
中国水稻科学 ›› 2022, Vol. 36 ›› Issue (3): 237-247.DOI: 10.16819/j.1001-7216.2022.210117
李兆伟1, 孙聪颖1, 零东兰1, 曾慧玲1, 张晓妹1, 范凯2, 林文雄1,*( )
)
收稿日期:2021-01-17
修回日期:2021-04-22
出版日期:2022-05-10
发布日期:2022-05-11
通讯作者:
林文雄
基金资助:
LI Zhaowei1, SUN Congying1, LING Donglan1, ZENG Huiling1, ZHANG Xiaomei1, FAN Kai2, LIN Wenxiong1,*( )
)
Received:2021-01-17
Revised:2021-04-22
Online:2022-05-10
Published:2022-05-11
Contact:
LIN Wenxiong
摘要:
【目的】探究OsARF7基因在水稻生长发育中的生物学功能及其对水稻农艺性状的影响。【方法】利用CRISPR/Cas9技术,在粳稻中花11背景下对OsARF7进行定向编辑,获得OsARF7基因突变植株,并考查其农艺性状。【结果】获得22株T0代转基因株系,经PCR扩增潮霉素基因鉴定出20株阳性植株。提取T2代转基因植株的基因组DNA,通过对OsARF7的2个编辑位点区域DNA片段进行PCR扩增及测序分析,筛选到15种突变类型的纯合基因型植株。基因表达分析显示,osarf7突变体的OsARF7和生长素转运关键基因OsPIN1b、OsPIN2和OsLAX2的表达量均显著低于野生型。田间调查发现,与野生型相比,osarf7突变体的分蘖成穗率显著降低,其中,分蘖数比野生型增多的osarf7突变体,有效穗数基本保持不变,而分蘖数与野生型相近的osarf7突变体,有效穗数少于野生型,最终表现为无效分蘖明显增多,分蘖成穗率降低。【结论】OsARF7基因与生长素调控水稻分蘖发生相关,并影响分蘖芽的生长发育及成穗;OsARF7基因突变会导致迟发分蘖芽不能正常发育成穗;粳稻OsARF7突变体的创建对揭示OsARF7基因生理功能和探索生长素促控分蘖发生与成穗机理等具有积极意义。
李兆伟, 孙聪颖, 零东兰, 曾慧玲, 张晓妹, 范凯, 林文雄. 利用CRISPR/Cas9创建osarf7突变体及其农艺性状调查[J]. 中国水稻科学, 2022, 36(3): 237-247.
LI Zhaowei, SUN Congying, LING Donglan, ZENG Huiling, ZHANG Xiaomei, FAN Kai, LIN Wenxiong. Construction of osarf7 Mutants in Rice Based on CRISPR/Cas9 Technology and Investigation on Their Agronomic Traits[J]. Chinese Journal OF Rice Science, 2022, 36(3): 237-247.
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) | 用途Usage |
|---|---|---|
| OsARF7-T1-fwd | gccgACACGAACCTGTGCCACCGC | 靶序列1构建 |
| OsARF7-T1-rev | aaacGCGGTGGCACAGGTTCGTGT | Construction of the target sequence 1 |
| OsARF7-T2-fwd | gttGAGCTATGGCATGCTTGCGC | 靶序列2构建 |
| OsARF7-T2-rev | aaacGCGCAAGCATGCCATAGCT | Construction of the target sequence 2 |
| Hyp-F | ACGGTGTCGTCCATCACAGTTTGCC | 阳性转基因植株鉴定 |
| Hyp-R | TTCCGGAAGTGCTTGACATTGGGGA | Identification of the positive transgenic plant |
| OsARF7-T1-F | ACCACCACCTCCTCCTCAC | 靶点1测序检测 |
| OsARF7-T1-R | TCGCACGAATCAGCACAAC | Sequencing for target 1 |
| OsARF7-T2-F | TTACGGATCTGAGAACCTG | 靶点2测序检测 |
| OsARF7-T2-R | TAACTACATTGCCGAAGAA | Sequencing for target 2 |
表1 本研究采用的gRNA靶点序列及相应的引物序列
Table 1. Sequences of gRNA targets and other primers used in this study.
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) | 用途Usage |
|---|---|---|
| OsARF7-T1-fwd | gccgACACGAACCTGTGCCACCGC | 靶序列1构建 |
| OsARF7-T1-rev | aaacGCGGTGGCACAGGTTCGTGT | Construction of the target sequence 1 |
| OsARF7-T2-fwd | gttGAGCTATGGCATGCTTGCGC | 靶序列2构建 |
| OsARF7-T2-rev | aaacGCGCAAGCATGCCATAGCT | Construction of the target sequence 2 |
| Hyp-F | ACGGTGTCGTCCATCACAGTTTGCC | 阳性转基因植株鉴定 |
| Hyp-R | TTCCGGAAGTGCTTGACATTGGGGA | Identification of the positive transgenic plant |
| OsARF7-T1-F | ACCACCACCTCCTCCTCAC | 靶点1测序检测 |
| OsARF7-T1-R | TCGCACGAATCAGCACAAC | Sequencing for target 1 |
| OsARF7-T2-F | TTACGGATCTGAGAACCTG | 靶点2测序检测 |
| OsARF7-T2-R | TAACTACATTGCCGAAGAA | Sequencing for target 2 |
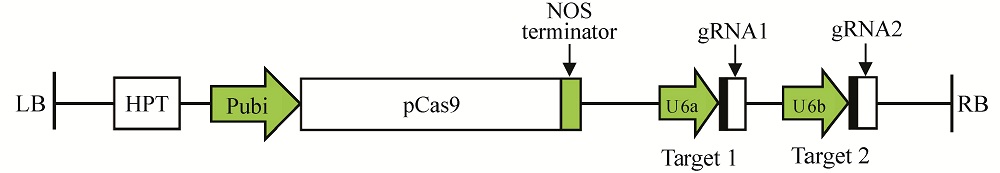
图2 pYLCRISPR/Cas9-ARF7-T12表达载体组装 2个靶位点gRNA表达盒与pYLCRISPR/Cas9-MT载体组装成重组表达载体。LB―左边界;RB―右边界;参照文献[24]略作修改。
Fig. 2. Diagrammatic sketch of pYLCRISPR/Cas9-ARF7-T12 recombinant vector. Cloning of two gRNA cassettes into the pYLCRISPR/Cas9-MT vector. LB, Left border; RB, Right border.
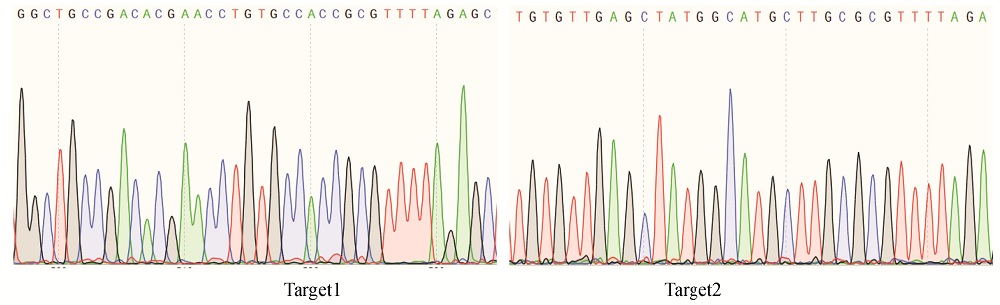
图3 pYLCRISPR/Cas9-ARF7-T12重组载体中2个靶点序列的测序结果
Fig. 3. Sequencing results for the two target sequences inserted in pYLCRISPR/Cas9-ARF7-T12 recombinant vector.
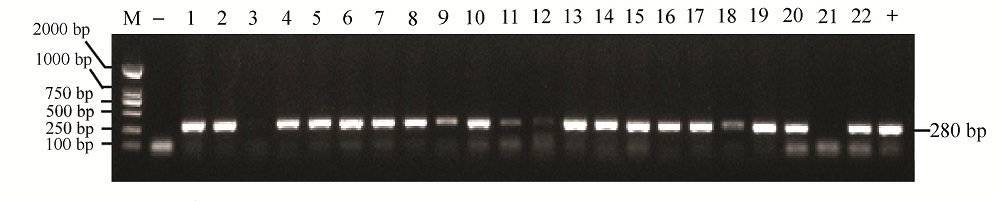
图4 PCR扩增鉴定阳性转基因植株 “—”代表阴性对照,“+”代表阳性对照。
Fig. 4. Identification of the positive transgenic plants by PCR amplification. –, Negative control; +, Positive control.
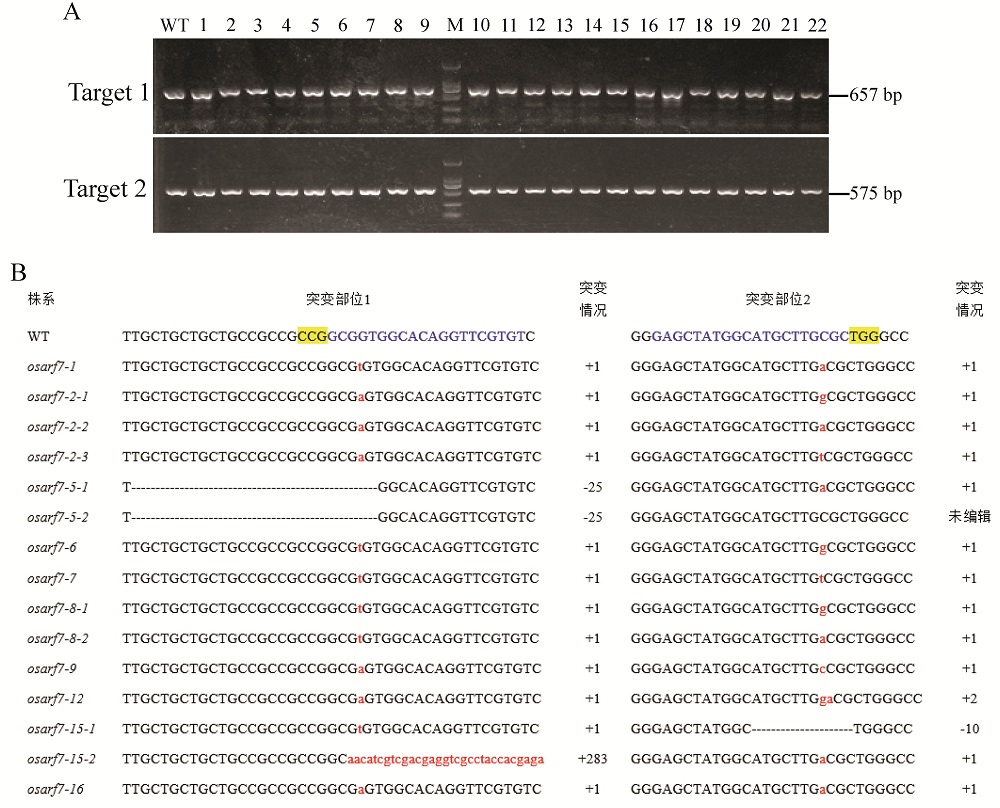
图5 osarf7突变体株系的PCR检测及其与野生型序列比对分析 A–部分T2代(1~22)转化水稻株系OsARF7编辑位点附近DNA片段的PCR检测,靶点1的扩增长度为657 bp,靶点2的扩增长度为575 bp,野生型(WT)为中花11;B–转化植株编辑位点的PCR产物测序序列与野生型(WT)序列的比对结果,蓝色字母为靶点序列,黄色高亮显示PAM序列,删除线表示缺失碱基,红色小写字母为插入碱基,–表示缺失,+表示插入,WT为野生型。
Fig. 5. PCR identification and sequence alignment of osarf7 mutants compared to the WT line. A, PCR amplification results of the DNA fragments near the OsARF7 locus for partial osarf7 T2 mutants, the fragment length of target 1 and 2 is 657 bp and 575 bp, respectively, and WT is ZH11; B, Sequence alignment of the edited locus for the osarf7 mutants and WT line. The blue letters are the target genome sequence; The yellow highlighted letters denote PAM; Dashes strikethrough indicate the deleted bases; Insertion nucleotides are shown in red lowercase letters; –, Deletion; +, Insertion; WT, Wild type.
| 编号 Number | 潜在脱靶位置 Putative off-target locus | 潜在脱靶序列 Sequence of the putative off-target site (5′-3′) | 错配碱基数 No. of mismatched bases | 测试株数 No. of plants tested | 突变株数 No. of plants with mutations |
|---|---|---|---|---|---|
| Off-target 1-1 | Chr08: 12898391-12898413 | ACCCTAACCTCTCCCACCGC | 4 | 9 | 0 |
| Off-target 1-2 | Chr10: 15856432-15856454 | AAACGGACGTGTGCCGCCGC | 4 | 9 | 0 |
| Off-target 1-3 | Chr02: 27855035-27855057 | AGACGGGCCTGTGCCAACGC | 4 | 9 | 0 |
| Off-target 1-4 | Chr12: 12787156-12787178 | CCAAGAACCTGCGCCGCCGC | 4 | 9 | 0 |
| Off-target 1-5 | Chr07: 2126716-2126738 | ACAGCAGCCGGTGCCACCGC | 4 | 9 | 0 |
| Off-target 2-1 | Chr12: 26007034-26007056 | GAGCTGTGGCATGCATGCGC | 2 | 10 | 0 |
| Off-target 2-2 | Chr02: 3488312-3488334 | GAGTTGTGGCATGCATGCGC | 3 | 9 | 0 |
| Off-target 2-3 | Chr04: 34285188-34285210 | GAGCTATGGCACGCCTGCGC | 2 | 8 | 0 |
| Off-target 2-4 | Chr06: 4926812-4926834 | GAGCTATGGCATGCTTGTGC | 1 | 10 | 0 |
| Off-target 2-5 | Chr04: 22007551-22007573 | GAGCTATGGCATGCTTGTGC | 1 | 10 | 0 |
| Off-target 2-6 | Chr01: 40696054-40696076 | GAGCTGTGGCACGCCTGCGC | 3 | 10 | 0 |
表2 CRISPR/Cas9-OsARF7-T12潜在脱靶位点检测
Table 2. Mutation detections in the putative CRISPR/Cas9-OsARF7-T12 off-target sites.
| 编号 Number | 潜在脱靶位置 Putative off-target locus | 潜在脱靶序列 Sequence of the putative off-target site (5′-3′) | 错配碱基数 No. of mismatched bases | 测试株数 No. of plants tested | 突变株数 No. of plants with mutations |
|---|---|---|---|---|---|
| Off-target 1-1 | Chr08: 12898391-12898413 | ACCCTAACCTCTCCCACCGC | 4 | 9 | 0 |
| Off-target 1-2 | Chr10: 15856432-15856454 | AAACGGACGTGTGCCGCCGC | 4 | 9 | 0 |
| Off-target 1-3 | Chr02: 27855035-27855057 | AGACGGGCCTGTGCCAACGC | 4 | 9 | 0 |
| Off-target 1-4 | Chr12: 12787156-12787178 | CCAAGAACCTGCGCCGCCGC | 4 | 9 | 0 |
| Off-target 1-5 | Chr07: 2126716-2126738 | ACAGCAGCCGGTGCCACCGC | 4 | 9 | 0 |
| Off-target 2-1 | Chr12: 26007034-26007056 | GAGCTGTGGCATGCATGCGC | 2 | 10 | 0 |
| Off-target 2-2 | Chr02: 3488312-3488334 | GAGTTGTGGCATGCATGCGC | 3 | 9 | 0 |
| Off-target 2-3 | Chr04: 34285188-34285210 | GAGCTATGGCACGCCTGCGC | 2 | 8 | 0 |
| Off-target 2-4 | Chr06: 4926812-4926834 | GAGCTATGGCATGCTTGTGC | 1 | 10 | 0 |
| Off-target 2-5 | Chr04: 22007551-22007573 | GAGCTATGGCATGCTTGTGC | 1 | 10 | 0 |
| Off-target 2-6 | Chr01: 40696054-40696076 | GAGCTGTGGCACGCCTGCGC | 3 | 10 | 0 |
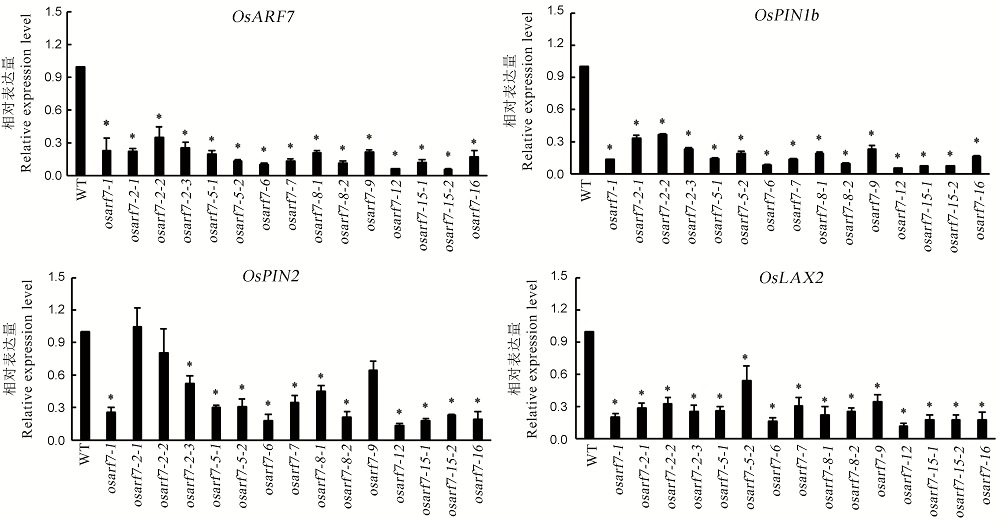
图6 osarf7突变体及其野生型水稻OsARF7、OsPIN1b、OsPIN2、OsLAX2相对表达量
Fig. 6. Relative expression levels of OsARF7, OsPIN1b, OsPIN2 and OsLAX2 in osarf7 mutant and its wild type.
| 株系 Line | 株高 Plant height / cm | 总分蘖数 Tiller number per plant | 有效穗数 Effective panicle per plant | 穗长 Length of main panicle /cm | 穗粒数 Grain number per panicle | 结实率 Seed setting rate / % | 千粒重 1000-grain weight / g | 单株产量 Grain yield per plant / g |
|---|---|---|---|---|---|---|---|---|
| osarf7-1 | 73.5±1.6 ab | 11.2±1.3 a | 24.6±2.1 a | 119.3±30.8 ab | 63.4±19.0 bc | 23.3±2.9 e | 8.6±4.6 ab | |
| osarf7-2-1 | 72.2±3.4 ab | 20.6±3.3 a | 7.4±1.5 abc | 22.8±1.5 ab | 135.9±34.9 ab | 74.5±14.7 abc | 25.5±0.2 bcde | 12.9±6.3 ab |
| osarf7-2-2 | 67.3±4.9 b | 11.0±2.6 e | 5.0±1.0 c | 23.5±2.5 ab | 131.8±35.0 ab | 82.5±8.8 ab | 23.5±0.3 de | 9.3±4.2 ab |
| osarf7-2-3 | 72.7±1.5 ab | 20.0±1.0 a | 7.3±3.5 abc | 23.7±1.2 ab | 167.5±13.8 a | 78.4±5.8 abc | 22.6±0.6 e | 9.0±3.3 ab |
| osarf7-5-1 | 67.3±3.2 b | 12.7±1.2 de | 5.0±0.9 c | 22.6±1.2 ab | 119.4±12.1 ab | 81.8±5.2 ab | 25.5±0.6 bcde | 10.4±0.5 ab |
| osarf7-5-2 | 70.8±3.2 ab | 12.8±1.5 de | 6.2±0.8 bc | 22.8±2.0 ab | 112.8±29.5 b | 75.9±9.5 abc | 25.2±1.3 cde | 9.7±1.2 ab |
| osarf7-6 | 68.5±3.6 b | 14.5±1.1 bcd | 7.6±1.8 bc | 23.0±1.5 ab | 126.2±23.0 ab | 69.9±11.9 abc | 24.4±1.2 de | 11.3±3.0 ab |
| osarf7-7 | 73.0±3.7 ab | 20.2±1.8 a | 10.2±1.9 ab | 23.2±1.4 ab | 135.9±21.2 ab | 73.7±14.6 abc | 25.0±1.5 cde | 14.5±4.2 ab |
| osarf7-8-1 | 74.8±2.2 a | 13.8±0.5 bcde | 6.5±2.1 bc | 23.7±2.0 ab | 144.8±34.6 ab | 76.5±10.0 abc | 25.4±1.2 bcde | 14.1±3.0 ab |
| osarf7-8-2 | 75.4±2.4 a | 16.0±0.7 bc | 7.0±2.6 bc | 23.1±1.8 ab | 140.5±32.2 ab | 83.21±9.3 a | 24.7±0.7 cde | 12.0±3.3 ab |
| osarf7-9 | 72.3±0.8 ab | 15.5±1.0 bc | 6.8±1.9 bc | 22.3±1.6 ab | 119.9±23.1 ab | 86.5±8.3 a | 26.7±1.6 bcd | 12.5±2.4 ab |
| osarf7-12 | 75.0±2.0 a | 17.3±0.6 ab | 6.3±1.5 bc | 24.0±1.0 ab | 139.8±36.0 ab | 90.8±3.3 a | 23.6±0.8 cde | 17.4±9.8 a |
| osarf7-15-1 | 75.8±2.4 a | 13.7±1.2 de | 5.1±1.7 c | 21.9±1.3 b | 117.2±22.5 b | 83.0±5.9 a | 28.1±1.1 ab | 10.0±3.2 ab |
| osarf7-15-2 | 74.7±1.4 a | 11.9±1.2 de | 4.9±2.3 c | 21.8±1.9 b | 122.0±24.5 ab | 79.1±15.0 ab | 27.4±1.3 abc | 9.4±3.1 ab |
| osarf7-16 | 72.0±2.3ab | 15.4±1.6 bc | 5.6±0.9 c | 21.8±1.3 b | 103.0±26.7 b | 57.2±26.4 c | 30.1±2.2 a | 13.0±3.9 b |
| WT | 71.2±1.9 ab | 11.3±0.6 de | 10.2±1.8 ab | 20.8±1.6 b | 111.8±15.0 b | 88.4±4.4 a | 24.3±0.7 de | 14.2±4.8 ab |
表3 不同类型osarf7突变体株系及其野生型水稻(WT)的农艺性状
Table 3. Agronomic traits of different types of osarf7 mutants and its corresponding wild type grown in paddy field.
| 株系 Line | 株高 Plant height / cm | 总分蘖数 Tiller number per plant | 有效穗数 Effective panicle per plant | 穗长 Length of main panicle /cm | 穗粒数 Grain number per panicle | 结实率 Seed setting rate / % | 千粒重 1000-grain weight / g | 单株产量 Grain yield per plant / g |
|---|---|---|---|---|---|---|---|---|
| osarf7-1 | 73.5±1.6 ab | 11.2±1.3 a | 24.6±2.1 a | 119.3±30.8 ab | 63.4±19.0 bc | 23.3±2.9 e | 8.6±4.6 ab | |
| osarf7-2-1 | 72.2±3.4 ab | 20.6±3.3 a | 7.4±1.5 abc | 22.8±1.5 ab | 135.9±34.9 ab | 74.5±14.7 abc | 25.5±0.2 bcde | 12.9±6.3 ab |
| osarf7-2-2 | 67.3±4.9 b | 11.0±2.6 e | 5.0±1.0 c | 23.5±2.5 ab | 131.8±35.0 ab | 82.5±8.8 ab | 23.5±0.3 de | 9.3±4.2 ab |
| osarf7-2-3 | 72.7±1.5 ab | 20.0±1.0 a | 7.3±3.5 abc | 23.7±1.2 ab | 167.5±13.8 a | 78.4±5.8 abc | 22.6±0.6 e | 9.0±3.3 ab |
| osarf7-5-1 | 67.3±3.2 b | 12.7±1.2 de | 5.0±0.9 c | 22.6±1.2 ab | 119.4±12.1 ab | 81.8±5.2 ab | 25.5±0.6 bcde | 10.4±0.5 ab |
| osarf7-5-2 | 70.8±3.2 ab | 12.8±1.5 de | 6.2±0.8 bc | 22.8±2.0 ab | 112.8±29.5 b | 75.9±9.5 abc | 25.2±1.3 cde | 9.7±1.2 ab |
| osarf7-6 | 68.5±3.6 b | 14.5±1.1 bcd | 7.6±1.8 bc | 23.0±1.5 ab | 126.2±23.0 ab | 69.9±11.9 abc | 24.4±1.2 de | 11.3±3.0 ab |
| osarf7-7 | 73.0±3.7 ab | 20.2±1.8 a | 10.2±1.9 ab | 23.2±1.4 ab | 135.9±21.2 ab | 73.7±14.6 abc | 25.0±1.5 cde | 14.5±4.2 ab |
| osarf7-8-1 | 74.8±2.2 a | 13.8±0.5 bcde | 6.5±2.1 bc | 23.7±2.0 ab | 144.8±34.6 ab | 76.5±10.0 abc | 25.4±1.2 bcde | 14.1±3.0 ab |
| osarf7-8-2 | 75.4±2.4 a | 16.0±0.7 bc | 7.0±2.6 bc | 23.1±1.8 ab | 140.5±32.2 ab | 83.21±9.3 a | 24.7±0.7 cde | 12.0±3.3 ab |
| osarf7-9 | 72.3±0.8 ab | 15.5±1.0 bc | 6.8±1.9 bc | 22.3±1.6 ab | 119.9±23.1 ab | 86.5±8.3 a | 26.7±1.6 bcd | 12.5±2.4 ab |
| osarf7-12 | 75.0±2.0 a | 17.3±0.6 ab | 6.3±1.5 bc | 24.0±1.0 ab | 139.8±36.0 ab | 90.8±3.3 a | 23.6±0.8 cde | 17.4±9.8 a |
| osarf7-15-1 | 75.8±2.4 a | 13.7±1.2 de | 5.1±1.7 c | 21.9±1.3 b | 117.2±22.5 b | 83.0±5.9 a | 28.1±1.1 ab | 10.0±3.2 ab |
| osarf7-15-2 | 74.7±1.4 a | 11.9±1.2 de | 4.9±2.3 c | 21.8±1.9 b | 122.0±24.5 ab | 79.1±15.0 ab | 27.4±1.3 abc | 9.4±3.1 ab |
| osarf7-16 | 72.0±2.3ab | 15.4±1.6 bc | 5.6±0.9 c | 21.8±1.3 b | 103.0±26.7 b | 57.2±26.4 c | 30.1±2.2 a | 13.0±3.9 b |
| WT | 71.2±1.9 ab | 11.3±0.6 de | 10.2±1.8 ab | 20.8±1.6 b | 111.8±15.0 b | 88.4±4.4 a | 24.3±0.7 de | 14.2±4.8 ab |
| [1] | 郭韬, 余泓, 邱杰, 李家洋, 韩斌, 林鸿宣. 中国水稻遗传学研究进展与分子设计育种[J]. 中国科学: 生命科学, 2019, 49: 1-28. |
| Guo T, Yu H, Qiu J, Li J Y, Han B, Lin H X. Advances in rice genetics and breeding by molecular design in China[J]. Scientia Sinica Vitae, 2019, 49: 1-28. (in Chinese with English abstract) | |
| [2] | Woodward A W, Bartel B. Auxin: Regulation, action, and interaction[J]. Annals of Botany, 2005, 95(5): 707-735. |
| [3] | Guilfoyle T J, Hagen G. Auxin response factors[J]. Current Opinion in Plant Biology, 2007, 10: 453-460. |
| [4] | Simonini S, Deb J, Moubayidin L, Stephenson P, Valluru M, Freire-Rios A, Sorefan K, Weijers D, Friml J, Østergaard L. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis[J]. Genes & Development, 2016, 30: 2286-2296. |
| [5] | Guilfoyle T, Hagen G, Ulmasov T, Murfett J. How does auxin turn on genes[J]. Plant Physiology, 1998, 118: 341-347. |
| [6] | Okushima Y, Overvoorde P J, Arima K, Alonso J M, Chan A, Chang C, Ecker J R, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19[J]. Plant Cell, 2005, 17(2): 444-463. |
| [7] | Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa)[J]. Gene, 2007, 394(1): 13-24. |
| [8] | Xing H, Pudake R N, Guo G, Xing G, Hu Z, Zhang Y, Sun Q, Ni Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize[J]. BMC Genomics, 2011, 12(1): 178. |
| [9] | Ha C V, Le D T, Nishiyama R, Watanabe Y, Sulieman S, Tran U T, Mochida K, Dong N V, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. The auxin response factor transcription factor family in soybean: genome-wide identification and expression analyses during development and water stress[J]. DNA Research, 2013, 20(5): 511-524. |
| [10] | Wu J, Wang F, Cheng L, Kong F, Peng Z, Liu S, Yu X, Lu G. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum[J]. Plant Cell Reports, 2011, 30(11): 2059-2073. |
| [11] | Liu S Q, Hu L F. Genome-wide analysis of the auxin response factor gene family in cucumber[J]. Genetics and Molecular Research, 2013, 12(4): 4317-4331. |
| [12] | 刘瑞娥, 胡长贵, 孙玉强. 植物生长素反应因子研究进展[J]. 植物生理学报, 2011, 47(7): 669-679. |
| Liu R E, Hu C G, Sun Y Q. Advances in plant auxin response factors[J]. Journal of Plant Physiology, 2011, 47(7): 669-679. (in Chinese with English abstract) | |
| [13] | 张赛娜. OsARF19调控水稻叶夹角的分子机制[D]. 杭州: 浙江大学, 2014: 32-33. |
| Zhang S N. Molecular mechanism of OsARF19 controlling leaf angle in rice (Oryza sativa)[D]. Hangzhou: Zhejiang University, 2014: 32-33. (in Chinese with English abstract) | |
| [14] | Ulmasov T, Hagen G, Guilfoyle T J. Activation and repression of transcription by auxin response factors[J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96: 5844-5849. |
| [15] | Waller F, Furuya M, Nick P. OsARF1, an auxin response factor from rice, is auxin-regulated and classifies as a primary auxin responsive gene[J]. Plant Molecular Biology, 2002, 50(3): 415-425. |
| [16] | 汪高航. 利用点突变研究水稻OsARF的功能[D]. 杭州: 浙江大学, 2010: 22-24. |
| Wang G H. Preliminary study of the functions of OsARFs by mutation[D]. Hangzhou: Zhejiang University, 2010: 22-24. (in Chinese with English abstract) | |
| [17] | 淳俊, 王文国, 王胜华, 陈放. 水稻(Oryza sativa L.)愈伤组织发育过程Auxin-miR167-ARF8信号通路的研究[J]. 四川大学学报, 2013, 50(4): 863-868. |
| Chun J, Wang W G, Wang S H, Chen F. The study on Auxin-miR167-ARF8 signal pathway during the growth and development process of rice callus[J]. Journal of Sichuan University, 2013, 50(4): 863-868. (in Chinese with English abstract) | |
| [18] | Shen C, Yue R, Yang Y, Zhang L, Sun T, Tie S, Wang H. OsARF16 is involved in cytokinin-mediated inhibition of phosphate transport and phosphate signaling in rice (Oryza sativa L.)[J]. PLoS One, 2014, 9(11): e112906. |
| [19] | Shen C, Yue R, Sun T, Zhang L, Yang Y, Wang H. OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.)[J]. Plant Science, 2015, 231: 148-158. |
| [20] | Sato Y, Nishimura A, Ito M, Ashikari M, Hirano H Y, Matsuoka M. Auxin response factor family in rice[J]. Genes & Genetic Systems, 2001, 76: 373-380. |
| [21] | Ellis C M, Nagpal P, Young J C, Hagen G, Guilfoyle T J, Reed J W. Auxin response factor1 and Auxin response factor1 regulate senescence and floral organ abscission in Arabidopsis thaliana[J]. Development, 2005, 132(20): 4563-4574. |
| [22] | Doudna J A, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096. |
| [23] | Belhaj K, Chaparro-Garcia A, Kamoun S, Patron N J, Nekrasov V. Editing plant genomes with CRISPR/ Cas9[J]. Current Opinion in Biotechnology, 2015, 32: 76-84. |
| [24] | 王加峰, 郑才敏, 刘维, 罗文龙, 王慧, 陈志强, 郭涛. 基于CRISPR/Cas9技术的水稻千粒重基因tgw6突变体的创建[J]. 作物学报, 2016, 42(8): 1160-1167. |
| Wang J F, Zheng C M, Liu W, Luo W L, Wang H, Chen Z Q, Guo T. Construction of tgw6 mutants in rice based on CRISPR/Cas9 technology[J]. Acta Agronomy Sinica, 2016, 42(8): 1160-1167. (in Chinese with English abstract) | |
| [25] | 周延彪, 赵新辉, 唐晓丹, 周在为, 庄楚雄, 杨远柱. 基于CRISPR/Cas9技术的水稻反光敏不育基因csa突变体的获得[J]. 杂交水稻, 2018, 33(6): 68-74. |
| Zhou Y B, Zhao X H, Tang X D, Zhou Z W, Zhuang C X, Yang Y Z. Acquisition of mutants of the reverse photoperiod-sensitive genic male sterility gene csa in rice based on CRISPR/Cas9 technology[J]. Hybrid Rice, 2018, 33(6): 68-74. (in Chinese with English abstract) | |
| [26] | 黄忠明, 周延彪, 唐晓丹, 赵新辉, 周在为, 符星学, 王凯, 史江伟, 李艳锋, 符辰建, 杨远柱. 基于CRISPR/Cas9技术的水稻温敏不育基因tms5突变体的构建[J]. 作物学报, 2018, 44(6): 844-851. |
| Huang Z M, Zhou Y B, Tang X D, Zhao X H, Zhou Z W, Fu X X, Wang K, Shi J W, Li Y F, Fu C J, Yang Y Z. Construction of tms5 mutants in rice based on CRISPR/Cas9 technology[J]. Acta Agronomy Sinica, 2018, 44(6): 844-851. (in Chinese with English abstract) | |
| [27] | Ma X L, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu Y G. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicotplants[J]. Molecular Plant, 2015, 8: 1274-1284. |
| [28] | Qi Y H, Wang S K, Shen C J, Zhang S N, Chen Y, Xu Y X, Liu Y, Wu Y R, Jiang D A. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa)[J]. New Phytologist, 2012, 193: 109-120. |
| [29] | Bibikova M, Beumer K, Trautman J K, Carroll D. Enhancing gene targeting with designed zinc finger nucleases[J]. Science, 2003, 300: 764. |
| [30] | Huang P, Xiao A, Zhou M G, Zhu Z Y, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs[J]. Nature Biotechnology, 2011, 29: 699-700. |
| [31] | 季新, 李飞, 晏云, 孙红正, 张静, 李俊周, 彭廷, 杜彦修, 赵全志. 基于CRISPR/Cas9系统的水稻光敏色素互作因子OsPIL15基因编辑[J]. 中国农业科学, 2017, 50(15): 2861-2871. |
| Ji X, Li F, Yan Y, Sun H Z, Zhang J, Li J Z, Peng T, Du Y X, Zhao Q Z. CRISPR/Cas9 system-based editing of phytochrome-interacting factor OsPIL15[J]. Scientia Agricultura Sinica, 2017, 50(15): 2861-2871. (in Chinese with English abstract) | |
| [32] | 唐丽, 李曜魁, 张丹, 毛毕刚, 吕启明, 胡远艺, 韶也, 彭彦, 赵炳然, 夏石头. 基于基因组编辑技术的水稻靶向突变特征及遗传分析[J]. 遗传, 2016, 38(8): 746-755. |
| Tang L, Li Y K, Zhang D, Mao B G, Lü Q M, Hu Y Y, Shao Y, Peng Y, Zhao B R, Xia S T. Characteristic and inheritance analysis of targeted mutagenesis mediated by genome editing in rice[J]. Hereditas, 2016, 38(8): 746-755. (in Chinese with English abstract) | |
| [33] | Xu M, Zhu L, Shou H, Wu P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice[J]. Plant Cell Physiology, 2005, 46(10): 1674-168. |
| [34] | Chen Y, Fan X, Song W, Zhang Y, Xu G. Over- expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1[J]. Plant Biotechnology Journal, 2012, 10(2): 139-149. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||