
中国水稻科学 ›› 2025, Vol. 39 ›› Issue (6): 711-730.DOI: 10.16819/j.1001-7216.2025.241005
• 综述与专论 • 下一篇
宋安琪1,2, 吴松权1, 马秋月1,2, 班宛宁1,2, 刘相国2,*( ), 金永梅2,*(
), 金永梅2,*( )
)
收稿日期:2024-10-16
修回日期:2024-11-28
出版日期:2025-11-10
发布日期:2025-11-19
通讯作者:
* email:lxgyyj@cjaas.com;ymjin0303@163.com
基金资助:
SONG Anqi1,2, WU Songquan1, MA Qiuyue1,2, BAN Wanning1,2, LIU Xiangguo2,*( ), JIN Yongmei2,*(
), JIN Yongmei2,*( )
)
Received:2024-10-16
Revised:2024-11-28
Online:2025-11-10
Published:2025-11-19
Contact:
* email:lxgyyj@cjaas.com;ymjin0303@163.com
摘要:
植物引导编辑技术(Plant Prime Editing,PPE)为植物基因组的精准改良提供了全新途径,克服了传统基因编辑方法中依赖双链断裂和外源供体DNA的局限性,不仅可以实现任意类型碱基替换,还可以进行小片段乃至大范围片段的精准插入和删除,并且脱靶率较低。本文详细阐述了PPE技术在国内外的最新研究进展,包括其发展历程与工作原理、在作物育种与性状改良中的应用,及PE技术在大规模基因组编辑和多重基因编辑等领域的拓展。针对PPE系统在植物遗传转化过程中遇到的瓶颈问题,提出了若干解决方案,并展望了PPE技术在植物遗传改良中的广泛应用前景及未来与人工智能(Artificial Intelligence,AI)相结合的研究方向。
宋安琪, 吴松权, 马秋月, 班宛宁, 刘相国, 金永梅. 植物引导编辑技术——作物育种的新方向[J]. 中国水稻科学, 2025, 39(6): 711-730.
SONG Anqi, WU Songquan, MA Qiuyue, BAN Wanning, LIU Xiangguo, JIN Yongmei. Plant Prime Editing: A New Direction in Crop Breeding[J]. Chinese Journal OF Rice Science, 2025, 39(6): 711-730.
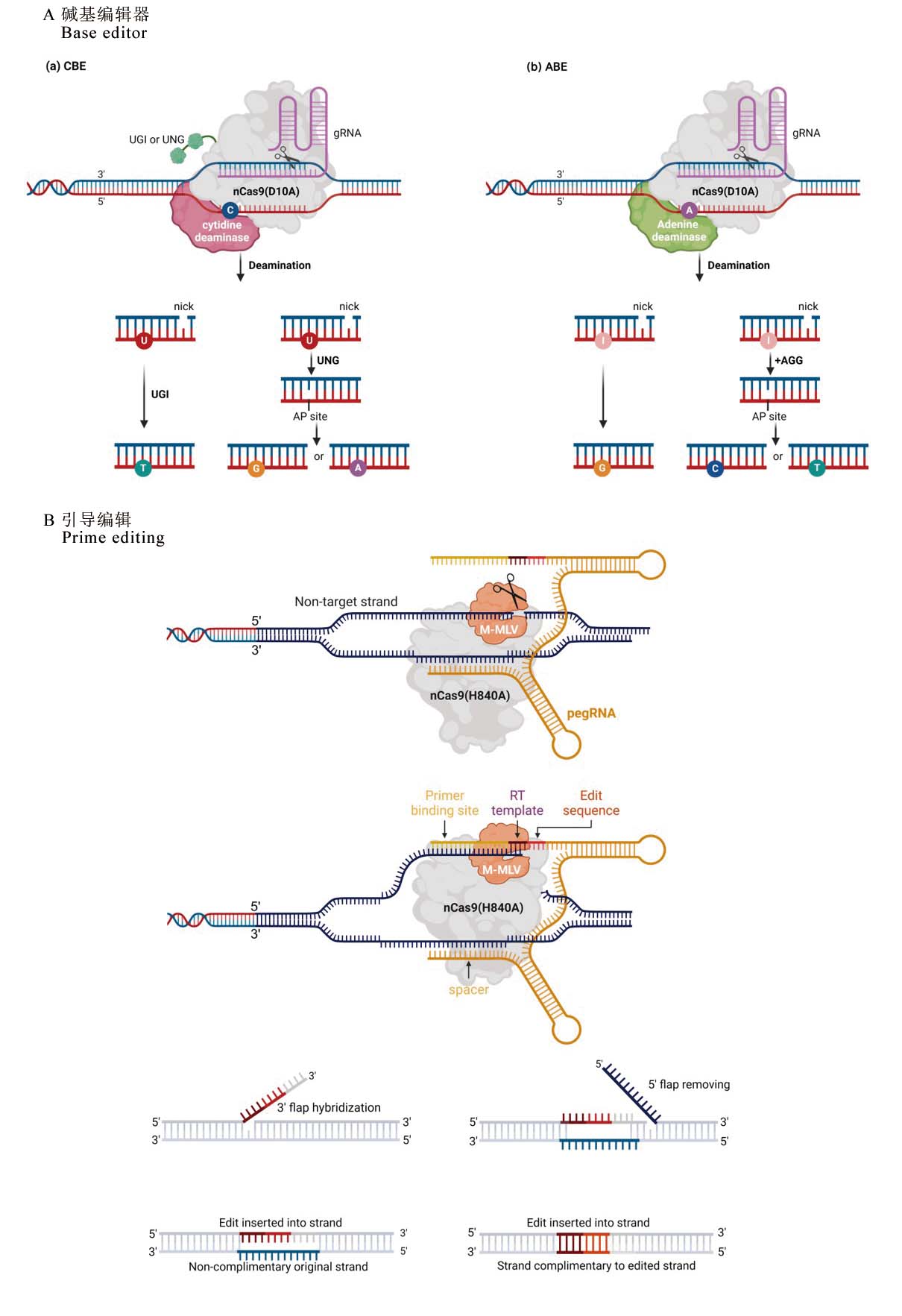
图1 碱基编辑器和引导编辑的工作模式 A, 碱基编辑器(Base editor)工作原理。(a)胞嘧啶碱基编辑(CBE):CBE系统使用nCas9缺刻酶(D10A突变)在目标DNA链上产生单链切口,暴露出单链DNA区域。胞苷脱氨酶作用于该区域,将胞嘧啶(C)转化为尿嘧啶(U)。尿嘧啶DNA糖基化酶抑制剂(UGI)可抑制细胞内尿嘧啶糖基化酶(UNG)对U的切除,使U在DNA修复过程中被识别为胸腺嘧啶(T),从而实现C→T的碱基替换。若未加入UGI或额外表达UNG,U将被切除形成无嘌呤/无嘧啶(AP)位点,进而可能导致C→G或C→A的替换。(b)腺嘌呤碱基编辑(ABE):ABE系统利用腺嘌呤脱氨酶将腺嘌呤(A)转化为肌苷(I),后者在DNA复制中被识别为鸟嘌呤(G),从而实现A→G的替换。若加入烷基腺嘌呤DNA糖基化酶(AAG),肌苷(I)可被切除形成AP位点,进而引发A→C或A→T的替换。B, 引导编辑工作原理。nCas9(H840A)诱导切割非靶链产生一个缺口。然后将引物结合位点(PBS)序列与缺口的5’端杂交,启动逆转录。逆转录酶以RTT为模板,合成新的带有编辑信息的3’-flap,该3’-flap在DNA修复过程中被整合。红色和深紫色代表了所需的编辑。
Fig. 1. Mechanisms of Base editor and Prime editing
| 系统名称 | 系统组件 | PAM | 启动子 | 编辑效率 (%) | 物种 | 参考文献 | ||
|---|---|---|---|---|---|---|---|---|
| Cas9 Nickcase | 逆转录酶 | pegRNA | ||||||
| PPE2 | nCas9(H840A) 密码子优化 | M-MLV RT密码子优 化(D200N/L603W/ T330P/T306K/W31S) | 原始的pegRNA | NGG | OsU3/TaU6, TaU3 | 0~21.8 (Pr, Re)* | 水稻, 小麦 | [ |
| PPE3 | pegRNA+nicking sgRNA | |||||||
| PPE3b | Nicking sgRNA中的spacer序列与被编辑后的序列互补 | |||||||
| PPE-CaMV | CaMV RT | 原始的pegRNA | NGG | 5.8、0.3 (Pr, Re) | 水稻 | |||
| PPE-ribozyme | nCas9(H840A) proteintranscript +polymerase II (Pol II) | Retron RT | Ribozyme-processed pegRNA | NGG | >9 (Pr, Re) | |||
| PPE3-V01, PPE3b-V01 | nCas9(H840A) 密码子优化 | M-MLV RT密码子优化 | pegRNA+nicking sgRNA | NGG | OsU3, OsU6 | 0.05~0.4 (Pr) | 水稻 | [ |
| PPE2-V02, PPE3-V02 | M-MLV RT密码子优化、 核定位信号配置优化 | pegRNA | NGG | OsU6 | <1.55 (Pr) | |||
| Prime editor basic (PPE3) | nCas9(H840A) C端+核定位信 号NLS | 在M-MLV RT上引入了 6个点突变(H9Y/ D200N/T306K/W313F/ T330P/L603W) | 使用多顺反子tRNA策略同时产生pegRNA和nicking sgRNA | NGG | Actin | 2.22~9.38 (Pr) | 水稻 | [ |
| PE-P1 | nCas9(H840A) 密码子优化 | M-MLV RT密码子优化、 核定位信号配置优化、 Link序列类型优化 | 原始的pegRNA | NGG | OsU3, OsU6a | 0%~1.4 (Re) | 水稻 | [ |
| PE-P2 | 在PE-P1的基础上,M- MLV 的C端添加了多 顺反子连接子:2A self-cleaving peptides(P2A) | pegRNA+由增强的esgRNA组成的pegRNA | 1.7~26 (Re) | |||||
| pPE2 | nCas9(H840A) 密码子优化 | M-MLV RT密码子 优化(D200N/L603W/ T330P/T306K/ W313F)、 Link序列类型优化 | 原始的pegRNA | NGG | OsU3 | 0~31.3 (Re) | 水稻 | [ |
| pPE3, pPE3b | pegRNA+nicking sgRNA | TaU3 | ||||||
| Surrogate pPE2 | 采用双pegRNA策略,“编辑HPT起始密码子pegRNA+带有编辑信息pegRNA” | NGG | OsU3 | |||||
| Sp-PE2 | nCas9(H840A) 密码子优化 | M-MLV RT密码子优 化(D200N/L603W/T306K/ W313F/T330P)、核定位信号配置优化 | 原始的pegRNA | NGG | OsU6 | 15.60 (Re) | 水稻 | [ |
表1 PPE系统的开发
Table 1. Development of the Plant Prime Editing system
| 系统名称 | 系统组件 | PAM | 启动子 | 编辑效率 (%) | 物种 | 参考文献 | ||
|---|---|---|---|---|---|---|---|---|
| Cas9 Nickcase | 逆转录酶 | pegRNA | ||||||
| PPE2 | nCas9(H840A) 密码子优化 | M-MLV RT密码子优 化(D200N/L603W/ T330P/T306K/W31S) | 原始的pegRNA | NGG | OsU3/TaU6, TaU3 | 0~21.8 (Pr, Re)* | 水稻, 小麦 | [ |
| PPE3 | pegRNA+nicking sgRNA | |||||||
| PPE3b | Nicking sgRNA中的spacer序列与被编辑后的序列互补 | |||||||
| PPE-CaMV | CaMV RT | 原始的pegRNA | NGG | 5.8、0.3 (Pr, Re) | 水稻 | |||
| PPE-ribozyme | nCas9(H840A) proteintranscript +polymerase II (Pol II) | Retron RT | Ribozyme-processed pegRNA | NGG | >9 (Pr, Re) | |||
| PPE3-V01, PPE3b-V01 | nCas9(H840A) 密码子优化 | M-MLV RT密码子优化 | pegRNA+nicking sgRNA | NGG | OsU3, OsU6 | 0.05~0.4 (Pr) | 水稻 | [ |
| PPE2-V02, PPE3-V02 | M-MLV RT密码子优化、 核定位信号配置优化 | pegRNA | NGG | OsU6 | <1.55 (Pr) | |||
| Prime editor basic (PPE3) | nCas9(H840A) C端+核定位信 号NLS | 在M-MLV RT上引入了 6个点突变(H9Y/ D200N/T306K/W313F/ T330P/L603W) | 使用多顺反子tRNA策略同时产生pegRNA和nicking sgRNA | NGG | Actin | 2.22~9.38 (Pr) | 水稻 | [ |
| PE-P1 | nCas9(H840A) 密码子优化 | M-MLV RT密码子优化、 核定位信号配置优化、 Link序列类型优化 | 原始的pegRNA | NGG | OsU3, OsU6a | 0%~1.4 (Re) | 水稻 | [ |
| PE-P2 | 在PE-P1的基础上,M- MLV 的C端添加了多 顺反子连接子:2A self-cleaving peptides(P2A) | pegRNA+由增强的esgRNA组成的pegRNA | 1.7~26 (Re) | |||||
| pPE2 | nCas9(H840A) 密码子优化 | M-MLV RT密码子 优化(D200N/L603W/ T330P/T306K/ W313F)、 Link序列类型优化 | 原始的pegRNA | NGG | OsU3 | 0~31.3 (Re) | 水稻 | [ |
| pPE3, pPE3b | pegRNA+nicking sgRNA | TaU3 | ||||||
| Surrogate pPE2 | 采用双pegRNA策略,“编辑HPT起始密码子pegRNA+带有编辑信息pegRNA” | NGG | OsU3 | |||||
| Sp-PE2 | nCas9(H840A) 密码子优化 | M-MLV RT密码子优 化(D200N/L603W/T306K/ W313F/T330P)、核定位信号配置优化 | 原始的pegRNA | NGG | OsU6 | 15.60 (Re) | 水稻 | [ |
| 系统名称 | 系统组件 | 编辑类型 | 编辑效率 (%) | 物种 | 参考文献 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cas9 Nickcase | 逆转录酶 | pegRNA | ||||||||||||
| MS2PE | nCas9(H840A)密码子优化 | M-MLV密码子优化(D200N/L603W/ T330P/T306K/W31S)与MS2 RNA 结合蛋白基因MCP融合 | 在原始pegRNA的3'端添加MS2 RNA结合蛋白基因MCP | 3-bp subs | 1.7~55.6 (Pr, Re) | 水稻 | [ | |||||||
| PE-P2 (水稻) | nCas9(H840A)密码子优化 | M-MLV依旧在nCas9的C端融合 | pegRNA+增强的esgRNA | 1-bp subs | 0~5.9(Re) | 水稻 | [ | |||||||
| PE-P3 (水稻) | M-MLV从nCas9 C端融合变为N 端融合 | 2.6~92.3(Re) | ||||||||||||
| PE-P2 (玉米) | M-MLV-02玉米密码子优化;依旧 在nCas9 C端融合 | Target+esgRNA和pegRNA | 0~7.7(Pr) | 玉米 | ||||||||||
| PE-P3 (玉米) | M-MLV-02玉米密码子优化,从 nCas9 C端融合变为N端融合 | 2.9~80(Pr) | ||||||||||||
| ePPE | nCas9(H840A)密码子优化 | 在原始PPE基础上,去除RT的 RNase H结构域并添加病毒核 衣壳(NC)蛋白 | 原始的pegRNA | 1-2-bp subs;3-bp ins;2-bp del; 15-90-bp ins or del | 0~31.5 (Re) | 水稻 | [ | |||||||
| PPE3- evopreQ1 | nCas9(H840A)密码子优化 | M-MLV密码子优化(D200N/ L603W/T330P/T306K/W31S) | 在PPE3基础上, 结构化pegRNA,在原始pegRNA的3'端添加8 bp linker和evopreQ1/ mpknot以提高稳定性 | 1-3-bp subs; 3-bp ins | 2.6~60.5 (Pr, Re) | 水稻 | [ | |||||||
| PPE3-mpknot | 0~6.3 (Pr, Re) | |||||||||||||
| pPEmax | nCas9(R221K/N394K/H840A) 密码子优化 | M-MLV密码子优化,N端添加 NLS-embedded linker,C端添加 异质串联NLS | epegRNA:在pegRNA 3'端添加evopreQ1 | 1-bp ins; 1-2-bp subs | 14.58~66.67 (Re) | 水稻 | [ | |||||||
| pPEmax- MLHdn | 在pPEmax的基础上,C端添 加MLH1dn,以抑制MMR途径 | 0.18~2.3 (Re) | ||||||||||||
| enpPE2 | 同pPEmax | 在U6复合启动子驱动下,构成tRNA(Gly-tRNA):: pegRNA::evopreQ1:: HDV表达盒 | 64.58~77(Re) | |||||||||||
| pINPE2 | nCas9(H840A)密码子优化, N端添加小肽 | M-MLV C端添加异质串联NLSs | epegRNA:在pegRNA 3'端添加evopreQ1 | 0.01~0.8 (Re) | ||||||||||
| phyPE2 | nCas9(H840A)密码子优化 | M-MLV N端添加DBD linker (hRad51-ssDBD) | <0.1(Re) | |||||||||||
| ePE3max | nCas9(R221K/N394K/H840A) 密码子优化 | M-MLV密码子优化(D200N/ L603W/T330P/T306K/W31S) | U6复合启动子驱动的表达盒中添加tRNA(Gly-tRNA)和HDV核酶,并添加nicking sgRNA | 1-bp ins; 1-3-bp subs | ~40(Re) | 水稻 | [ | |||||||
| ePE5max | M-MLV C端添加MLH1dn,以 抑制MMR途径 | ~40(纯合率提高)(Re) | ||||||||||||
| PrimeRoot | nCas9(H840A)密码子优化 | M-MLV去除RNase H结构域, 并添加重组酶 | 采用“双ePPE”策略,使用了两个相邻的epegRNA,每个模板都包含一个仅与另一个epegRNA模板具有同源性的RT模板 | 720-bp,1.4-kb,4.9-kb, 7.7-kb,11.1-kb ins | 0~8.3 (Pr, Re) | 水稻 | [ | |||||||
| ePPE* | nCas9(H840A)密码子优化 | M-MLV密码子优化(D200N/ V223A/T306K/W313F/T330P/ L603W),去除RNase H结构域 并添加病毒核衣壳(NC)蛋白;优 化核定位信号 | epegRNA:在pegRNA 3’端添加evopreQ1 | 1-3-bp subs; 3-6-bp del; 4-bp ins | 0~18.9 (Pr, Re) | 小麦 | [ | |||||||
| ePPEmax | ||||||||||||||
| ePPEmax* | nCas9(R221K/N394K/H840A) 密码子优化 | |||||||||||||
| ePPEplus | ||||||||||||||
| CMPE- ePPEplus | Csy4核糖核酸内切酶 | 串联多个(e)pegRNA | 6- bp del; 1-bp subs | 2~21.6; 19.6~86.3 (Pr, Re) | ||||||||||
| ePE5max (玉米) | nCas9(R221K/N394K/H840A) 密码子优化 | M-MLV C端添加MLH1dn玉米同 源物,以抑制MMR途径 | U6复合启动子驱动的双pegRNA表达盒中添加tRNA(Gly-tRNA)和HDV核酶 | 1-bp subs | Homo: 1.4~3 hetero: 15~75 (Re) | 玉米 | [ | |||||||
| pCXPE01, pCXPE02, pCXPE03 | nCas9(H840A)密码子优化 | M-MLV植物密码子优化 | 原始的pegRNA | 2-bp subs; 1-3-bp del | 0.025~1.66; 2.6 (Pr) | 番茄 | [ | |||||||
| pPPED | nCas9(H840A)密码子优化 | M-MLV植物密码子优linker序 列类型优化 | 原始的pegRNA | 2-bp subs | 0.06 (Pr) | 本氏烟草 | [ | |||||||
| 系统名称 | 系统组件 | 编辑类型 | 编辑效率 (%) | 物种 | 参考文献 | |||||||||
| Cas9 Nickcase | 逆转录酶 | pegRNA | ||||||||||||
| pPPEDs | 2-bp subs; 25-66-bp ins | 0.07±0.12 (Re) | 拟南芥 | |||||||||||
| pPPEM | 2-bp subs;25-66-bp ins | 0.7~2.2 (Pr) | 水稻 | |||||||||||
| pAct-PPE | nCas9(H840A)密码子优化 | M-MLV植物密码子优化 | 原始的pegRNA | 3-bp subs | 0~5 (Re) | 小立碗藓、 马铃薯 | [ | |||||||
| PPE2 PPE3 PPE3b | nCas9(H840A)密码子优化 | M-MLV植物密码子优化 | 原始的pegRNA和nicking sgRNA | 1-bp subs | 0.2~0.5 (Pr) | 花生, 鹰嘴豆, 豇豆 | [ | |||||||
| PE-Nt2 | nCas9(H840A)密码子优化 | M-MLV通过linker在nCas9 C 端融合 | pegRNA与nesgRNA (用于切割非编辑链的增强sgRNA)通过tRNA序列连接 | 1-bp subs | 0~1.4 (Re) | 烟草 | [ | |||||||
| PE-Nt3 | M-MLV通过linker在nCas9 N 端融合 | 1.1~7.5 (Re) | ||||||||||||
| PE-Nt4 | 1.3~16.3 (Re) | |||||||||||||
| PE2 (v2) | nCas9(H840A)密码子优化 | 在nCas9-M-MLV 融合蛋白N端 添加T5外切酶 | 原始的pegRNA、 双pegRNA优化、 PBS的长度优化 | 4-bp ins; 4-bp del; 1−2-bp subs | 3.5~48.65 (Pr, Re) | 水稻 | [ | |||||||
| PE3-HS/AS/ DS | nCas9(H840A)密码子优化 | M-MLV植物密码子优化;潮霉素 代理系统、除草剂代理系统和潮 霉素+除草剂双代理系统 | tRNA-pegRNA(n)- tRNA-sgRNA(n)- tRNA | 1-7-bp subs | 1.3~54.2 (Re) | 水稻 | [ | |||||||
| PE-DSM vector | 潮霉素+除草剂双代理多基因编辑 系统 | 1.8~45.8 (Re) | ||||||||||||
| DPE, TPE, QPE | nCas9(R221K/N394K/H840A)密码子优化 | 同ePE3max | pegRNA(n)-ngRNA(n)通过Gateway连接 | 2-3-bp subs, 28-bp ins | 57.14 (Re) | 水稻 | [ | |||||||
| - | nCas9(R221K/N394K/H840A) 密码子优化 | 同ePE5max | epegRNA:在pegRNA 3’端添加evopreQ1 | 4-bp ins | 9.7 (Re) | 番茄、 拟南芥 | [ | |||||||
| GRAND | nCas9(H840A)密码子优化 | M-MLV植物密码子优化 | RTT部分对齐,但与双pegRNA中的目标序列非同源 | 46-bp subs | 0.59~9.88 (Pr, Re) | 水稻 | [ | |||||||
| PE5max | nCas9(R221K/N394K/H840A) 密码子优化 | M-MLV C端添加MLH1dn,以 抑制MMR途径 | U6复合启动子驱动的双pegRNA表达盒中添加tRNA(Gly-tRNA)和HDV核酶 | 30-bp ins | 47 (Re) | 水稻 | [ | |||||||
表2 PPE系统的优化
Table 2. Optimization of the Plant Prime Editing system
| 系统名称 | 系统组件 | 编辑类型 | 编辑效率 (%) | 物种 | 参考文献 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cas9 Nickcase | 逆转录酶 | pegRNA | ||||||||||||
| MS2PE | nCas9(H840A)密码子优化 | M-MLV密码子优化(D200N/L603W/ T330P/T306K/W31S)与MS2 RNA 结合蛋白基因MCP融合 | 在原始pegRNA的3'端添加MS2 RNA结合蛋白基因MCP | 3-bp subs | 1.7~55.6 (Pr, Re) | 水稻 | [ | |||||||
| PE-P2 (水稻) | nCas9(H840A)密码子优化 | M-MLV依旧在nCas9的C端融合 | pegRNA+增强的esgRNA | 1-bp subs | 0~5.9(Re) | 水稻 | [ | |||||||
| PE-P3 (水稻) | M-MLV从nCas9 C端融合变为N 端融合 | 2.6~92.3(Re) | ||||||||||||
| PE-P2 (玉米) | M-MLV-02玉米密码子优化;依旧 在nCas9 C端融合 | Target+esgRNA和pegRNA | 0~7.7(Pr) | 玉米 | ||||||||||
| PE-P3 (玉米) | M-MLV-02玉米密码子优化,从 nCas9 C端融合变为N端融合 | 2.9~80(Pr) | ||||||||||||
| ePPE | nCas9(H840A)密码子优化 | 在原始PPE基础上,去除RT的 RNase H结构域并添加病毒核 衣壳(NC)蛋白 | 原始的pegRNA | 1-2-bp subs;3-bp ins;2-bp del; 15-90-bp ins or del | 0~31.5 (Re) | 水稻 | [ | |||||||
| PPE3- evopreQ1 | nCas9(H840A)密码子优化 | M-MLV密码子优化(D200N/ L603W/T330P/T306K/W31S) | 在PPE3基础上, 结构化pegRNA,在原始pegRNA的3'端添加8 bp linker和evopreQ1/ mpknot以提高稳定性 | 1-3-bp subs; 3-bp ins | 2.6~60.5 (Pr, Re) | 水稻 | [ | |||||||
| PPE3-mpknot | 0~6.3 (Pr, Re) | |||||||||||||
| pPEmax | nCas9(R221K/N394K/H840A) 密码子优化 | M-MLV密码子优化,N端添加 NLS-embedded linker,C端添加 异质串联NLS | epegRNA:在pegRNA 3'端添加evopreQ1 | 1-bp ins; 1-2-bp subs | 14.58~66.67 (Re) | 水稻 | [ | |||||||
| pPEmax- MLHdn | 在pPEmax的基础上,C端添 加MLH1dn,以抑制MMR途径 | 0.18~2.3 (Re) | ||||||||||||
| enpPE2 | 同pPEmax | 在U6复合启动子驱动下,构成tRNA(Gly-tRNA):: pegRNA::evopreQ1:: HDV表达盒 | 64.58~77(Re) | |||||||||||
| pINPE2 | nCas9(H840A)密码子优化, N端添加小肽 | M-MLV C端添加异质串联NLSs | epegRNA:在pegRNA 3'端添加evopreQ1 | 0.01~0.8 (Re) | ||||||||||
| phyPE2 | nCas9(H840A)密码子优化 | M-MLV N端添加DBD linker (hRad51-ssDBD) | <0.1(Re) | |||||||||||
| ePE3max | nCas9(R221K/N394K/H840A) 密码子优化 | M-MLV密码子优化(D200N/ L603W/T330P/T306K/W31S) | U6复合启动子驱动的表达盒中添加tRNA(Gly-tRNA)和HDV核酶,并添加nicking sgRNA | 1-bp ins; 1-3-bp subs | ~40(Re) | 水稻 | [ | |||||||
| ePE5max | M-MLV C端添加MLH1dn,以 抑制MMR途径 | ~40(纯合率提高)(Re) | ||||||||||||
| PrimeRoot | nCas9(H840A)密码子优化 | M-MLV去除RNase H结构域, 并添加重组酶 | 采用“双ePPE”策略,使用了两个相邻的epegRNA,每个模板都包含一个仅与另一个epegRNA模板具有同源性的RT模板 | 720-bp,1.4-kb,4.9-kb, 7.7-kb,11.1-kb ins | 0~8.3 (Pr, Re) | 水稻 | [ | |||||||
| ePPE* | nCas9(H840A)密码子优化 | M-MLV密码子优化(D200N/ V223A/T306K/W313F/T330P/ L603W),去除RNase H结构域 并添加病毒核衣壳(NC)蛋白;优 化核定位信号 | epegRNA:在pegRNA 3’端添加evopreQ1 | 1-3-bp subs; 3-6-bp del; 4-bp ins | 0~18.9 (Pr, Re) | 小麦 | [ | |||||||
| ePPEmax | ||||||||||||||
| ePPEmax* | nCas9(R221K/N394K/H840A) 密码子优化 | |||||||||||||
| ePPEplus | ||||||||||||||
| CMPE- ePPEplus | Csy4核糖核酸内切酶 | 串联多个(e)pegRNA | 6- bp del; 1-bp subs | 2~21.6; 19.6~86.3 (Pr, Re) | ||||||||||
| ePE5max (玉米) | nCas9(R221K/N394K/H840A) 密码子优化 | M-MLV C端添加MLH1dn玉米同 源物,以抑制MMR途径 | U6复合启动子驱动的双pegRNA表达盒中添加tRNA(Gly-tRNA)和HDV核酶 | 1-bp subs | Homo: 1.4~3 hetero: 15~75 (Re) | 玉米 | [ | |||||||
| pCXPE01, pCXPE02, pCXPE03 | nCas9(H840A)密码子优化 | M-MLV植物密码子优化 | 原始的pegRNA | 2-bp subs; 1-3-bp del | 0.025~1.66; 2.6 (Pr) | 番茄 | [ | |||||||
| pPPED | nCas9(H840A)密码子优化 | M-MLV植物密码子优linker序 列类型优化 | 原始的pegRNA | 2-bp subs | 0.06 (Pr) | 本氏烟草 | [ | |||||||
| 系统名称 | 系统组件 | 编辑类型 | 编辑效率 (%) | 物种 | 参考文献 | |||||||||
| Cas9 Nickcase | 逆转录酶 | pegRNA | ||||||||||||
| pPPEDs | 2-bp subs; 25-66-bp ins | 0.07±0.12 (Re) | 拟南芥 | |||||||||||
| pPPEM | 2-bp subs;25-66-bp ins | 0.7~2.2 (Pr) | 水稻 | |||||||||||
| pAct-PPE | nCas9(H840A)密码子优化 | M-MLV植物密码子优化 | 原始的pegRNA | 3-bp subs | 0~5 (Re) | 小立碗藓、 马铃薯 | [ | |||||||
| PPE2 PPE3 PPE3b | nCas9(H840A)密码子优化 | M-MLV植物密码子优化 | 原始的pegRNA和nicking sgRNA | 1-bp subs | 0.2~0.5 (Pr) | 花生, 鹰嘴豆, 豇豆 | [ | |||||||
| PE-Nt2 | nCas9(H840A)密码子优化 | M-MLV通过linker在nCas9 C 端融合 | pegRNA与nesgRNA (用于切割非编辑链的增强sgRNA)通过tRNA序列连接 | 1-bp subs | 0~1.4 (Re) | 烟草 | [ | |||||||
| PE-Nt3 | M-MLV通过linker在nCas9 N 端融合 | 1.1~7.5 (Re) | ||||||||||||
| PE-Nt4 | 1.3~16.3 (Re) | |||||||||||||
| PE2 (v2) | nCas9(H840A)密码子优化 | 在nCas9-M-MLV 融合蛋白N端 添加T5外切酶 | 原始的pegRNA、 双pegRNA优化、 PBS的长度优化 | 4-bp ins; 4-bp del; 1−2-bp subs | 3.5~48.65 (Pr, Re) | 水稻 | [ | |||||||
| PE3-HS/AS/ DS | nCas9(H840A)密码子优化 | M-MLV植物密码子优化;潮霉素 代理系统、除草剂代理系统和潮 霉素+除草剂双代理系统 | tRNA-pegRNA(n)- tRNA-sgRNA(n)- tRNA | 1-7-bp subs | 1.3~54.2 (Re) | 水稻 | [ | |||||||
| PE-DSM vector | 潮霉素+除草剂双代理多基因编辑 系统 | 1.8~45.8 (Re) | ||||||||||||
| DPE, TPE, QPE | nCas9(R221K/N394K/H840A)密码子优化 | 同ePE3max | pegRNA(n)-ngRNA(n)通过Gateway连接 | 2-3-bp subs, 28-bp ins | 57.14 (Re) | 水稻 | [ | |||||||
| - | nCas9(R221K/N394K/H840A) 密码子优化 | 同ePE5max | epegRNA:在pegRNA 3’端添加evopreQ1 | 4-bp ins | 9.7 (Re) | 番茄、 拟南芥 | [ | |||||||
| GRAND | nCas9(H840A)密码子优化 | M-MLV植物密码子优化 | RTT部分对齐,但与双pegRNA中的目标序列非同源 | 46-bp subs | 0.59~9.88 (Pr, Re) | 水稻 | [ | |||||||
| PE5max | nCas9(R221K/N394K/H840A) 密码子优化 | M-MLV C端添加MLH1dn,以 抑制MMR途径 | U6复合启动子驱动的双pegRNA表达盒中添加tRNA(Gly-tRNA)和HDV核酶 | 30-bp ins | 47 (Re) | 水稻 | [ | |||||||
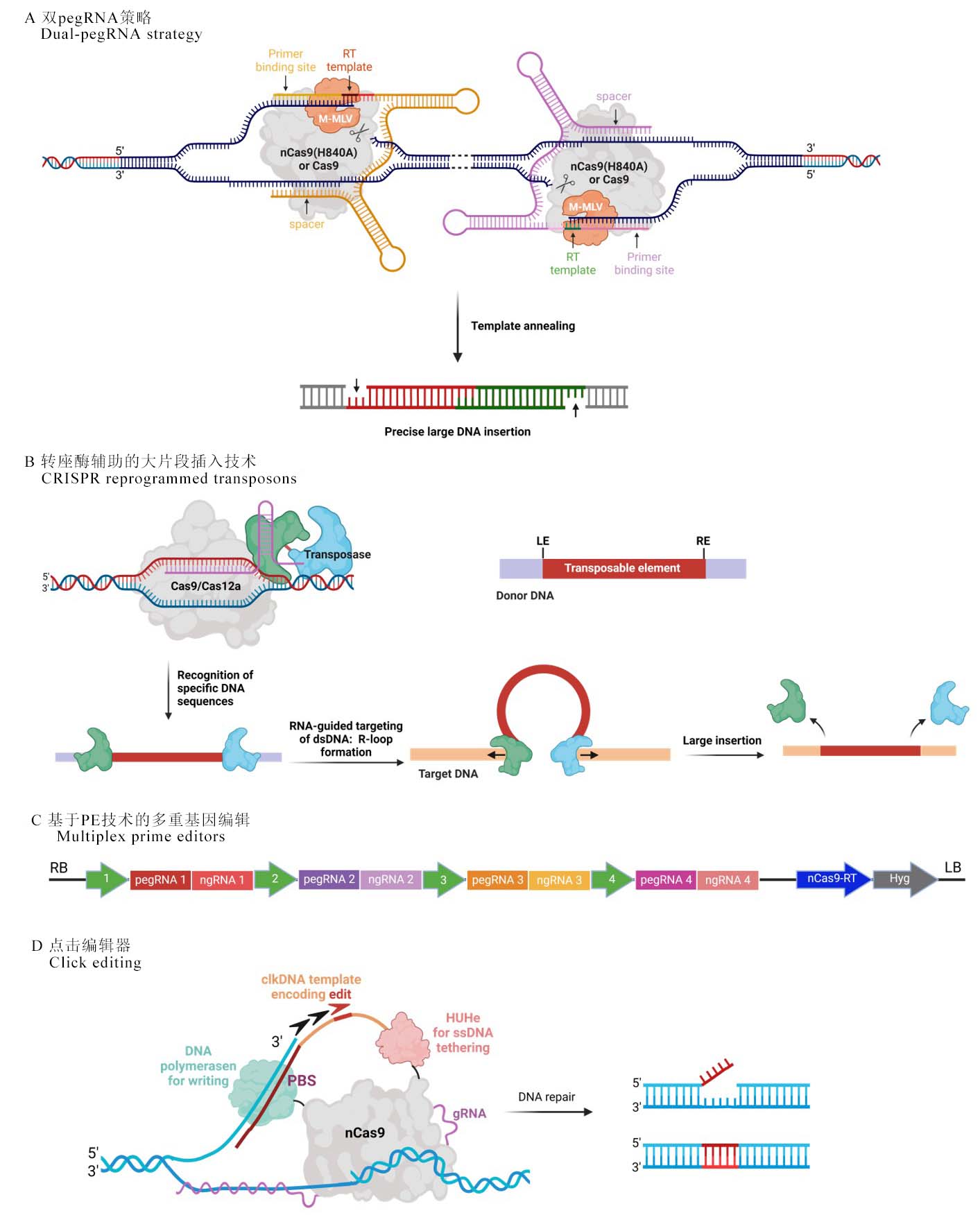
图2 PPE技术的拓展 A, 双pegRNA策略。可用于插入和删除大DNA片段(1 kb碱基及以上)。逆转录序列(RT template)可以精确替换两个目标切割位点之间的序列,红色和绿色表示插入的序列。B, 转座酶辅助的大片段插入技术。通过利用转座酶的切割和重组功能,能够将目标大片段DNA插入到宿主基因组中的特定位点。转座酶(Transposase)能够识别特定的DNA序列,并在该序列处进行切割。接着,将含有目标DNA片段的供体与转座酶一同引入宿主细胞。转座酶在识别到靶位点后,切割宿主基因组,并形成一个R环,然后将目标DNA片段插入到这个位置中。红色的序列代表所需的供体片段;橙色的序列代表宿主细胞的靶位点DNA;LE(左端)和RE(右端)代表转座酶识别位点。C, 基于PE技术的多重基因编辑。可进行多路复用pegRNAs,并编辑多达4个基因位点。D, 点击编辑器(Click editing)的示意图。由nCas9核酸酶、DNA聚合酶和ssDNA的栓链结构域HUHe融合蛋白与gRNA配对组成。
Fig. 2. Expansion of plant prime editing technology
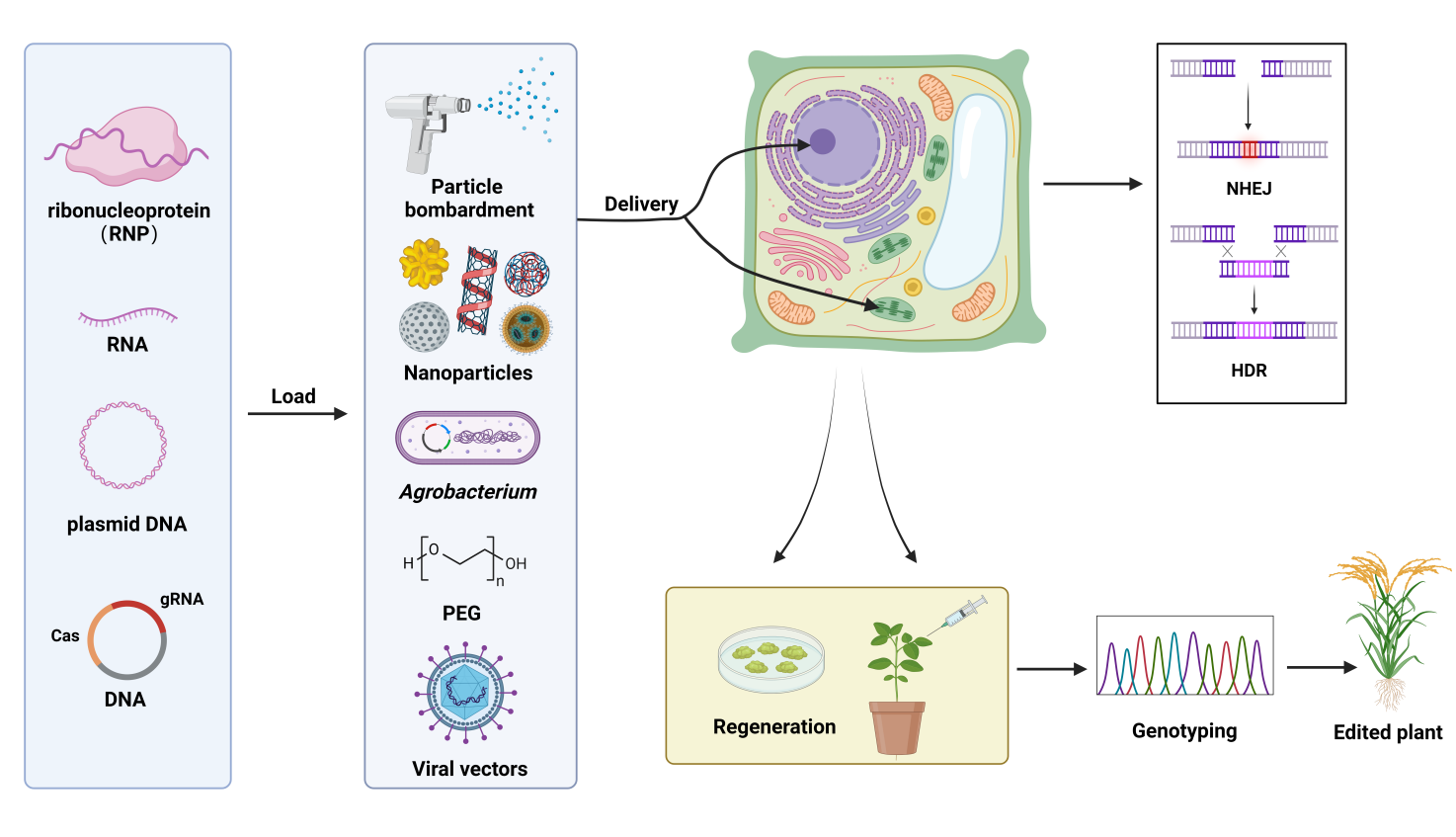
图3 CRISPR-Cas系统递送方法示意图 CRISPR-Cas系统可以通过不同的方式递送到植物细胞中:例如由Cas蛋白和sgRNA组成的核糖核蛋白(RNPs),或者编码 CRISPR-Cas元件的RNA分子,或者包含CRISPR-Cas序列的DNA分子和质粒。这些分子可以通过粒子轰击、农杆菌介导的遗传转化、聚乙二醇(PEG)介导的原生质体转化或植物病毒载体进入植物细胞。一旦进入植物细胞核,CRISPR-Cas系统就会产生位点特异性双链断裂(DSB),这些断裂可以通过非同源末端连接(NHEJ)或同源定向修复(HDR)途径进行修复,最终获得再生植株。
Fig. 3. Diagram of CRISPR-Cas delivery methods
| Cas9变体 | Cas蛋白大小(kb) | 预测的PE载体大小(kb) | 参考文献 |
|---|---|---|---|
| SpCas9 | 4.2 | 6.2 | [ |
| SaCas9 | 3.2 | 5.2 | [ |
| CjCas9 | 3.0 | 5.0 | [ |
| Casϕ | 2.1~2.4 | 4.1~4.4 | [ |
| Cas12f | 1.2~1.8 | 3.2~3.8 | [ |
| CasX | <3.0 | <5.0 | [ |
| Cas13j | 1.2~1.5 | 3.2~3.5 | [ |
表3 预测不同Cas蛋白的PE载体大小
Table 3. Prediction of PE size with various Cas enzymes
| Cas9变体 | Cas蛋白大小(kb) | 预测的PE载体大小(kb) | 参考文献 |
|---|---|---|---|
| SpCas9 | 4.2 | 6.2 | [ |
| SaCas9 | 3.2 | 5.2 | [ |
| CjCas9 | 3.0 | 5.0 | [ |
| Casϕ | 2.1~2.4 | 4.1~4.4 | [ |
| Cas12f | 1.2~1.8 | 3.2~3.8 | [ |
| CasX | <3.0 | <5.0 | [ |
| Cas13j | 1.2~1.5 | 3.2~3.5 | [ |
| [1] | Townsend J A, Wright D A, Winfrey R J, Fu F L, Maeder M L, Joung J K, Voytas D F. High-frequency modification of plant genes using engineered zinc-finger nucleases[J]. Nature, 2009, 459(7245): 442-445. |
| [2] | Bogdanove A J, Voytas D F. TAL effectors: Customizable proteins for DNA targeting[J]. Science, 2011, 333(6051): 1843-1846. |
| [3] | Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(39): E2579-E2586. |
| [4] | Mali P, Yang L H, Esvelt K M, Aach J, Guell M, DiCarlo J E, Norville J E, Church G M. RNA-guided human genome engineering via cas9[J]. Science, 2013, 339(6121): 823-826. |
| [5] | Lu Y M, Tian Y F, Shen R D, Yao Q, Wang M G, Chen M, Dong J S, Zhang T G, Li F, Lei M G, Zhu J K. Targeted, efficient sequence insertion and replacement in rice[J]. Nature Biotechnology, 2020, 38(12): 1402-1407. |
| [6] | Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh O O, Gootenberg J S, Makarova K S, Wolf Y I, Severinov K, Zhang F, Koonin E V. Diversity and evolution of class 2 CRISPR-Cas systems[J]. Nature Reviews Microbiology, 2017, 15(3): 169-182. |
| [7] | Zetsche B, Gootenberg J S, Abudayyeh O O, Slaymaker I M, Makarova K S, Essletzbichler P, Volz S, Joung J, van der Oost J, Regev A, Koonin E V, Zhang F. Cpf1 is a single-RNA-guided endonuclease of a Class 2 CRISPR-Cas system[J]. Cell, 2015, 163(3): 759-771. |
| [8] | Barrangou R, Gersbach C A. Expanding the CRISPR Toolbox: Targeting RNA with Cas13b[J]. Molecular Cell, 2017, 65(4): 582-584. |
| [9] | Komor A C, Kim Y B, Packer M S, Zuris J A, Liu D R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
| [10] | Gaudelli N M, Komor A C, Rees H A, Packer M S, Badran A H, Bryson D I, Liu D R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| [11] | Huang T P, Newby G A, Liu D V R. Precision genome editing using cytosine and adenine base editors in mammalian cells[J]. Nature Protocols, 2021, 16(2): 1089-1128. |
| [12] | Rees H A, Liu D R. Base editing: precision chemistry on the genome and transcriptome of living cells[J]. Nature Reviews Genetics, 2018, 19(12): 770-788. |
| [13] | Ren Q R, Sretenovic S, Liu G Q, Zhong Z H, Wang J H, Huang L, Tang X, Guo Y C, Liu L, Wu Y C, Zhou J, Zhao Y X, Yang H, He Y, Liu S S, Yin D S, Mayorga R, Zheng X L, Zhang T, Qi Y P, Zhang Y. Improved plant cytosine base editors with high editing activity, purity, and specificity[J]. Plant Biotechnology Journal, 2021, 19(10): 2052-2068. |
| [14] | Monsur M B I, Cao N, Wei X J, Xie L H, Jiao G A, Tang S Q, Sreenivasulu N, Shao G N, Hu P S. Improved eating and cooking quality of indica rice cultivar YK17 via adenine base editing of Wxa allele of granule-bound starch synthase I (GBSS I)[J]. Rice Science, 2021, 28(5): 427-430. |
| [15] | Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, Wang D, Qiu J L, Zhang F, Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice[J]. Science, 2019, 364(6437): 292-295. |
| [16] | Anzalone A V, Randolph P B, Davis J R, Sousa A A, Koblan L W, Levy J M, Chen P J, Wilson C, Newby G A, Raguram A, Liu D R. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157. |
| [17] | Gehrke J M, Cervantes O, Clement M K, Wu Y X, Zeng J, Bauer D E, Pinello L, Joung J K. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities[J]. Nature Biotechnology, 2018, 36(10): 977-982. |
| [18] | Kim Y B, Komor A C, Levy J M, Packer M S, Zhao K T, Liu D R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions[J]. Nature Biotechnology, 2017, 35(4): 371-376. |
| [19] | Nishimasu H, Shi X, Ishiguro S, Gao L Y, Hirano S, Okazaki S, Noda T, Abudayyeh O O, Gootenberg J S, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O. Engineered CRISPR-Cas9 nuclease with expanded targeting space[J]. Science, 2018, 361(6408): 1259-1262. |
| [20] | Huang T P, Zhao K T, Miller S M, Gaudelli N M, Oakes B L, Fellmann C, Savage D F, Liu D R. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors[J]. Nature Biotechnology, 2019, 37(6): 626-631. |
| [21] | Walton R T, Christie K A, Whittaker M N, Kleinstiver B P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants[J]. Science, 2020, 368(6488): 290-296. |
| [22] | Miller S M, Wang T, Randolph P B, Arbab M, Shen M W, Huang T P, Matuszek Z, Newby G A, Rees H A, Liu D R. Continuous evolution of SpCas9 variants compatible with non-G PAMs[J]. Nature Biotechnology, 2020, 38(4): 471-481. |
| [23] | Doman J L, Raguram A, Newby G A, Liu D R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors[J]. Nature Biotechnology, 2020, 38(5): 620-628. |
| [24] | Rees H A, Wilson C, Doman J L, Liu D R. Analysis and minimization of cellular RNA editing by DNA adenine base editors[J]. Science Advances, 2019, 5(5): eaax5717. |
| [25] | Grunewald J, Zhou R H, Iyer S, Lareau C A, Garcia S P, Aryee M J, Joung J K. CRISPR DNA base editors with reduced RNA off-target and self-editing activities[J]. Nature Biotechnology, 2019, 37(9): 1041-1048. |
| [26] | Liu T T, Zou J P, Yang X, Wang K J, Rao Y C, Wang C. Development and application of prime editing in plants[J]. Rice Science, 2023, 30(6): 509-522. |
| [27] | Gao R Z, Fu Z C, Li X Y, Wang Y, Wei J, Li G Y, Wang L J, Wu J, Huang X X, Yang L, Chen J. Genomic and transcriptomic analyses of prime editing guide RNA-independent off-target effects by prime editors[J]. CRISPR Journal, 2022, 5(2): 276-293. |
| [28] | Schene I F, Joore I P, Oka R, Mokry M, van Vugt A H M, van Boxtel R, Verstegen M M A, van Hasselt P M, Nieuwenhuis E E S, Fuchs S A. Prime editing for functional repair in patient-derived disease models[J]. Nature Communications, 2020, 11(1): 5352. |
| [29] | Geurts M H, de Poel E, Pleguezuelos-Manzano C, Oka R, Carrillo L, Andersson-Rolf A, Boretto M, Brunsveld J E, van Boxtel R, Beekman J M, Clevers H. Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids[J]. Life Science Alliance, 2021, 4(10): e202000940. |
| [30] | Park S J, Jeong T Y, Shin S K, Yoon D E, Lim S Y, Kim S P, Choi J, Lee H, Hong J I, Ahn J, Seong J K, Kim K. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor[J]. Genome Biology, 2021, 22(1): 170. |
| [31] | Liu Y, Li X Y, He S T, Huang S H, Li C, Chen Y L, Liu Z, Huang X X, Wang X L. Efficient generation of mouse models with the prime editing system[J]. Cell Discovery, 2020, 6(1): 27. |
| [32] | Gao P, Lyu Q, Ghanam A R, Lazzarotto C R, Newby G A, Zhang W, Choi M, Slivano O J, Holden K, Walker J A, Kadina A P, Munroe R J, Abratte C M, Schimenti J C, Liu D R, Tsai S Q, Long X C, Miano J M. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression[J]. Genome Biology, 2021, 22(1): 83. |
| [33] | Lin J X, Liu X C, Lu Z Y, Huang S S, Wu S S, Yu W X, Liu Y, Zheng X G, Huang X X, Sun Q, Qiao Y B, Liu Z. Modeling a cataract disorder in mice with prime editing[J]. Molecular Therapy Nucleic Acids, 2021, 25: 494-501. |
| [34] | Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone A V, Raguram A, Doman J L, Liu D R, Gao C. Prime genome editing in rice and wheat[J]. Nature Biotechnology, 2020, 38(5): 582-585. |
| [35] | Tang X, Sretenovic S, Ren Q R, Jia X Y, Li M K, Fan T T, Yin D S, Xiang S Y, Guo Y H, Liu L, Zheng X L, Qi Y P, Zhang Y. Plant prime editors enable precise gene editing in rice cells[J]. Molecular Plant, 2020, 13(5): 667-670. |
| [36] | Li H Y, Li J Y, Chen J L, Yan L, Xia L Q. Precise modifications of both exogenous and endogenous genes in rice by prime editing[J]. Molecular Plant, 2020, 13(5): 671-674. |
| [37] | Xu W, Zhang C W, Yang Y X, Zhao S, Kang G T, He X Q, Song J L, Yang J X. Versatile nucleotides substitution in plant using an improved prime editing system[J]. Molecular Plant, 2020, 13(5): 675-678. |
| [38] | Xu R F, Li J, Liu X S, Shan T F, Qin R Y, Wei P C. Development of plant prime-editing systems for precise genome editing[J]. Plant Communications, 2020, 1(3): 100043. |
| [39] | Hua K, Jiang Y W, Tao X P, Zhu J K. Precision genome engineering in rice using prime editing system[J]. Plant Biotechnology Journal, 2020, 18(11): 2167-2169. |
| [40] | Jin S, Lin Q P, Luo Y F, Zhu Z X, Liu G W, Li Y J, Chen K L, Qiu J L, Gao C X. Genome-wide specificity of prime editors in plants[J]. Nature Biotechnology, 2021, 39(10): 1292-1299. |
| [41] | Chai Y, Jiang Y, Wang J, Qiao D, Zhang Y, Xin C, Zhou Y, Wang X C, Chen Q. MS2 RNA aptamer greatly enhances prime editing in rice[J]. Research Square, 2021. |
| [42] | Xu W, Yang Y X, Yang B Y, Krueger C J, Xiao Q L, Zhao S, Zhang L, Kang G T, Wang F P, Yi H M, Ren W, Li L, He X Q, Zhang C M, Zhang B, Zhao J R, Yang J X. A design optimized prime editor with expanded scope and capability in plants[J]. Nature Plants, 2022, 8(1): 45-52. |
| [43] | Zong Y, Liu Y J, Xue C X, Li B S, Li X Y, Wang Y P, Li J, Liu G W, Huang X X, Cao X F, Gao C X. An engineered prime editor with enhanced editing efficiency in plants[J]. Nature Biotechnology, 2022, 40(9): 1394-1402. |
| [44] | Zou J P, Meng X B, Liu Q, Shang M Q, Wang K J, Li J Y, Yu H, Wang C. Improving the efficiency of prime editing with epegRNAs and high-temperature treatment in rice[J]. Science China-Life Sciences, 2022, 65(11): 2328-2331. |
| [45] | Li J, Chen L K, Liang J, Xu R F, Jiang Y L, Li Y Z, Ding J, Li M, Qin R Y, Wei P C. Development of a highly efficient prime editor 2 system in plants[J]. Genome Biology, 2022, 23(1): 161. |
| [46] | Jiang Y, Chai Y, Qiao D, Wang J, Xin C, Sun W, Cao Z, Zhang Y, Zhou Y, Wang X C. Optimized prime editing efficiently generates glyphosate-resistant rice plants carrying homozygous TAP-IVS mutation in EPSPS[J]. Molecular plant, 2022, 15(11): 1646-1649. |
| [47] | Sun C, Lei Y, Li B S, Gao Q, Li Y J, Cao W, Yang C, Li H C, Wang Z W, Li Y, Wang Y P, Liu J, Zhao K T, Gao C X. Precise integration of large DNA sequences in plant genomes using PrimeRoot editors[J]. Nature Biotechnology, 2024, 42(2): 316-327. |
| [48] | Ni P, Zhao Y D, Zhou X M, Liu Z H, Huang Z W, Ni Z F, Sun Q X, Zong Y. Efficient and versatile multiplex prime editing in hexaploid wheat[J]. Genome Biology, 2023, 24(1): 156. |
| [49] | Qiao D, Wang J, Lu M H, Xin C, Chai Y, Jiang Y, Sun W, Cao Z, Guo S, Wang X C. Optimized prime editing efficiently generates heritable mutations in maize[J]. Journal of Integrative Plant Biology, 2023, 65(4): 900-906. |
| [50] | Lu Y M, Tian Y F, Shen R D, Yao Q, Zhong D T, Zhang X N, Zhu J K. Precise genome modification in tomato using an improved prime editing system[J]. Plant Biotechnology Journal, 2021, 19(3): 415-417. |
| [51] | Wang L, Kaya H B, Zhang N, Rai R, Willmann M R, Carpenter S C D, Read A C, Martin F, Fei Z J, Leach J E, Martin G B, Bogdanove A J. Spelling changes and fluorescent tagging with prime editing vectors for plants[J]. Frontiers in Genome Editing, 2021, 3: 617553. |
| [52] | Perroud P F, Guyon-Debast A, Veillet F, Kermarrec M P, Chauvin L, Chauvin J E, Gallois J L, Nogué F. Prime Editing in the model plant Physcomitrium patens and its potential in the tetraploid potato[J]. Plant Science, 2022, 316: 111162. |
| [53] | Biswas S, Bridgeland A, Irum S, Thomson M J, Septiningsih E M. Optimization of prime editing in rice, peanut, chickpea, and cowpea protoplasts by restoration of GFP activity[J]. International Journal of Molecular Sciences, 2022, 23(17): 9809. |
| [54] | Zhang J D, Zhang L, Zhang C W, Yang Y X, Liu H Y, Li L, Zhang S X, Li X G, Liu X X, Liu Y, Wang J, Yang G Y, Xia Q Y, Wang W G, Yang J X. Developing an efficient and visible prime editing system to restore tobacco 8-hydroxy-copalyl diphosphate gene for labdane diterpene Z-abienol biosynthesis[J]. Science China-Life Sciences, 2023, 66(12): 2910-2921. |
| [55] | Liang Z, Wu Y Q, Guo Y J, Wei S. Addition of the T5 exonuclease increases the prime editing efficiency in plants[J]. Journal of Genetics and Genomics, 2023, 50(8): 582-588. |
| [56] | Li H Y, Zhu Z W, Li S Y, Li J Y, Yan L, Zhang C, Ma Y Z, Xia L Q. Multiplex precision gene editing by a surrogate prime editor in rice[J]. Molecular Plant, 2022, 15(7): 1077-1080. |
| [57] | Gupta A, Liu B, Raza S, Chen Q J, Yang B. Modularly assembled multiplex prime editors for simultaneous editing of agronomically important genes in rice[J]. Plant Communications, 2024, 5(2): 100741. |
| [58] | Vu T V, Nguyen N T, Kim J, Song Y J, Nguyen T H, Kim J-Y. Optimized dicot prime editing enables heritable desired edits in tomato and Arabidopsis[J]. Nature Plants, 2024, 10(10): 1502-1513. |
| [59] | Liu X X, Wang Y Y, Wang H Z, He Y X, Song Y J, Li Z R, Li M, Wei C, Dong Y H, Xue L, Zhang J S, Zhu J K, Wang M G. Generating herbicide resistant and dwarf rice germplasms through precise sequence insertion or replacement[J]. Plant Biotechnology Journal, 2024, 22(2): 293-295. |
| [60] | Gupta A, Liu B, Chen Q J, Yang B. High-efficiency prime editing enables new strategies for broad-spectrum resistance to bacterial blight of rice[J]. Plant Biotechnology Journal, 2023, 21(7): 1454-1464. |
| [61] | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna J A, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| [62] | Hu J H, Miller S M, Geurts M H, Tang W X, Chen L W, Sun N, Zeina C M, Gao X, Rees H A, Lin Z, Liu D R. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity[J]. Nature, 2018, 556(7699): 57-63. |
| [63] | Kleinstiver B P, Prew M S, Tsai S Q, Topkar V V, Nguyen N T, Zheng Z L, Gonzales A P W, Li Z Y, Peterson R T, Yeh J R J, Aryee M J, Joung J K. Engineered CRISPR-Cas9 nucleases with altered PAM specificities[J]. Nature, 2015, 523(7561): 481-485. |
| [64] | Christie K A, Guo J A, Silverstein R A, Doll R M, Mabuchi M, Stutzman H E, Lin J, Ma L, Walton R T, Pinello L, Robb G B, Kleinstiver B P. Precise DNA cleavage using CRISPR-SpRYgests[J]. Nature Biotechnology, 2023, 41(3): 409-416. |
| [65] | Ren Q R, Sretenovic S, Liu S S, Tang X, Huang L, He Y, Liu L, Guo Y C, Zhong Z H, Liu G Q, Cheng Y H, Zheng X L, Pan C T, Yin D S, Zhang Y X, Li W F, Qi L W, Li C H, Qi Y P, Zhang Y. PAM-less plant genome editing using a CRISPR-SpRY toolbox[J]. Nature Plants, 2021, 7(1): 25-33. |
| [66] | Liu Y, Yang G, Huang S H, Li X Y, Wang X, Li G L, Chi T, Chen Y L, Huang X X, Wang X L. Enhancing prime editing by Csy4-mediated processing of pegRNA[J]. Cell Research, 2021, 31(10): 1134-1136. |
| [67] | Nelson J W, Randolph P B, Shen S P, Everette K A, Chen P J, Anzalone A V, An M R, Newby G A, Chen J C, Hsu A, Liu D R. Engineered pegRNAs improve prime editing efficiency[J]. Nature Biotechnology, 2022, 40(3): 402-410. |
| [68] | Chow R D, Chen J S, Shen J, Chen S. A web tool for the design of prime-editing guide RNAs[J]. Nature Biomedical Engineering, 2021, 5(2): 190-194. |
| [69] | Hsu J Y, Grünewald J, Szalay R, Shih J, Anzalone A V, Lam K C, Shen M W, Petri K, Liu D R, Joung J K. PrimeDesign software for rapid and simplified design of prime editing guide RNAs[J]. Nature Communications, 2021, 12(1): 1034. |
| [70] | Standage-Beier K, Tekel S J, Brafman D A, Wang X. Prime editing guide RNA design automation using PINE-CONE[J]. ACS Synthetic Biology, 2021, 10(2): 422-427. |
| [71] | Hwang G H, Jeong Y K, Habib O, Hong S A, Lim K, Kim J S, Bae S. PE-Designer and PE-Analyzer: Web-based design and analysis tools for CRISPR prime editing[J]. Nucleic Acids Research, 2021, 49(W1): W499-W504. |
| [72] | Siegner S M, Karasu M E, Schröder M S, Kontarakis Z, Corn J E. PnB Designer: A web application to design prime and base editor guide RNAs for animals and plants[J]. BMC Bioinformatics, 2021, 22(1): 101. |
| [73] | Anderson M V, Haldrup J, Thomsen E A, Wolff J H, Mikkelsen J G. pegIT-a web-based design tool for prime editing[J]. Nucleic Acids Research, 2021, 49(W1): W505-W509. |
| [74] | Kim H K, Yu G, Park J, Min S, Lee S, Yoon S, Kim H H. Predicting the efficiency of prime editing guide RNAs in human cells[J]. Nature Biotechnology, 2021, 39(2): 198-206. |
| [75] | Lin Q, Jin S, Zong Y, Yu H, Zhu Z, Liu G, Kou L, Wang Y, Qiu J L, Li J. High-efficiency prime editing with optimized, paired pegRNAs in plants[J]. Nature Biotechnology, 2021, 39(8): 923-927. |
| [76] | Chen P J, Hussmann J A, Yan J, Knipping F, Ravisankar P, Chen P F, Chen C, Nelson J W, Newby G A, Sahin M. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes[J]. Cell, 2021, 184(22): 5635-5652. |
| [77] | Ding X, Seebeck T, Feng Y, Jiang Y, Davis G D, Chen F. Improving CRISPR-Cas9 genome editing efficiency by fusion with chromatin-modulating peptides[J]. The CRISPR Journal, 2019, 2(1): 51-63. |
| [78] | Wang J, He Z, Wang G, Zhang R, Duan J, Gao P, Lei X, Qiu H, Zhang C, Zhang Y. Efficient targeted insertion of large DNA fragments without DNA donors[J]. Nature Methods, 2022, 19(3): 331-340. |
| [79] | Choi J, Chen W, Suiter C C, Lee C, Chardon F M, Yang W, Leith A, Daza R M, Martin B, Shendure J. Precise genomic deletions using paired prime editing[J]. Nature Biotechnology, 2022, 40(2): 218-226. |
| [80] | Jiang T, Zhang X O, Weng Z, Xue W. Deletion and replacement of long genomic sequences using prime editing[J]. Nature Biotechnology, 2022, 40(2): 227-234. |
| [81] | Liu P, Panda K, Edwards S A, Swanson R, Yi H, Pandesha P, Hung Y H, Klaas G, Ye X, Collins M V. Transposase-assisted target-site integration for efficient plant genome engineering[J]. Nature, 2024, 631(8021): 593-600. |
| [82] | Zou J, Meng X, Hong Z, Rao Y, Wang K, Li J, Yu H, Wang C. Cas9-PE: A robust multiplex gene editing tool for simultaneous precise editing and site-specific random mutation in rice[J]. Trends in Biotechnology, 2025, 43(2): 433-446. |
| [83] | Ferreira da Silva J, Tou C J, King E M, Eller M L, Rufino-Ramos D, Ma L, Cromwell C R, Metovic J, Benning F M, Chao L H. Click editing enables programmable genome writing using DNA polymerases and HUH endonucleases[J]. Nature Biotechnology, 2025, 43(6): 923-935. |
| [84] | Ghogare R, Ludwig Y, Bueno G M, Slamet-Loedin I H, Dhingra A. Genome editing reagent delivery in plants[J]. Transgenic Research, 2021, 30(4): 321-335. |
| [85] | Li B, Sun C, Li J, Gao C. Targeted genome-modification tools and their advanced applications in crop breeding[J]. Nature Reviews Genetics, 2024, 25(9): 603-622. |
| [86] | Woo J W, Kim J, Kwon S I, Corvalán C, Cho S W, Kim H, Kim S G, Kim S T, Choe S, Kim J S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins[J]. Nature Biotechnology, 2015, 33(11): 1162-1164. |
| [87] | Svitashev S, Schwartz C, Lenderts B, Young J K, Mark Cigan A. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes[J]. Nature Communications, 2016, 7(1): 13274. |
| [88] | Kim H, Kim S T, Ryu J, Kang B C, Kim J S, Kim S G. CRISPR/Cpf1-mediated DNA-free plant genome editing[J]. Nature Communications, 2017, 8(1): 14406. |
| [89] | Liang Z, Chen K, Zhang Y, Liu J, Yin K, Qiu J L, Gao C. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins[J]. Nature Protocols, 2018, 13(3): 413-430. |
| [90] | Murovec J, Guček K, Bohanec B, Avbelj M, Jerala R. DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR-Cas9 ribonucleoprotein complexes[J]. Frontiers in Plant Science, 2018, 9: 1594. |
| [91] | Osakabe Y, Liang Z, Ren C, Nishitani C, Osakabe K, Wada M, Komori S, Malnoy M, Velasco R, Poli M. CRISPR-Cas9-mediated genome editing in apple and grapevine[J]. Nature Protocols, 2018, 13(12): 2844-2863. |
| [92] | Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho M J, Scelonge C, Lenderts B, Chamberlin M, Cushatt J. Morphogenic regulators Baby boom and Wuschel improve monocot transformation[J]. The Plant Cell, 2016, 28(9): 1998-2015. |
| [93] | Pan C, Li G, Malzahn A A, Cheng Y, Leyson B, Sretenovic S, Gurel F, Coleman G D, Qi Y. Boosting plant genome editing with a versatile CRISPR-Combo system[J]. Nature Plants, 2022, 8(5): 513-525. |
| [94] | Debernardi J M, Tricoli D M, Ercoli M F, Hayta S, Ronald P, Palatnik J F, Dubcovsky J. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants[J]. Nature Biotechnology, 2020, 38(11): 1274-1279. |
| [95] | Chen Z, Debernardi J M, Dubcovsky J, Gallavotti A. Recent advances in crop transformation technologies[J]. Nature Plants, 2022, 8(12): 1343-1351. |
| [96] | Maher M F, Nasti R A, Vollbrecht M, Starker C G, Clark M D, Voytas D F. Plant gene editing through de novo induction of meristems[J]. Nature Biotechnology, 2020, 38(1): 84-89. |
| [97] | Oh Y, Kim H, Kim S G. Virus-induced plant genome editing[J]. Current Opinion in Plant Biology, 2021, 60: 101992. |
| [98] | Ellison E E, Nagalakshmi U, Gamo M E, Huang P J, Dinesh-Kumar S, Voytas D F. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs[J]. Nature Plants, 2020, 6(6): 620-624. |
| [99] | Čermák T, Curtin S J, Gil-Humanes J, Čegan R, Kono T J, Konečná E, Belanto J J, Starker C G, Mathre J W, Greenstein R L. A multipurpose toolkit to enable advanced genome engineering in plants[J]. The Plant Cell, 2017, 29(6): 1196-1217. |
| [100] | Li T, Hu J, Sun Y, Li B, Zhang D, Li W, Liu J, Li D, Gao C, Zhang Y. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture[J]. Molecular Plant, 2021, 14(11): 1787-1798. |
| [101] | Nguyen Tran M T, Mohd Khalid M K N, Wang Q, Walker J K, Lidgerwood G E, Dilworth K L, Lisowski L, Pébay A, Hewitt A W. Engineering domain-inlaid SaCas9 adenine base editors with reduced RNA off-targets and increased on-target DNA editing[J]. Nature Communications, 2020, 11(1): 4871. |
| [102] | Ma X, Zhang X, Liu H, Li Z. Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9[J]. Nature Plants, 2020, 6(7): 773-779. |
| [103] | Liu Q, Zhao C, Sun K, Deng Y, Li Z. Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes[J]. Molecular Plant, 2023, 16(3): 616-631. |
| [104] | Anzalone A V, Koblan L W, Liu D R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nature Biotechnology, 2020, 38(7): 824-844. |
| [105] | Kantor A, McClements M E, MacLaren R E. CRISPR-Cas9 DNA base-editing and prime-editing[J]. International Journal of Molecular Sciences, 2020, 21(17): 6240. |
| [106] | Li G, Cheng Y, Yu J, Zhu Y, Ma H, Zhou Y, Pu Z, Zhu G, Yuan Y, Zhang Z. Compact RNA editors with natural miniature Cas13j nucleases[J]. Nature Chemical Biology, 2025, 21(2): 280-290. |
| [107] | Xu C, Lu P, Gamal El-Din T M, Pei X Y, Johnson M C, Uyeda A, Bick M J, Xu Q, Jiang D, Bai H. Computational design of transmembrane pores[J]. Nature, 2020, 585(7823): 129-134. |
| [108] | Glasscock C J, Pecoraro R J, McHugh R, Doyle L A, Chen W, Boivin O, Lonnquist B, Na E, Politanska Y, Haddox H K. Computational design of sequence-specific DNA-binding proteins[J]. Nature Structural & Molecular Biology, 2025: 1-10. |
| [109] | Watson J L, Juergens D, Bennett N R, Trippe B L, Yim J, Eisenach H E, Ahern W, Borst A J, Ragotte R J, Milles L F. De novo design of protein structure and function with RFdiffusion[J]. Nature, 2023, 620(7976): 1089-1100. |
| [110] | Yeh A H-W, Norn C, Kipnis Y, Tischer D, Pellock S J, Evans D, Ma P, Lee G R, Zhang J Z, Anishchenko I. De novo design of luciferases using deep learning[J]. Nature, 2023, 614(7949): 774-780. |
| [111] | Vora Z, Pandya J, Sangh C, Vaikuntapu P R. The evolving landscape of global regulations on genome-edited crops[J]. Journal of Plant Biochemistry and Biotechnology, 2023, 32(4): 831-845. |
| [112] | Ahmad A, Jamil A, Munawar N. GMOs or non-GMOs? The CRISPR conundrum[J]. Frontiers in Plant Science, 2023, 14: 1232938. |
| [1] | 陈伟, 叶元妹, 赵剑华, 冯志明, 陈宗祥, 胡珂鸣, 左示敏. 利用CRISPR/Cas9技术改良南粳46抽穗期[J]. 中国水稻科学, 2025, 39(6): 760-770. |
| [2] | 杨佳欣, 管玉圣, 杜润, 李贤勇, 蔡座坤, 王楚桃, 阳启样, 何永歆, 朱子超, 张毅. 利用CRISPR/Cas9技术创制无芽鞘紫线的香型环境非敏感隐性核雄性不育种质[J]. 中国水稻科学, 2025, 39(5): 643-649. |
| [3] | 何勇, 刘耀威, 熊翔, 祝丹晨, 王爱群, 马拉娜, 王廷宝, 张健, 李建雄, 田志宏. 利用CRISPR/Cas9技术编辑OsOFP30基因创制水稻粒型突变体[J]. 中国水稻科学, 2024, 38(5): 507-515. |
| [4] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [5] | 李景芳, 温舒越, 赵利君, 陈庭木, 周振玲, 孙志广, 刘艳, 陈海元, 张云辉, 迟铭, 邢运高, 徐波, 徐大勇, 王宝祥. 基于CRISPR/Cas9技术创制耐盐香稻[J]. 中国水稻科学, 2023, 37(5): 478-485. |
| [6] | 李刚, 高清松, 李伟, 张雯霞, 王健, 程保山, 王迪, 高浩, 徐卫军, 陈红旗, 纪剑辉. 定向敲除SD1基因提高水稻的抗倒性和稻瘟病抗性[J]. 中国水稻科学, 2023, 37(4): 359-367. |
| [7] | 段敏, 谢留杰, 高秀莹, 唐海娟, 黄善军, 潘晓飚. 利用CRISPR/Cas9技术创制广亲和水稻温敏雄性不育系[J]. 中国水稻科学, 2023, 37(3): 233-243. |
| [8] | 王石光, 陆展华, 刘维, 卢东柏, 王晓飞, 方志强, 巫浩翔, 何秀英. 应用CRISPR/Cas9技术与分子标记辅助选择创制广东丝苗米新种质[J]. 中国水稻科学, 2023, 37(1): 29-36. |
| [9] | 张元野, 尹丽颖, 李荣田, 何明良, 刘欣欣, 潘婷婷, 田晓杰, 卜庆云, 李秀峰. 利用CRISPR/Cas9技术创制Rc基因恢复红稻[J]. 中国水稻科学, 2022, 36(6): 572-578. |
| [10] | 尹丽颖, 张元野, 李荣田, 何明良, 王芳权, 许扬, 刘欣欣, 潘婷婷, 田晓杰, 卜庆云, 李秀峰. 利用CRISPR/Cas9技术创制高效抗除草剂水稻[J]. 中国水稻科学, 2022, 36(5): 459-466. |
| [11] | 周永林, 申小磊, 周立帅, 林巧霞, 王朝露, 陈静, 冯慧捷, 张振文, 陈晓婷, 鲁国东. OsLOX10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2022, 36(4): 348-356. |
| [12] | 梁敏敏, 张华丽, 陈俊宇, 戴冬青, 杜成兴, 王惠梅, 马良勇. 利用CRISPR/Cas9技术创制抗稻瘟病香型早籼温敏核不育系[J]. 中国水稻科学, 2022, 36(3): 248-258. |
| [13] | 李兆伟, 孙聪颖, 零东兰, 曾慧玲, 张晓妹, 范凯, 林文雄. 利用CRISPR/Cas9创建osarf7突变体及其农艺性状调查[J]. 中国水稻科学, 2022, 36(3): 237-247. |
| [14] | 周天顺, 余东, 刘玲, 欧阳宁, 袁贵龙, 段美娟, 袁定阳. 利用CRISPR/Cas9技术编辑AFP1基因提高水稻耐逆性[J]. 中国水稻科学, 2021, 35(1): 11-18. |
| [15] | 李孟珠, 王高鹏, 巫月, 任怡, 李刚华, 刘正辉, 丁艳锋, 陈琳. 水稻蔗糖转运蛋白OsSUT4参与蔗糖转运的功能研究[J]. 中国水稻科学, 2020, 34(6): 491-498. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||