
中国水稻科学 ›› 2024, Vol. 38 ›› Issue (3): 256-265.DOI: 10.16819/j.1001-7216.2024.231012
尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学*( ), 王文明*(
), 王文明*( )
)
收稿日期:2023-10-30
修回日期:2023-12-12
出版日期:2024-05-10
发布日期:2024-05-13
通讯作者:
*email: zhixuezhao@sicau.edu.cn;
j316wenmingwang@163.com
基金资助:
YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue*( ), WANG Wenming*(
), WANG Wenming*( )
)
Received:2023-10-30
Revised:2023-12-12
Online:2024-05-10
Published:2024-05-13
Contact:
*email: zhixuezhao@sicau.edu.cn;
j316wenmingwang@163.com
摘要:
【目的】信号肽对效应因子的分泌至关重要,影响病原微生物的致病进程。前期鉴定到在稻曲病菌侵染水稻花器官过程中显著上调表达的10个效应因子基因,但这些效应因子是否含有功能性信号肽及其表达模式尚不明确。【方法】利用SignaIP-5.0数据库预测、烟草瞬时表达系统和酵母信号肽分泌实验,分析并鉴定这些效应因子的信号肽。利用实时荧光定量PCR技术,检测效应因子基因的表达模式。【结果】UV_44、UV_1548、UV_1567等10个稻曲病菌效应因子均含有信号肽。烟草瞬时表达结果显示,含有信号肽的eYFP融合蛋白在细胞质壁分离后均能分泌到质外体空间。酵母菌信号肽分泌结果显示,分别携带这10个信号肽的酵母菌株可在以棉子糖为唯一碳源的培养基上生长,且与TTC发生显色反应。实时荧光定量结果表明,除了UV_6754,其余9个效应因子基因的表达模式不尽相同,其中UV_44、UV_1567、UV_3667、UV_4213、UV_4989、UV_5215和UV_8184的表达量在接种后显著升高。【结论】这10个稻曲病菌效应因子均含有分泌功能的信号肽,编码效应因子的基因在稻曲病菌侵染过程中具有不同的表达模式。该结果为深入研究以上稻曲病菌效应因子的功能奠定了基础。
尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265.
YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens[J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265.
| 引物名称 Primer name | 引物序列(5′-3′) Primer sequence(5′-3′) | 用途 Usage | |
|---|---|---|---|
| Psuc2-44-F | AATTCATGAAGGCCTCGACGCTGCTGGCTCTCCTGCCCTTTGCCGCGGCCC | 酵母系统鉴定信号肽功能 Functional validation of the signal peptides using yeast | |
| Psuc2-44-R | TCGAGGGCCGCGGCAAAGGGCAGGAGAGCCAGCAGCGTCGAGGCCTTCATG | ||
| Psuc2-1548-F | AATTCATGAACCCAATCCTGCTTCTCCTCGCCGCATCAGTCCCCGCCCGCAGCC | ||
| Psuc2-1548-R | TCGAGGCTGCGGGCGGGGACTGATGCGGCGAGGAGAAGCAGGATTGGGTTCATG | ||
| Psuc2-1567-F | AATTCATGCAGTTGTCCTACCTTCTCTCGCTCGTCTCCCTTGTGGCCGCTC | ||
| Psuc2-1567-R | TCGAGAGCGGCCACAAGGGAGACGAGCGAGAGAAGGTAGGACAACTGCATG | ||
| Psuc2-1990-F | AATTCATGCGCCCAGCAACCGTCATCTTCGCACTGGCCGTCTCCGTCAAGGCTC | ||
| Psuc2-1990-R | TCGAGAGCCTTGACGGAGACGGCCAGTGCGAAGATGACGGTTGCTGGGCGCATG | ||
| Psuc2-3667-F | AATTCATGAAGTACACTGCTGCTCTCGTCGCCCTGGCTGCTGCCGTCATGGCCC | ||
| Psuc2-3667-R | TCGAGGGCCATGACGGCAGCAGCCAGGGCGACGAGAGCAGCAGTGTACTTCATG | ||
| Psuc2-4213-F | AATTCATGATTCTGCTGCTCCTCTTGCTATGCACGACAACATGTTTAGGAC | ||
| Psuc2-4213-R | TCGAGTCCTAAACATGTTGTCGTGCATAGCAAGAGGAGCAGCAGAATCATG | ||
| Psuc2-4989-F | AATTCATGCACCAGCTCCTTGCACTACTCCTCGCCAGCCTCACCGCTGCCC | ||
| Psuc2-4989-R | TCGAGGGCAGCGGTGAGGCTGGCGAGGAGTAGTGCAAGGAGCTGGTGCATG | ||
| Psuc2-5215-F | AATTCATGAAGTTTATTTCCCTCGTAGCCGCGGCCGGTGTGGCGTATGCCC | ||
| Psuc2-5215-R | TCGAGGGCATACGCCACACCGGCCGCGGCTACGAGGGAAATAAACTTCATG | ||
| Psuc2-6754-F | AATTCATGGCTGCAGCATCAGCCCTTTTCTTCCTCACGGCGGCTGTCTCCTGGACAC | ||
| Psuc2-6754-R | TCGAGTGTCCAGGAGACAGCCGCCGTGAGGAAGAAAAGGGCTGATGCTGCAGCCATG | ||
| Psuc2-8184-F | AATTCATGCAAATCGTCCTCCCTCTCCTGGCCTGCGCAGCCGTGGCTGCCGCCC | ||
| Psuc2-8184-R | TCGAGGGCGGCAGCCACGGCTGCGCAGGCCAGGAGAGGGAGGACGATTTGCATG | ||
| P1300-44-F | CATGAAGGCCTCGACGCTGCTGGCTCTCCTGCCCTTTGCCGCGGCCGGTAC | 烟草瞬时表达鉴定信号肽功能 Functional validation of the signal peptides using N. benthamiana | |
| P1300-44-R | CGGCCGCGGCAAAGGGCAGGAGAGCCAGCAGCGTCGAGGCCTTCATGGTAC | ||
| P1300-1548-F | CATGAACCCAATCCTGCTTCTCCTCGCCGCATCAGTCCCCGCCCGCAGCGGTAC | ||
| P1300-1548-R | CGCTGCGGGCGGGGACTGATGCGGCGAGGAGAAGCAGGATTGGGTTCATGGTAC | ||
| P1300-1567-F | CATGCAGTTGTCCTACCTTCTCTCGCTCGTCTCCCTTGTGGCCGCTGGTAC | ||
| P1300-1567-R | CAGCGGCCACAAGGGAGACGAGCGAGAGAAGGTAGGACAACTGCATGGTAC | ||
| P1300-1990-F | CATGCGCCCAGCAACCGTCATCTTCGCACTGGCCGTCTCCGTCAAGGCTGGTAC | ||
| P1300-1990-R | CAGCCTTGACGGAGACGGCCAGTGCGAAGATGACGGTTGCTGGGCGCATGGTAC | ||
| P1300-3667-F | CATGAAGTACACTGCTGCTCTCGTCGCCCTGGCTGCTGCCGTCATGGCCGGTAC | ||
| P1300-3667-R | CGGCCATGACGGCAGCAGCCAGGGCGACGAGAGCAGCAGTGTACTTCATGGTAC | ||
| P1300-4213-F | CATGATTCTGCTGCTCCTCTTGCTATGCACGACAACATGTTTAGGAGGTAC | ||
| P1300-4213-R | CTCCTAAACATGTTGTCGTGCATAGCAAGAGGAGCAGCAGAATCATGGTAC | ||
| P1300-4989-F | CATGCACCAGCTCCTTGCACTACTCCTCGCCAGCCTCACCGCTGCCGGTAC | ||
| P1300-4989-R | CGGCAGCGGTGAGGCTGGCGAGGAGTAGTGCAAGGAGCTGGTGCATGGTAC | ||
| P1300-5215-F | CATGAAGTTTATTTCCCTCGTAGCCGCGGCCGGTGTGGCGTATGCCGGTAC | ||
| P1300-5215-R | CGGCATACGCCACACCGGCCGCGGCTACGAGGGAAATAAACTTCATGGTAC | ||
| P1300-6754-F | CATGGCTGCAGCATCAGCCCTTTTCTTCCTCACGGCGGCTGTCTCCTGGACAGGTAC | ||
| P1300-6754-R | CTGTCCAGGAGACAGCCGCCGTGAGGAAGAAAAGGGCTGATGCTGCAGCCATGGTAC | ||
| P1300-8184-F | CATGCAAATCGTCCTCCCTCTCCTGGCCTGCGCAGCCGTGGCTGCCGCCGGTAC | ||
| P1300-8184-R | CGGCGGCAGCCACGGCTGCGCAGGCCAGGAGAGGGAGGACGATTTGCATGGTAC | ||
| Uv_Tublin2α-2F/R | ACCAGCTCGTTGAGAACTCG / AATCAGAGTTGAGCTGGCCG | 实时荧光定量PCR RT-qPCR | |
| UV_44-RT-F/R | GCGTTGCCAAGAAGACCAAG / ATTCCAGCAATCACGGCAGA | ||
| UV_1548-RT- F/R | GACAACGGGCAGACAAACTG / CTAGCAATCCCAGCAGTCGT | ||
| UV_1567-RT-F/R | TGCTAGCGCTATCGTTCCTG / GTCGGCATGGTTTTGCAACT | ||
| UV_1990-RT-F/R | ACTCACTTCCAAACACGGCA / GCCTTGTCTTCTCAACACGC | ||
| UV_3667-RT-F/R | GCCTCCTCTGCAATGTGGAA / CTTGAGGCAGTTGAGTCCGA | ||
| UV_4213-RT-F/R | ATGCGAGAACTTGGTCGTGT / AACCAACGCCTCCAACTTGA | ||
| UV_4989-RT-F/R | CCTGCCTCCTTCGTCCAAAT / GGTCGATCTGGCAAAGTGGA | ||
| UV_5215-RT-F/R | TAGCGTACTGCGACGGTAAC / GAAACAGCGCCCAATTCCTG | ||
| UV_6754-RT-F/R | AAATGCGAGGACTAGGGCAC / CTTTTTGCCGGAGGAGGTCT | ||
| UV_8184-RT-F/R | CATCGCTGCGAAGACAAAGG / GCGCCTGTGAACTTTTCCTG | ||
表1 本研究所需引物
Table 1. Primers used in this study
| 引物名称 Primer name | 引物序列(5′-3′) Primer sequence(5′-3′) | 用途 Usage | |
|---|---|---|---|
| Psuc2-44-F | AATTCATGAAGGCCTCGACGCTGCTGGCTCTCCTGCCCTTTGCCGCGGCCC | 酵母系统鉴定信号肽功能 Functional validation of the signal peptides using yeast | |
| Psuc2-44-R | TCGAGGGCCGCGGCAAAGGGCAGGAGAGCCAGCAGCGTCGAGGCCTTCATG | ||
| Psuc2-1548-F | AATTCATGAACCCAATCCTGCTTCTCCTCGCCGCATCAGTCCCCGCCCGCAGCC | ||
| Psuc2-1548-R | TCGAGGCTGCGGGCGGGGACTGATGCGGCGAGGAGAAGCAGGATTGGGTTCATG | ||
| Psuc2-1567-F | AATTCATGCAGTTGTCCTACCTTCTCTCGCTCGTCTCCCTTGTGGCCGCTC | ||
| Psuc2-1567-R | TCGAGAGCGGCCACAAGGGAGACGAGCGAGAGAAGGTAGGACAACTGCATG | ||
| Psuc2-1990-F | AATTCATGCGCCCAGCAACCGTCATCTTCGCACTGGCCGTCTCCGTCAAGGCTC | ||
| Psuc2-1990-R | TCGAGAGCCTTGACGGAGACGGCCAGTGCGAAGATGACGGTTGCTGGGCGCATG | ||
| Psuc2-3667-F | AATTCATGAAGTACACTGCTGCTCTCGTCGCCCTGGCTGCTGCCGTCATGGCCC | ||
| Psuc2-3667-R | TCGAGGGCCATGACGGCAGCAGCCAGGGCGACGAGAGCAGCAGTGTACTTCATG | ||
| Psuc2-4213-F | AATTCATGATTCTGCTGCTCCTCTTGCTATGCACGACAACATGTTTAGGAC | ||
| Psuc2-4213-R | TCGAGTCCTAAACATGTTGTCGTGCATAGCAAGAGGAGCAGCAGAATCATG | ||
| Psuc2-4989-F | AATTCATGCACCAGCTCCTTGCACTACTCCTCGCCAGCCTCACCGCTGCCC | ||
| Psuc2-4989-R | TCGAGGGCAGCGGTGAGGCTGGCGAGGAGTAGTGCAAGGAGCTGGTGCATG | ||
| Psuc2-5215-F | AATTCATGAAGTTTATTTCCCTCGTAGCCGCGGCCGGTGTGGCGTATGCCC | ||
| Psuc2-5215-R | TCGAGGGCATACGCCACACCGGCCGCGGCTACGAGGGAAATAAACTTCATG | ||
| Psuc2-6754-F | AATTCATGGCTGCAGCATCAGCCCTTTTCTTCCTCACGGCGGCTGTCTCCTGGACAC | ||
| Psuc2-6754-R | TCGAGTGTCCAGGAGACAGCCGCCGTGAGGAAGAAAAGGGCTGATGCTGCAGCCATG | ||
| Psuc2-8184-F | AATTCATGCAAATCGTCCTCCCTCTCCTGGCCTGCGCAGCCGTGGCTGCCGCCC | ||
| Psuc2-8184-R | TCGAGGGCGGCAGCCACGGCTGCGCAGGCCAGGAGAGGGAGGACGATTTGCATG | ||
| P1300-44-F | CATGAAGGCCTCGACGCTGCTGGCTCTCCTGCCCTTTGCCGCGGCCGGTAC | 烟草瞬时表达鉴定信号肽功能 Functional validation of the signal peptides using N. benthamiana | |
| P1300-44-R | CGGCCGCGGCAAAGGGCAGGAGAGCCAGCAGCGTCGAGGCCTTCATGGTAC | ||
| P1300-1548-F | CATGAACCCAATCCTGCTTCTCCTCGCCGCATCAGTCCCCGCCCGCAGCGGTAC | ||
| P1300-1548-R | CGCTGCGGGCGGGGACTGATGCGGCGAGGAGAAGCAGGATTGGGTTCATGGTAC | ||
| P1300-1567-F | CATGCAGTTGTCCTACCTTCTCTCGCTCGTCTCCCTTGTGGCCGCTGGTAC | ||
| P1300-1567-R | CAGCGGCCACAAGGGAGACGAGCGAGAGAAGGTAGGACAACTGCATGGTAC | ||
| P1300-1990-F | CATGCGCCCAGCAACCGTCATCTTCGCACTGGCCGTCTCCGTCAAGGCTGGTAC | ||
| P1300-1990-R | CAGCCTTGACGGAGACGGCCAGTGCGAAGATGACGGTTGCTGGGCGCATGGTAC | ||
| P1300-3667-F | CATGAAGTACACTGCTGCTCTCGTCGCCCTGGCTGCTGCCGTCATGGCCGGTAC | ||
| P1300-3667-R | CGGCCATGACGGCAGCAGCCAGGGCGACGAGAGCAGCAGTGTACTTCATGGTAC | ||
| P1300-4213-F | CATGATTCTGCTGCTCCTCTTGCTATGCACGACAACATGTTTAGGAGGTAC | ||
| P1300-4213-R | CTCCTAAACATGTTGTCGTGCATAGCAAGAGGAGCAGCAGAATCATGGTAC | ||
| P1300-4989-F | CATGCACCAGCTCCTTGCACTACTCCTCGCCAGCCTCACCGCTGCCGGTAC | ||
| P1300-4989-R | CGGCAGCGGTGAGGCTGGCGAGGAGTAGTGCAAGGAGCTGGTGCATGGTAC | ||
| P1300-5215-F | CATGAAGTTTATTTCCCTCGTAGCCGCGGCCGGTGTGGCGTATGCCGGTAC | ||
| P1300-5215-R | CGGCATACGCCACACCGGCCGCGGCTACGAGGGAAATAAACTTCATGGTAC | ||
| P1300-6754-F | CATGGCTGCAGCATCAGCCCTTTTCTTCCTCACGGCGGCTGTCTCCTGGACAGGTAC | ||
| P1300-6754-R | CTGTCCAGGAGACAGCCGCCGTGAGGAAGAAAAGGGCTGATGCTGCAGCCATGGTAC | ||
| P1300-8184-F | CATGCAAATCGTCCTCCCTCTCCTGGCCTGCGCAGCCGTGGCTGCCGCCGGTAC | ||
| P1300-8184-R | CGGCGGCAGCCACGGCTGCGCAGGCCAGGAGAGGGAGGACGATTTGCATGGTAC | ||
| Uv_Tublin2α-2F/R | ACCAGCTCGTTGAGAACTCG / AATCAGAGTTGAGCTGGCCG | 实时荧光定量PCR RT-qPCR | |
| UV_44-RT-F/R | GCGTTGCCAAGAAGACCAAG / ATTCCAGCAATCACGGCAGA | ||
| UV_1548-RT- F/R | GACAACGGGCAGACAAACTG / CTAGCAATCCCAGCAGTCGT | ||
| UV_1567-RT-F/R | TGCTAGCGCTATCGTTCCTG / GTCGGCATGGTTTTGCAACT | ||
| UV_1990-RT-F/R | ACTCACTTCCAAACACGGCA / GCCTTGTCTTCTCAACACGC | ||
| UV_3667-RT-F/R | GCCTCCTCTGCAATGTGGAA / CTTGAGGCAGTTGAGTCCGA | ||
| UV_4213-RT-F/R | ATGCGAGAACTTGGTCGTGT / AACCAACGCCTCCAACTTGA | ||
| UV_4989-RT-F/R | CCTGCCTCCTTCGTCCAAAT / GGTCGATCTGGCAAAGTGGA | ||
| UV_5215-RT-F/R | TAGCGTACTGCGACGGTAAC / GAAACAGCGCCCAATTCCTG | ||
| UV_6754-RT-F/R | AAATGCGAGGACTAGGGCAC / CTTTTTGCCGGAGGAGGTCT | ||
| UV_8184-RT-F/R | CATCGCTGCGAAGACAAAGG / GCGCCTGTGAACTTTTCCTG | ||
| 基因名称 Gene name | 蛋白大小(氨基酸) Length of protein (aa) | 信号肽长度(氨基酸) Length of signal peptide (aa) | 信号肽评分 Signal peptide scoring(Sec/SPI) |
|---|---|---|---|
| UV_44 | 386 | 15 | 0.8501 |
| UV_1548 | 104 | 16 | 0.9929 |
| UV_1567 | 388 | 15 | 0.9722 |
| UV_1990 | 279 | 16 | 0.9845 |
| UV_3667 | 89 | 16 | 0.9995 |
| UV_4213 | 216 | 15 | 0.9642 |
| UV_4989 | 231 | 15 | 0.9924 |
| UV_5215 | 88 | 15 | 0.9981 |
| UV_6754 | 211 | 17 | 0.4707 |
| UV_8184 | 106 | 16 | 0.9982 |
表2 10个稻瘟病菌效应因子的信号肽预测
Table 2. Signal peptide prediction of ten effectors from U. virens
| 基因名称 Gene name | 蛋白大小(氨基酸) Length of protein (aa) | 信号肽长度(氨基酸) Length of signal peptide (aa) | 信号肽评分 Signal peptide scoring(Sec/SPI) |
|---|---|---|---|
| UV_44 | 386 | 15 | 0.8501 |
| UV_1548 | 104 | 16 | 0.9929 |
| UV_1567 | 388 | 15 | 0.9722 |
| UV_1990 | 279 | 16 | 0.9845 |
| UV_3667 | 89 | 16 | 0.9995 |
| UV_4213 | 216 | 15 | 0.9642 |
| UV_4989 | 231 | 15 | 0.9924 |
| UV_5215 | 88 | 15 | 0.9981 |
| UV_6754 | 211 | 17 | 0.4707 |
| UV_8184 | 106 | 16 | 0.9982 |
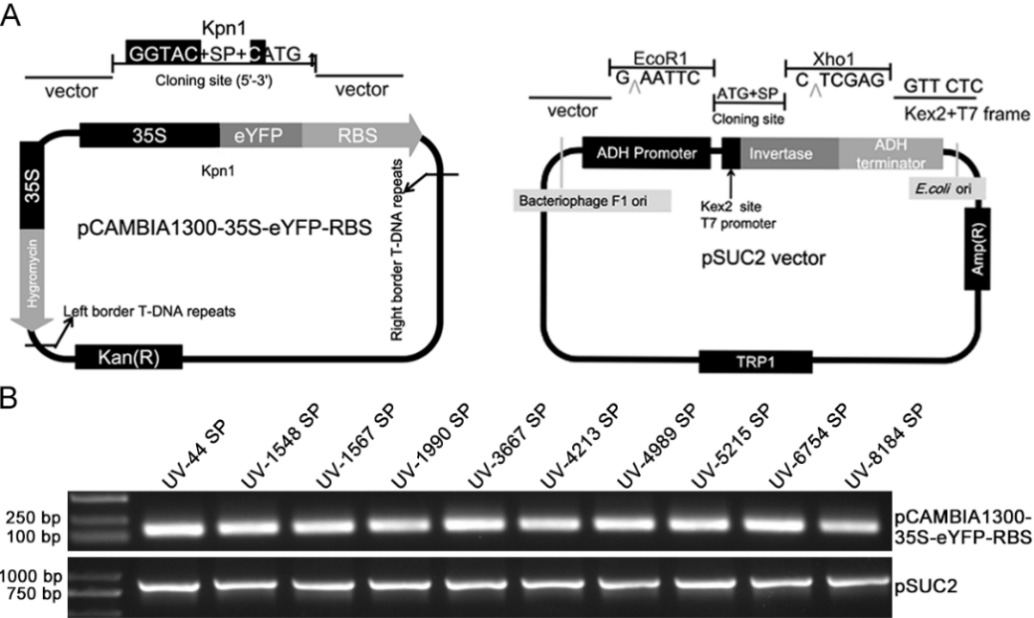
图1 载体的构建及阳性克隆的鉴定 A: pCAMBIA1300-35S-eYFP-RBS和pSUC2质粒示意图;B: PCR和琼脂糖凝胶电泳鉴定阳性克隆。
Fig. 1. Construction of vectors and identification of the positive clones A, Diagrams of the pCAMBIA1300-35S-eYFP-RBS and pSUC2 plasmids; B, Positive clones were identified by PCR and agarose gel electrophoresis.
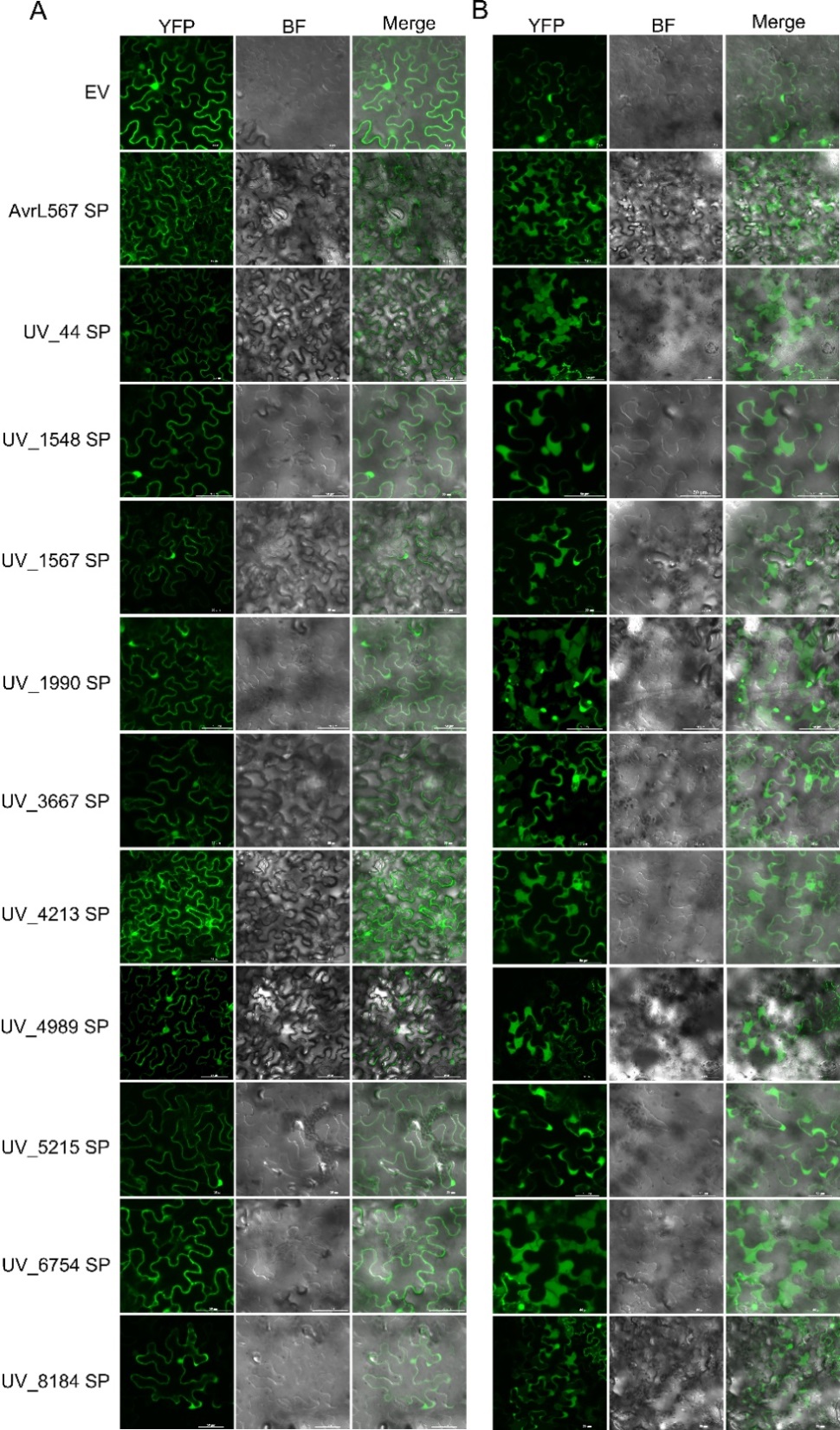
图2 烟草瞬时表达分析10个稻瘟病菌效应因子信号肽的亚细胞定位0.8 mol/L甘露醇诱导质壁分离前(A)和分离后(B)融合蛋白的亚细胞定位。注射农杆菌36 h后拍摄。EV、AvrL567SP分别为阴性和阳性对照。
Fig. 2. Subcellular localization of the transiently expressed signal peptides of ten effectors from U. virens in N. benthamiana Subcellular localization of the fusion proteins containing the predicted signal peptide before (A) and after (B) plasmolysis induced by 0.8 mol/L mannitol. The images were captured at 36 h after infiltration. EV was used as a negative control, and AvrL567SP was used as a positive control.

图3 利用酵母转化酶实验验证信号肽的分泌功能 A: 样品位置示意图;B, C, D: 酵母信号肽筛选实验验证信号肽的分泌功能;E: TTC显色实验验证信号肽的分泌功能。pSUC2-Avr1b为阳性对照,pSUC2-Mg87和YTK12为阴性对照。
Fig. 3. Functional evaluation of the signal peptides using the yeast invertase assay A, Diagrams showing the sample location on Petri plates; B, C and D, Detection of the secretion of signal peptides by the yeast signal trap assay; E, TTC assay of the signal peptides. pSUC2-Avr1b was used as a positive control. YTK12 and pSUC2-Mg87 were used as the negative controls.
| [1] | Brooks S A, Anders M M, Yeater K M. Effect of cultural management practices on the severity of false smut and kernel smut of rice[J]. Plant Disease, 2009, 93(11): 1202-1208. |
| [2] | Tang Y X, Jin J, Hu D W, Yong M L, Xu Y, He L P. Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets[J]. Plant Pathology, 2013, 62(1): 1-8. |
| [3] | Cooke M C. Some extra-european fungi[J]. Grevillea, 1878, 7: 13-15. |
| [4] | Sun W, Fan J, Fang A, Li Y, Tariqjaveed M, Li D, Hu D, Wang W M. Ustilaginoidea virens: Insights into an emerging rice pathogen[J]. Annual Review of Phytopathology, 2020, 58(1): 363-385. |
| [5] | Fan J, Liu J, Gong Z Y, Xu P Z, Hu X H, Wu J L, Li G B, Yang J, Wang Y Q, Zhou Y F, Li S C, Wang L, Chen X Q. The false smut pathogen Ustilaginoidea virens requires rice stamens for false smut ball formation[J]. Environmental Microbiology, 2020, 22(2): 646-659. |
| [6] | Fan J, Guo X Y, Li L, Huang F, Sun W X, Li Y, Huang Y Y, Xu Y J, Shi J, Zheng A P, Wang W M. Infection of Ustilaginoidea virens intercepts rice seed formation but activates grain-filling-related genes[J]. Journal of Integrative Plant Biology, 2015, 57(6): 577-590. |
| [7] | Fan J, Yang J, Wang Y Q, Li G B, Li Y, Wang W M. Current understanding on Villosiclava virens, a unique flower-infecting fungus causing rice false smut disease[J]. Molecular Plant Pathology, 2016, 17(9): 1321-1330. |
| [8] | Wang B, Liu L, Li Y, Zou J, Li D, Zhao D, Li W, Sun W. Ustilaginoidin D induces hepatotoxicity and behaviour aberrations in zebrafish larvae[J]. Toxicology, 2021, 456: 152786. |
| [9] | Lin X, Bian Y, Mou R, Cao Z, Cao Z, Zhu Z, Chen M. Isolation, identification, and characterization of Ustilaginoidea virens from rice false smut balls with high ustilotoxin production potential[J]. Journal of Basic Microbiology, 2018, 58(8): 670-678. |
| [10] | Rafiqi M, Ellis J G, Ludowici V A, Hardham A R, Dodds P N. Challenges and progress towards understanding the role of effectors in plant-fungal interactions[J]. Current Opinion in Plant Biology, 2012, 15(4): 477-482. |
| [11] | Zhang K, Zhao Z, Zhang Z, Li Y, Li S, Yao N, Hsiang T. Insights into genomic evolution from chromosomal and mitochondrial genomes of Ustilaginoidea virens[J]. Phytopathology Research, 2021, 3(1): 9. |
| [12] | Zhang N, Yang J, Fang A, Wang J, Li D, Li Y, Wang S, Cui F, Yu J, Liu Y, Peng Y L, Sun W. The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region[J]. Molecular Plant Pathology, 2020, 21(4): 445-459. |
| [13] | Zheng X, Fang A, Qiu S, Zhao G, Wang J, Wang S, Wei J, Gao H, Yang J, Mou B, Cui F, Zhang J, Liu J, Sun W. Ustilaginoidea virens secretes a family of phosphatases that stabilize the negative immune regulator OsMPK6 and suppress plant immunity[J]. The Plant Cell, 2022, 34(8): 3088-3109. |
| [14] | Chen X, Duan Y, Qiao F, Liu H, Huang J, Luo C, Chen X, Li G, Xie K, Hsiang T, Zheng L. A secreted fungal effector suppresses rice immunity through host histone hypoacetylation[J]. New Phytologist, 2022, 235(5): 1977. |
| [15] | Li G B, He J X, Wu J L, Wang H, Zhang X, Liu J, Hu X H, Zhu Y, Shen S, Bai Y F, Wang W M, Fan J. Overproduction of OsRACK1A, an effector-targeted scaffold protein promoting OsRBOHB-mediated ROS production, confers rice floral resistance to false smut disease without yield penalty[J]. Molecular Plant, 2022, 15(11): 1790-1806. |
| [16] | Chen X, Li X, Duan Y, Pei Z, Liu H, Yin W, Huang J, Luo C, Chen X, Li G, Xie K, Hsiang T, Zheng L. A secreted fungal subtilase interferes with rice immunity via degradation of SUPPRESSOR OF G2 ALLELE OF skp1[J]. Plant Physiology, 2022, 190(2): 1474-1489. |
| [17] | Owji H, Nezafat N, Negahdaripour M, Hajiebrahimi A, Ghasemi Y. A comprehensive review of signal peptides: Structure, roles, and applications[J]. European Journal of Cell Biology, 2018, 97(6): 422-441. |
| [18] | von Heijne G. Signal sequences: Limits of variation[J]. Journal of Molecular Biology, 1985, 184(1): 99-105. |
| [19] | Rafiqi M, Gan P H P, Ravensdale M, Lawrence G J, Ellis J G, Jones D A, Hardham A R, Dodds P N. Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen[J]. The Plant Cell, 2010, 22(6): 2017-2032. |
| [20] | Yin W, Wang Y, Chen T, Lin Y, Luo C. Functional evaluation of the signal peptides of secreted proteins[J]. Bio-protocol, 2018, 8(9): e2839. |
| [21] | Gu B, Kale S D, Wang Q, Wang D, Pan Q, Cao H, Meng Y, Kang Z, Tyler B M, Shan W. Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells[J]. PLOS ONE, 2011, 6(11): e27217. |
| [22] | Bao J, Wang R, Gao S, Wang Z, Fang Y, Wu L, Wang M. High-Quality genome sequence resource of a rice false smut fungus Ustilaginoidea virens isolate, UV-FJ-1[J]. Phytopathology, 2021, 111(10): 1889-1892. |
| [23] | Zhang Y, Zhang K, Fang A, Han Y, Yang J, Xue M, Bao J, Hu D, Zhou B, Sun X, Li S, Wen M, Yao N, Ma L J, Liu Y, Zhang M, Huang F, Luo C, Zhou L, Li J, Chen Z. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics[J]. Nature Communications, 2014, 5(1): 3849. |
| [24] | Wang H, Yang X, Wei S, Wang Y. Proteomic analysis of mycelial exudates of Ustilaginoidea virens[J]. Pathogens, 2021, 10(3): 364. |
| [25] | Fang A, Han Y, Zhang N, Zhang M, Liu L, Li S, Lu F, Sun W. Identification and characterization of plant cell death-inducing secreted proteins from Ustilaginoidea virens[J]. Molecular Plant-Microbe Interactions, 2016, 29(5): 405-416. |
| [26] | Wei S, Wang Y, Zhou J, Xiang S, Sun W, Peng X, Li J, Hai Y, Wang Y, Li S. The conserved effector UvHrip1 interacts with OsHGW and infection of Ustilaginoidea virens regulates defense- and heading date-related signaling pathway[J]. International Journal of Molecular Sciences, 2020, 21(9): 3376. |
| [27] | Qiu S, Fang A, Zheng X, Wang S, Wang J, Fan J, Sun Z, Gao H, Yang J, Zeng Q, Cui F, Wang W M, Chen J, Sun W. Ustilaginoidea virens nuclear effector SCRE4 suppresses rice immunity via inhibiting expression of a positive immune regulator OsARF17[J]. International Journal of Molecular Sciences, 2022, 23(18): 10527. |
| [28] | Fang A, Gao H, Zhang N, Zheng X, Qiu S, Li Y, Zhou S, Cui F, Sun W. A novel effector gene SCRE2 contributes to full virulence of Ustilaginoidea virens to rice[J]. Frontiers in Microbiology, 2019, 10. |
| [29] | Presti L L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R. Fungal Effectors and plant susceptibility[J]. Annual Review of Plant Biology, 2015, 66(1): 513-545. |
| [30] | Fan J, Du N, Li L, Li G B, Wang Y Q, Zhou Y F, Hu X H, Liu J, Zhao J Q, Li Y, Huang F, Wang W M. A core effector UV_1261 promotes Ustilaginoidea virens infection via spatiotemporally suppressing plant defense[J]. Phytopathology Research, 2019, 1(1): 11. |
| [1] | 汪邑晨, 朱本顺, 周磊, 朱骏, 杨仲南. 光/温敏核不育系的不育机理及两系杂交稻的发展与展望 [J]. 中国水稻科学, 2024, 38(5): 463-474. |
| [2] | 许用强, 徐军, 奉保华, 肖晶晶, 王丹英, 曾宇翔, 符冠富. 水稻花粉管生长及其对非生物逆境胁迫的响应机理研究进展 [J]. 中国水稻科学, 2024, 38(5): 495-506. |
| [3] | 何勇, 刘耀威, 熊翔, 祝丹晨, 王爱群, 马拉娜, 王廷宝, 张健, 李建雄, 田志宏. 利用CRISPR/Cas9技术编辑OsOFP30基因创制水稻粒型突变体 [J]. 中国水稻科学, 2024, 38(5): 507-515. |
| [4] | 吕阳, 刘聪聪, 杨龙波, 曹兴岚, 王月影, 童毅, Mohamed Hazman, 钱前, 商连光, 郭龙彪. 全基因组关联分析(GWAS)鉴定水稻氮素利用效率候选基因 [J]. 中国水稻科学, 2024, 38(5): 516-524. |
| [5] | 杨好, 黄衍焱, 王剑, 易春霖, 石军, 谭楮湉, 任文芮, 王文明. 水稻中八个稻瘟病抗性基因特异分子标记的开发及应用 [J]. 中国水稻科学, 2024, 38(5): 525-534. |
| [6] | 杨铭榆, 陈志诚, 潘美清, 张汴泓, 潘睿欣, 尤林东, 陈晓艳, 唐莉娜, 黄锦文. 烟-稻轮作下减氮配施生物炭对水稻茎鞘同化物转运和产量 形成的影响 [J]. 中国水稻科学, 2024, 38(5): 555-566. |
| [7] | 熊家欢, 张义凯, 向镜, 陈惠哲, 徐一成, 王亚梁, 王志刚, 姚坚, 张玉屏. 覆膜稻田施用炭基肥对水稻产量及氮素利用的影响 [J]. 中国水稻科学, 2024, 38(5): 567-576. |
| [8] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [9] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [10] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [11] | 伏荣桃, 陈诚, 王剑, 赵黎宇, 陈雪娟, 卢代华. 转录组和代谢组联合分析揭示稻曲病菌的致病因子[J]. 中国水稻科学, 2024, 38(4): 375-385. |
| [12] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [13] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [14] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [15] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||