.7±2.1 | 23.9±1.3 |
| 结实率Seed setting rate/% | 87.2±5.8 | 90.0±2.4 |
表3 野生型和htd3的主要农艺性状比较
Table 3 Comparison of agronomic traits between WT and htd3.
| 农艺性状Agronomic trait | 野生型WT | 突变体htd3 |
|---|---|---|
| 株高Plant height /cm | 97.08±2.78 | 83.76±1.54* |
| 有效穗数Effective panicles | 11.0±1.0 | 28.0±2.0** |
| 穗长Panicle length/cm | 21.52±0.98 | 20.82±0.62 |
| 一次枝梗数 Primary branches | 12.0±2.0 | 9.0±1.0* |
| 二次枝梗数Secondary branches | 26.0±3.0 | 23.0±4.0 |
| 每穗粒数Grains per panicle | 121.0±11.0 | 104.0±16.0* |
| 每株总粒数Total grains per plant | 1331±28 | 2018±36** |
| 千粒重1000-grain weight/g | 23.7±2.1 | 23.9±1.3 |
| 结实率Seed setting rate/% | 87.2±5.8 | 90.0±2.4 |
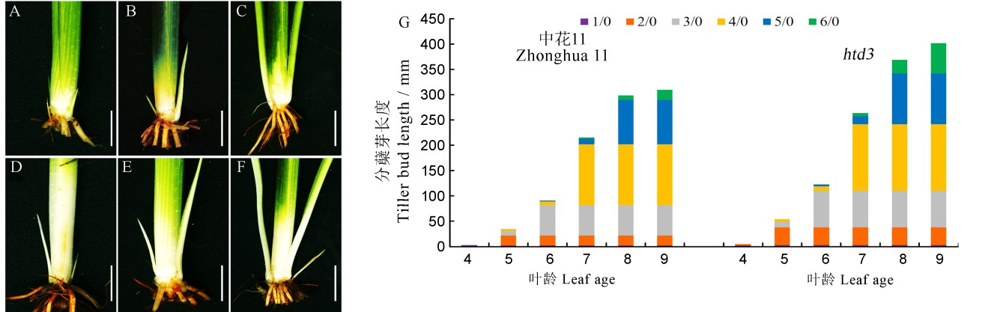
图2 野生型和htd3分蘖芽表型观察 A~F-野生型和突变体htd3在5叶期、6叶期、7叶期的分蘖芽表型, A~C为野生型, D~F为突变体, 标尺为1 cm;G-野生型和突变体htd3在各叶期的分蘖长度, n/0中n表示叶位, 0表示主茎, 即1/0表示主茎第一片叶对应的分蘖芽, 2/0表示主茎第2片叶对应的分蘖芽, 以此类推。
Fig. 2. Phenotypic observation of tillering buds of WT and htd3. A~F, Phenotype of tillering buds of WT and htd3 at five-, six- and seven-leaf stage; A-C represents WT; D-F represents htd3; bars=1 cm; G, Tillering bud length of WT and htd3 at each leaf stage, in n/0, n represents the leaf position, 0 represents the main stem, 1/0 represents the tiller bud corresponding to the first leaf of the main stem, 2/0 represents the tiller bud corresponding to the second leaf of the main stem, and so on.
| 组合Cross | F1表型 Phenotype of F1 plants | F2群体 F2 population | χ2 (3:1) | χ20. 05 | ||
|---|---|---|---|---|---|---|
| 总株数 No. of plants | 正常分蘖株数 No. of normal plants | 多分蘖株数 No. of high-tillering plants | ||||
| 中花11/ TN1 Zhonghua 11/TN1 | 正常分蘖Normal tiller | 864 | 666 | 198 | 2 | 3. 84 |
表4 水稻矮秆多分蘖突变体htd3的遗传分析
Table 4 Genetic analysis of rice high-tillering mutant htd3.
| 组合Cross | F1表型 Phenotype of F1 plants | F2群体 F2 population | χ2 (3:1) | χ20. 05 | ||
|---|---|---|---|---|---|---|
| 总株数 No. of plants | 正常分蘖株数 No. of normal plants | 多分蘖株数 No. of high-tillering plants | ||||
| 中花11/ TN1 Zhonghua 11/TN1 | 正常分蘖Normal tiller | 864 | 666 | 198 | 2 | 3. 84 |
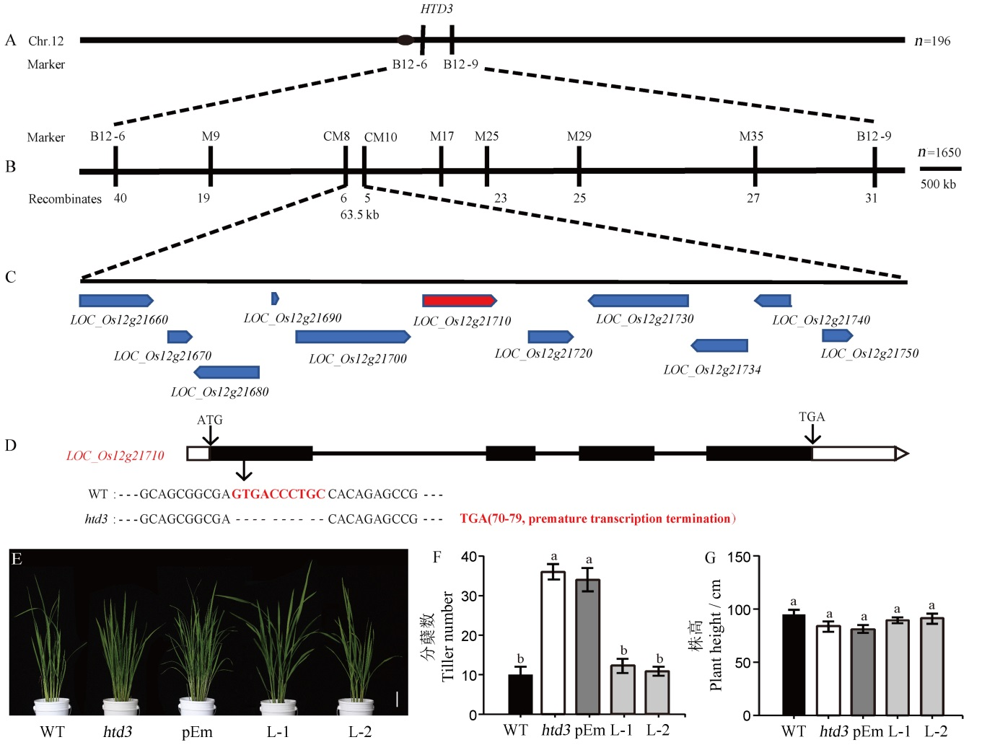
图3 HTD3的图位克隆和互补转基因验证 A-HTD3的初定位;B-HTD3的精细定位;C-定位区间内预测的ORF;D-候选基因LOC_Os12g21710的基因结构及在野生型和htd3中的序列差异,黑色方框代表外显子,白色方框代表UTR,黑色线条代表内含子;E-野生型、htd3和互补转基因T2代植株分蘖期表型,标尺为6 cm;F~G-野生型、htd3和互补转基因T2代植株分蘖期的分蘖数(F)和株高(G)统计,数据为平均值±标准差(n=5)。用新复极差法进行多重比较,相同小写字母表示在0.05水平上无显著差异。
Fig. 3. Map-based cloning of HTD3 and verification of complementary transgene. A, Initial location of HTD3; B, Fine mapping of HTD3; C, Predicted ORFs in the location interval; D, Gene structure of the candidate gene LOC_Os12g21710 and the sequence difference between WT and htd3, in which black boxes, white boxes, black lines represent exons, UTR and introns, respectively; E, Phenotype of WT, htd3 and complementary transgenic T2 generation plants in the tilling stage, bar=6 cm; F~G, Number of tillers (F) and plant height (G) of WT, htd3 and complementary transgenic T2 generation plants in the tillering stage, the data are means ± standard deviation (n=5). Significant difference by Duncan’s multiple range test. The same letters indicate no significant difference at P<0.05.
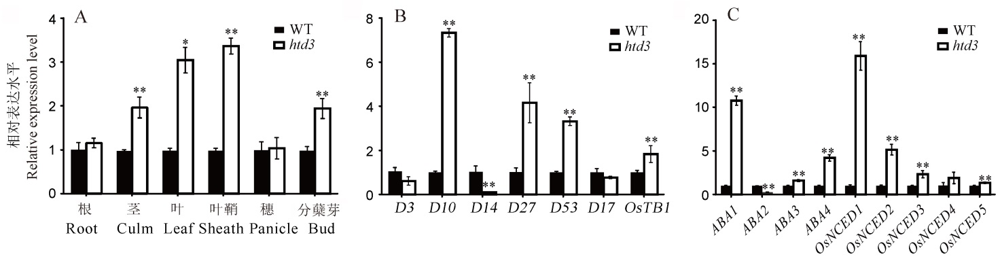
图4 HTD3和激素相关基因的表达水平分析 A-HTD3基因在野生型和突变体htd3各组织中相对表达水平;B~C-独脚金内酯(B)和ABA(C)相关基因在野生型和突变体htd3叶片中的相对表达水平。以相应基因在野生型中的表达水平作为参考并设为1, 数据为平均值±标准差, 3次生物学重复;采用t测验, *表示在0. 05水平上差异显著, **表示在0. 01水平上差异显著。
Fig. 4. Expression analysis of HTD3 and hormone related genes. A, HTD3 relative expression level in various tissues of WT and htd3; B-C, Relative expression of SLs (B) and ABA (C) related genes in WT and htd3. RNA was isolated from WT and htd3 leaves in B and C. Expression levels are represented as relative to the corresponding genes in WT (set as reference value of 1), and data are shown as means ± SD from three biological replicates. Asterisks indicate statistical significance between the WT and the mutant, as determined by Student’s t-test (*P < 0.05; **P < 0.01).
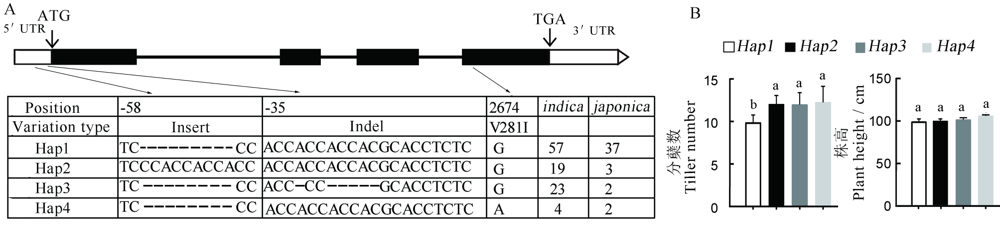
图5 HTD3的单倍型分析 A-HTD3的基因结构及147份水稻品种中HTD3的5'-UTR和编码区上核苷酸多态性, 黑色方块代表外显子, 白色方框代表UTR, 黑色方框之间的黑色线条代表内含子, Hap代表单倍型, 数字代表相对起始密码子ATG的碱基位置;B-每种单倍型对应的分蘖数和株高。用新复极差法进行多重比较, 相同小写字母表示在0.05水平上无显著差异。
Fig. 5. Analysis of the HTD3 gene haplotypes. A, Gene structure of the HTD3, and nucleotide polymorphisms in the HTD3 5'-UTR and coding region, exons are indicated by black boxes, UTRs are indicated by white boxes, and the black lines between black boxes represent introns, hap means haplotypes, the number represents the base position relative to the start codon ATG. B, Tiller number and plant height of each haplotype. Significant difference by Duncan’s multiple range test. The same letters indicate no significant difference at P<0.05.
| [1] | 李学勇, 钱前, 李家洋. 水稻分蘖的分子机理研究[J]. 中国科学院院刊, 2003, 18(4): 274-276. |
| Li X Y, Qian Q, Li J Y.Progress in elucidating the molecular mechanism of rice tillering[J]. Bulletin of the Chinese Academy of Sciences, 2003, 18(4): 274-276. (in Chinese with English abstract) | |
| [2] | Wu T, Shen Y, Zheng M, Yang C, Chen Y, Feng Z, Liu X, Liu S, Chen Z, Lei C, Wang J, Jiang L, Wan J.Gene SGL, encoding a kinesin-like protein with transactivation activity, is involved in grain length and plant height in rice[J]. Plant Cell Reports, 2014, 33(2): 235-244. |
| [3] | Li X Y, Qian Q, Fu Z M, Wang Y H, Xiong G S, Zeng D L, Wang X Q, Liu X F, Teng S, Hiroshi F, Yuan M, Luo D, Han B, Li J Y.Control of tillering in rice[J]. Nature, 2003, 422(6932): 618-621. |
| [4] | Xu C, Wang Y, Yu Y, Duan J, Liao Z, Xiong G, Meng X, Liu G, Qian Q, Li J.Degradation of MONOCULM 1 by APC/C(TAD1) regulates rice tillering[J]. Nature Communications, 2012, 3(1): 1-9. |
| [5] | Liang W H, Shang F, Lin Q T, Lou C, Zhang J.Tillering and panicle branching genes in rice[J]. Gene, 2014, 537(1): 1-5. |
| [6] | Kim H, Hwang H, Hong J W, Lee Y N, Ahn I P, Yoon I S, Yoo S D, Lee S, Lee S C, Kim B G.A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth[J]. Journal of Experimental Botany, 2012, 63(2): 1013-1024. |
| [7] | Ljung K, Bhalerao R P, Sandberg G.Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J, 2001, 28(4):465-474. |
| [8] | Lee M, Jung J H, Han D Y, Seo P J, Park W J, Park C M.Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis[J]. Planta, 2012, 235(5): 923-938. |
| [9] | Xu M, Zhu L, Shou H, Wu P.A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice[J]. Plant Cell Physiology, 2005, 46(10): 1674-1681. |
| [10] | Xu J X, Ding C Q, Ding Y F, Tang S, Zha M R, Luo B J, Wang S H.A proteomic approach to analyze differential regulation of proteins during bud outgrowth under apical dominance based on the auxin transport canalization model in rice (Oryza sativa L.)[J]. Journal of Plant Growth Regulation, 2015, 34(1): 122-136. |
| [11] | Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, Wang Y.DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth[J]. Plant Cell, 2009, 21(5): 1512-1525. |
| [12] | Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L.The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds[J]. Plant Journal, 2006, 48(5): 687-698. |
| [13] | Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J.DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice[J]. Plant Journal, 2007, 51(6): 1019-1029. |
| [14] | Fang Z, Ji Y, Hu J, Guo R, Sun S, Wang X.Strigolactones and brassinosteroids antagonistically regulate the stability of the D53-OsBZR1 complex codetermine FC1 expression in rice tillering[J]. Molecular Plant, 2020, 13(4): 586-597. |
| [15] | Zhao J, Wang T, Wang M, Zhao J, Wang T, Wang M, Liu Y, Yuan S, Gao Y, Yin L, Sun W, Wan J, Li X.DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching[J]. Plant Cell Physiology, 2014, 55(6): 1096-1109. |
| [16] | Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J.d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers[J]. Plant Cell Physiology, 2009, 50(8): 1416-1424. |
| [17] | Liu W, Wu C, Fu Y, Hu G, Si H, Zhu L, Luan W, He Z, Sun Z.Identification and characterization of HTD2: A novel gene negatively regulating tiller bud outgrowth in rice[J]. Planta, 2009, 230(4): 649-658. |
| [18] | Gao Z, Qian Q, Liu X, Yan M, Feng Q, Dong G, Liu J, Han B.Dwarf 88, a novel putative esterase gene affecting architecture of rice plant[J]. Plant Molecular Biology, 2009, 71(3): 265-276. |
| [19] | Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J.Suppression of tiller bud activity in tillering dwarf mutants of rice[J]. Plant Cell Physiology, 2005, 46(1): 79-86. |
| [20] | De S A, Clavé G, Badet-Denisot M A, Pillot J P, Cornu D, Le Caer J P, Burger M, Pelissier F, Retailleau P, Turnbull C, Bonhomme S, Chory J, Rameau C, Boyer F D. An histidine covalent receptor and butenolide complex mediates strigolactone perception[J]. Nature Chemical Biology, 2016, 12(10): 787-794. |
| [21] | Sharma R, De V D, Sharma M K, Ronald P C.Recent advances in dissecting stress-regulatory crosstalk in rice[J]. Molecular Plant, 2013, 6(2): 250-260. |
| [22] | Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Li Y, Yan C, Miao D, Sun Z, Yan J, Sun Y, Wang L, Chu J, Fan S, He W, Deng H, Nan F, Li J, Rao Z, Lou Z, Xie D.DWARF14 is a non-canonical hormone receptor for strigolactone[J]. Nature, 2016, 536(7617): 469-473. |
| [23] | Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, Ma W.D14-SCF(D3)- dependent degradation of D53 regulates strigolactone signaling[J]. Nature, 2013, 504(7480): 406-410. |
| [24] | Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi- Tanaka M, Ashikari M, Matsuoka M, Ueguchi C.The OsTB1 gene negatively regulates lateral branching in rice[J]. Plant Journal, 2003, 33(3): 513-520. |
| [25] | Umehara M, Hanada A, Magome H, Takeda K N, Yamaguchi S.Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice[J]. Plant Cell Physiology, 2010, 51(7): 1118-1126. |
| [26] | Wang Y, Shang L, Yu H, Zeng L, Hu J, Ni S, Rao Y, Li S, Chu J, Meng X, Wang L, Hu P, Yan J, Kang S, Qu M, Lin H, Wang T, Wang Q, Hu X, Chen H, Wang B, Gao Z, Guo L, Xiong G, Li J, Qian Q.A strigolactone biosynthesis gene contributed to the green revolution in rice[J]. Molecular Plant, 2020, 13(6): 923-932. |
| [27] | Liu X, Hu Q, Yan J, Sun K, Liang Y, Jia M, Meng X, Fang S, Wang Y, Jing Y, Liu G, Wu D, Chu C, Smith S M, Chu J, Wang Y, Li J, Wang B.ζ-carotene isomerase suppresses tillering in rice through the coordinated biosynthesis of strigolactone and abscisic acid[J]. Molecular Plant, 2020, 13(12): 1784-1801. |
| [28] | Liu L, Ren M, Peng P, Chun Y, Li L, Zhao J, Fang J, Peng L, Yan J, Chu J, Wang Y, Yuan S, Li X.MIT1, encoding a 15-cis-ζ-carotene isomerase, regulates tiller number and stature in rice[J]. Journal of Genet Genomics, 2021, 48(1): 88-91. |
| [29] | 陈彩艳, 邹军煌, 张淑英, 朱立煌. 独角金内酯能抑制植物的分枝并介导植物与枞枝真菌及寄生植物间的相互作用[J]. 中国科学: 生命科学, 2009(6): 525-533. |
| Chen C, Zou J, Zhang S, Zhu L.Strigolactones are a new-defined class of plant hormones which inhibit shoot branching and mediate the interaction of plant-AM fungi and plant-parasitic weeds[J]. Science in China: Life Sciences, 2009(6): 525-533. (in Chinese with English abstract) | |
| [30] | Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Sun X, Deng X.Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice[J]. Plant Physiology, 2010, 153(3): 994-1003. |
| [31] | Cline M G, Oh C.A reappraisal of the role of abscisic acid and its interaction with auxin in apical dominance[J]. Annual Botany, 2006, 98(4): 891-897. |
| [32] | 刘杨. 水稻分蘖芽萌发与休眠相互转换的激素学机制[D]. 南京: 南京农业大学, 2011. |
| Liu Y.The Mechanism of hormonal regulation of the transformation between germination and dormancy of rice tiller buds[D]. Nanjing: Nanjing Agricultural University.(in Chinese with English abstract) | |
| [33] | Zang G, Zou H, Zhang Y.The De-Etiolated 1 homolog of Arabidopsis modulates the ABA signaling pathway and ABA biosynthesis in rice[J]. Plant Physiology, 2016, 171(2): 1259-1276. |
| [34] | Bang S W, Park S H, Jeong J S, Kim Y S, Jung H, Ha S H, Kim J K.Characterization of the stress-inducible OsNCED3 promoter in different transgenic rice organs and over three homozygous generations[J]. Planta, 2013, 237(1): 211-224. |
| [1] | 汪邑晨, 朱本顺, 周磊, 朱骏, 杨仲南. 光/温敏核不育系的不育机理及两系杂交稻的发展与展望 [J]. 中国水稻科学, 2024, 38(5): 463-474. |
| [2] | 许用强, 徐军, 奉保华, 肖晶晶, 王丹英, 曾宇翔, 符冠富. 水稻花粉管生长及其对非生物逆境胁迫的响应机理研究进展 [J]. 中国水稻科学, 2024, 38(5): 495-506. |
| [3] | 何勇, 刘耀威, 熊翔, 祝丹晨, 王爱群, 马拉娜, 王廷宝, 张健, 李建雄, 田志宏. 利用CRISPR/Cas9技术编辑OsOFP30基因创制水稻粒型突变体 [J]. 中国水稻科学, 2024, 38(5): 507-515. |
| [4] | 吕阳, 刘聪聪, 杨龙波, 曹兴岚, 王月影, 童毅, Mohamed Hazman, 钱前, 商连光, 郭龙彪. 全基因组关联分析(GWAS)鉴定水稻氮素利用效率候选基因 [J]. 中国水稻科学, 2024, 38(5): 516-524. |
| [5] | 杨好, 黄衍焱, 王剑, 易春霖, 石军, 谭楮湉, 任文芮, 王文明. 水稻中八个稻瘟病抗性基因特异分子标记的开发及应用 [J]. 中国水稻科学, 2024, 38(5): 525-534. |
| [6] | 杨铭榆, 陈志诚, 潘美清, 张汴泓, 潘睿欣, 尤林东, 陈晓艳, 唐莉娜, 黄锦文. 烟-稻轮作下减氮配施生物炭对水稻茎鞘同化物转运和产量 形成的影响 [J]. 中国水稻科学, 2024, 38(5): 555-566. |
| [7] | 熊家欢, 张义凯, 向镜, 陈惠哲, 徐一成, 王亚梁, 王志刚, 姚坚, 张玉屏. 覆膜稻田施用炭基肥对水稻产量及氮素利用的影响 [J]. 中国水稻科学, 2024, 38(5): 567-576. |
| [8] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [9] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [10] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [11] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [12] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [13] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [14] | 胡丽, 杨范敏, 陈薇兰, 袁华. 水稻SPL家族转录因子的生物学功能研究进展[J]. 中国水稻科学, 2024, 38(3): 223-232. |
| [15] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||