
中国水稻科学 ›› 2022, Vol. 36 ›› Issue (4): 348-356.DOI: 10.16819/j.1001-7216.2022.210604
周永林1,2, 申小磊1,2, 周立帅1,2, 林巧霞1,2, 王朝露1,2, 陈静1,2, 冯慧捷1, 张振文1, 陈晓婷1,2,3,*( ), 鲁国东2
), 鲁国东2
收稿日期:2021-06-10
修回日期:2021-09-30
出版日期:2022-07-10
发布日期:2022-07-12
通讯作者:
陈晓婷
基金资助:
ZHOU Yonglin1,2, SHEN Xiaolei1,2, ZHOU Lishuai1,2, LIN Qiaoxia1,2, WANG Zhaolu1,2, CHEN Jing1,2, FENG Huijie1, ZHANG Zhenwen1, CHEN Xiaoting1,2,3,*( ), LU Guodong2
), LU Guodong2
Received:2021-06-10
Revised:2021-09-30
Online:2022-07-10
Published:2022-07-12
Contact:
CHEN Xiaoting
摘要:
【目的】由稻瘟病菌(Magnaporthe oryzae)引起的稻瘟病和由水稻黄单胞菌(Xanthomonas oryzae pv. oryzae,Xoo)引起的白叶枯病严重影响水稻的产量和品质。创制转OsLOX10基因水稻材料,进行稻瘟菌和白叶枯菌的抗病性分析,有助于揭示其调控水稻对稻瘟病和白叶枯病的抗性机制。【方法】采用CRISPR/Cas9系统构建OsLOX10的敲除载体,利用限制性内切酶XcmⅠ线性化pCXUN-HA,TA连接构建OsLOX10的过表达载体,遗传转化获得OsLOX10转基因水稻,筛选过表达株系和纯合敲除株系进行真菌和细菌的抗病性分析。在稻瘟菌(Guy11)侵染水稻后,对水杨酸(salicylic acid,SA)、茉莉酸(jasmonic acid,JA)途径的标志基因进行qRT-PCR分析;在几丁质(chitin)和flg22诱导下,观测水稻活性氧(reactive oxygen species,ROS)的暴发情况。【结果】 qRT-PCR分析表明,接种稻瘟菌和白叶枯菌24 h后,OsLOX10表达量上调;OsLOX10的纯合敲除和过表达水稻转基因株系接种稻瘟病菌Guy11孢子悬浮液,与野生型(日本晴)相比,OsLOX10敲除株系更易感病,过表达株系则无典型的病斑症状;接种 6、12、24和36 h时,3个病程相关蛋白基因OsPBZ1、OsPR1a、OsPR1b和SA通路基因OsPAL1,以及JA合成通路上的2个基因OsAOS2、OsLOX5的转录水平在敲除转基因株系中显著下调,而在过表达转基因株系中显著上调。对转OsLOX10基因水稻接种白叶枯菌(PXO99A),发现敲除OsLOX10的转基因水稻对白叶枯菌更易感病。qRT-PCR分析OsPR1b和OsPAL1以及JA合成通路上的3个基因OsAOS2、OsAOC和OsJAZ在OsLOX10过表达基因水稻中表达量明显上调,而在敲除OsLOX10的转基因水稻中却保持在较低水平,在接种7 d后表现出显著性差异。在几丁质和flg22诱导下,OsLOX10敲除株系的ROS水平显著性降低,而且在几丁质诱导下,ROS的起峰时间推迟。【结论】稻瘟病菌和白叶枯病菌能够诱导OsLOX10的表达,OsLOX10通过病原菌分子模式触发的免疫途径(PTI)参与抗病反应,其在水稻抵御稻瘟病和白叶枯病中起着正调控作用。同时,OsLOX10可能通过调节SA和JA介导的信号通路来正调控水稻对稻瘟病和白叶枯病的抗性。
周永林, 申小磊, 周立帅, 林巧霞, 王朝露, 陈静, 冯慧捷, 张振文, 陈晓婷, 鲁国东. OsLOX10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2022, 36(4): 348-356.
ZHOU Yonglin, SHEN Xiaolei, ZHOU Lishuai, LIN Qiaoxia, WANG Zhaolu, CHEN Jing, FENG Huijie, ZHANG Zhenwen, CHEN Xiaoting, LU Guodong. OsLOX10 Positively Regulates Defense Responses of Rice to Rice Blast and Bacterial Blight[J]. Chinese Journal OF Rice Science, 2022, 36(4): 348-356.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Purpose | 片段长度Length/bp |
|---|---|---|---|
| OsLOX10-F | ATGCAGCAGCCGCAGGCGAG | 过表达载体构建 Construction of OE vector | 2607 |
| OsLOX10-R | TCAGATGGAAGCGCTGTTGG | ||
| UbiP-seq | TTTTAGCCCTGCCTTCATACGC | 过表达载体测序 Sequencing of OE vector | 2795 |
| NosR-seq | AGACCGGCAACAGGATTCAATC | ||
| OsLOX10-U3-F | ggcaGCTACACCCAACTCCCTACG | CRISPR/Cas9第1靶点验证 Verification of first-target site of CRISPR/Cas9 | 259 |
| OsLOX10-U3-R | aaacCGTAGGGAGTTGGGTGTAGC | ||
| OsLOX10-U6a-F | gccgCGCCAGAGTCTGATCAACGC | CRISPR/Cas9第2靶点验证 Verification of second-target site of CRISPR/Cas9 | 329 |
| OsLOX10-U6a-R | aaacGCGTTGATCAGACTCTGGCG | ||
| U-F | CTCCGTTTTACCTGTGGAATCG | 敲除靶点第1轮扩增 First round amplification of knockout targets | 534 |
| gRNA-R | CGGAGGAAAATTCCATCCAC | ||
| Uctcg-B1' | TTCAGAggtctcTctcgCACTGGAATCGGCAGCAAAGG | CRISPR/Cas9第1靶点扩增 Amplification of first-target site | 564 |
| gRctga-B2 | AGCGTGggtctcGtcagGGTCCATCCACTCCAAGCTC | ||
| Uctga-B2' | TTCAGAggtctcTctgaCACTGGAATCGGCAGCAAAGG | CRISPR/Cas9第2靶点扩增 Amplification of second-target site target site | 629 |
| gRcggt-BL | AGCGTGggtctcGaccgGGTCCATCCACTCCAAGCTC | ||
| Actin-QF | GAGTATGATGAGTCGGGTCCAG | Actin定量PCR qRT-PCR of Actin | 143 |
| Actin-QR | ACACCAACAATCCCAAACAGAG | ||
| OsPR1a-QF | CGTCTTCATCACCTGCAACTACTC | OsPR1a定量PCR qRT-PCR of OsPR1a | 132 |
| OsPR1a-QR | CATGCATAAACACGTAGCATAGCA | ||
| OsPR1b-QF | ACGGGCGTACGTACTGGCTA | OsPR1b定量PCR qRT-PCR of OsPR1b | 105 |
| OsPR1b-QR | CTCGGTATGGACCGTGAAG | ||
| OsAOS2-QF | CAATACGTGTACTGGTCGAATGG | OsAOS2定量PCR qRT-PCR of OsAOS2 | 134 |
| OsAOS2-QR | AAGGTGTCGTACCGGAGGAA | ||
| OsPAL1-QF | AGGAGCTCGGCTGCGTATT | OsPAL1定量PCR qRT-PCR of OsPAL1 | 79 |
| OsPAL1-QR | ATGCCGAGGAACACCTTGTT | ||
| OsLOX5-QF | CTGATGAGGAGTTTGCACGA | OsLOX5定量PCR qRT-PCR of OsLOX5 | 542 |
| OsLOX5-QR | TCGTCCTTCAGGAGCAGAAT | ||
| OsAOC-QF | CCAAGGTGCAGGAGATGTT | OsAOC定量PCR qRT-PCR of OsAOC | 150 |
| OsAOC-QR | TACAGCTTGTTGGTGAAGGG | ||
| OsJAZ-QF | GAAGGCTCAACAGCTGACCAT | OsJAZ定量PCR qRT-PCR of OsJAZ | 69 |
| OsJAZ-QR | TTGGTGGACGGGAAGTTCTC | ||
| OsUG-F | TTCTGGTCCTTCCACTTTCAG | 泛素融合蛋白定量PCR qRT-PCR of ubiquitin fusion protein OsUG | 92 |
| OsUG-R | ACGATTGATTTAACCAGTCCATGA | ||
| MoPot2-F | ACGACCCGTCTTTACTTATTTGG | MoPot2定量PCR qRT-PCR of MoPot2 | 99 |
| MoPot2-R | AAGTAGCGTTGGTTTTGTTGGAT |
表1 本研究所使用引物
Table 1. Primers used in this study.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Purpose | 片段长度Length/bp |
|---|---|---|---|
| OsLOX10-F | ATGCAGCAGCCGCAGGCGAG | 过表达载体构建 Construction of OE vector | 2607 |
| OsLOX10-R | TCAGATGGAAGCGCTGTTGG | ||
| UbiP-seq | TTTTAGCCCTGCCTTCATACGC | 过表达载体测序 Sequencing of OE vector | 2795 |
| NosR-seq | AGACCGGCAACAGGATTCAATC | ||
| OsLOX10-U3-F | ggcaGCTACACCCAACTCCCTACG | CRISPR/Cas9第1靶点验证 Verification of first-target site of CRISPR/Cas9 | 259 |
| OsLOX10-U3-R | aaacCGTAGGGAGTTGGGTGTAGC | ||
| OsLOX10-U6a-F | gccgCGCCAGAGTCTGATCAACGC | CRISPR/Cas9第2靶点验证 Verification of second-target site of CRISPR/Cas9 | 329 |
| OsLOX10-U6a-R | aaacGCGTTGATCAGACTCTGGCG | ||
| U-F | CTCCGTTTTACCTGTGGAATCG | 敲除靶点第1轮扩增 First round amplification of knockout targets | 534 |
| gRNA-R | CGGAGGAAAATTCCATCCAC | ||
| Uctcg-B1' | TTCAGAggtctcTctcgCACTGGAATCGGCAGCAAAGG | CRISPR/Cas9第1靶点扩增 Amplification of first-target site | 564 |
| gRctga-B2 | AGCGTGggtctcGtcagGGTCCATCCACTCCAAGCTC | ||
| Uctga-B2' | TTCAGAggtctcTctgaCACTGGAATCGGCAGCAAAGG | CRISPR/Cas9第2靶点扩增 Amplification of second-target site target site | 629 |
| gRcggt-BL | AGCGTGggtctcGaccgGGTCCATCCACTCCAAGCTC | ||
| Actin-QF | GAGTATGATGAGTCGGGTCCAG | Actin定量PCR qRT-PCR of Actin | 143 |
| Actin-QR | ACACCAACAATCCCAAACAGAG | ||
| OsPR1a-QF | CGTCTTCATCACCTGCAACTACTC | OsPR1a定量PCR qRT-PCR of OsPR1a | 132 |
| OsPR1a-QR | CATGCATAAACACGTAGCATAGCA | ||
| OsPR1b-QF | ACGGGCGTACGTACTGGCTA | OsPR1b定量PCR qRT-PCR of OsPR1b | 105 |
| OsPR1b-QR | CTCGGTATGGACCGTGAAG | ||
| OsAOS2-QF | CAATACGTGTACTGGTCGAATGG | OsAOS2定量PCR qRT-PCR of OsAOS2 | 134 |
| OsAOS2-QR | AAGGTGTCGTACCGGAGGAA | ||
| OsPAL1-QF | AGGAGCTCGGCTGCGTATT | OsPAL1定量PCR qRT-PCR of OsPAL1 | 79 |
| OsPAL1-QR | ATGCCGAGGAACACCTTGTT | ||
| OsLOX5-QF | CTGATGAGGAGTTTGCACGA | OsLOX5定量PCR qRT-PCR of OsLOX5 | 542 |
| OsLOX5-QR | TCGTCCTTCAGGAGCAGAAT | ||
| OsAOC-QF | CCAAGGTGCAGGAGATGTT | OsAOC定量PCR qRT-PCR of OsAOC | 150 |
| OsAOC-QR | TACAGCTTGTTGGTGAAGGG | ||
| OsJAZ-QF | GAAGGCTCAACAGCTGACCAT | OsJAZ定量PCR qRT-PCR of OsJAZ | 69 |
| OsJAZ-QR | TTGGTGGACGGGAAGTTCTC | ||
| OsUG-F | TTCTGGTCCTTCCACTTTCAG | 泛素融合蛋白定量PCR qRT-PCR of ubiquitin fusion protein OsUG | 92 |
| OsUG-R | ACGATTGATTTAACCAGTCCATGA | ||
| MoPot2-F | ACGACCCGTCTTTACTTATTTGG | MoPot2定量PCR qRT-PCR of MoPot2 | 99 |
| MoPot2-R | AAGTAGCGTTGGTTTTGTTGGAT |
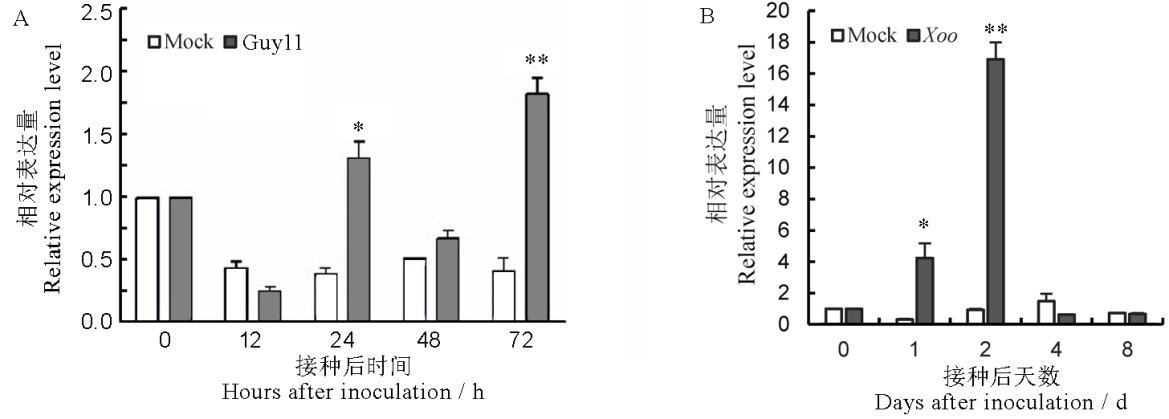
图1 OsLOX10在水稻接种稻瘟菌(A)和白叶枯菌(B)后表达量的动态变化 图中数据为平均值±标准误,*、**分别代表处理间在P<0.05和P<0.01水平差异达显著水平(t-检验)。
Fig. 1. Expression profiles of OsLOX10 after inoculation with M. oryzae (A) and Xoo (B) in rice. Data in the figure are Mean±SE, and *, ** represent significant difference at the 0.05 and 0.01 levels (t-test), respectively.

图2 OsLOX3编辑纯合突变体的鉴定 A-OsLOX10结构和编辑位点;B-OsLOX10 T0代18号植株的突变位点检测;C-OsLOX10 T0代24号植株的突变位点检测。
Fig. 2. Identification of homozygous mutant of OsLOX10 by gene editing. A, Schematic presentation of the OsLOX10 structure and gene editing site; B, Mutation site in OsLOX10 gene editing line-18; C, Mutation sites in OsLOX10 gene editing line-24.
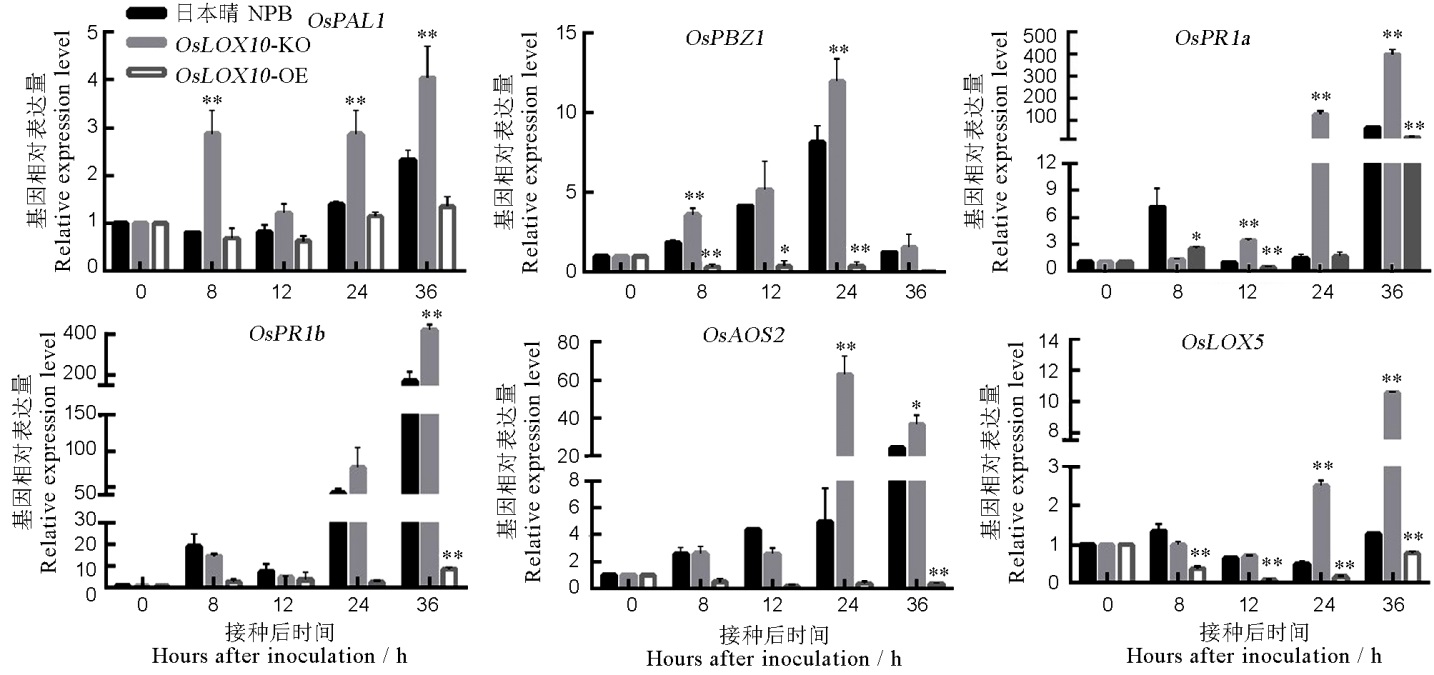
图6 OsLOX10转基因水稻相关抗病基因在稻瘟病菌侵染早期的qRT-PCR分析
Fig. 6. qRT-PCR analysis of disease resistance-related genes in OsLOX10 transgenic lines during the early infection of rice blast fungus.
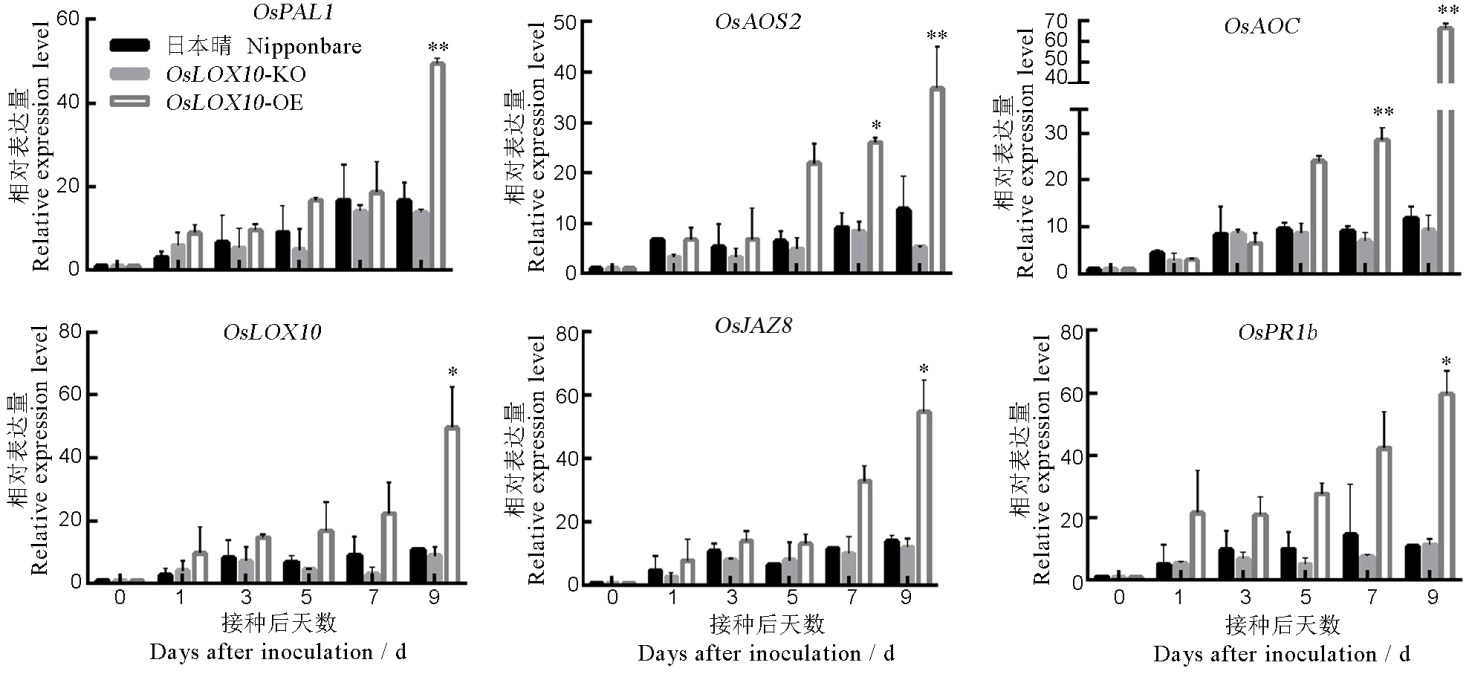
图7 OsLOX10转基因水稻相关抗病基因在白叶枯病菌侵染早期的qRT-PCR分析
Fig. 7. qRT-PCR analysis of disease resistance-related genes in OsLOX10 transgenic lines during the early infection of bacterial blight.

图8 转基因水稻OsLOX10-KO对几丁质和flg22诱导的响应 A-几丁质诱导的转基因水稻OsLOX10-KO和日本晴中活性氧的暴发情况,水稻叶片用400 nmol/L的几丁质和水处理;B-flg22诱导的转基因水稻OsLOX10-KO和日本晴中活性氧的暴发情况,水稻叶片用500 nmol/L的flg22和水处理。
Fig. 8. Response of OsLOX10-KO transgenic rice and wild-type Nipponbare to flg22 and chitin. A, Chitin-induced ROS burst in the transgenic rice OsLOX10-KO and Nipponbare (rice leaf disks were exposed to 500 nmol/L chitin and water); B, flg22-induced ROS burst in the transgenic rice OsLOX10-KO and NPB (rice leaf disks were exposed to 400 nmol/L flg22 and water).
| [1] | Khush G S. What it will take to feed 5.0 billion rice consumers in 2030[J]. Plant Molecular Biology, 2005, 59(1): 1-6. |
| [2] | Ke Y, Deng H, Wang S. Advances in understanding broad-spectrum resistance to pathogens in rice[J]. The Plant Journal, 2017, 90: 738-748. |
| [3] | Liu W, Wang G L. Plant innate immunity in rice: A defense against pathogen infection[J]. National Science Review, 2016, 3: 295-308 |
| [4] | Ngou B P M, Ahn H K, Ding P, Jones J D G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors[J]. Nature, 2021: 1-6. |
| [5] | Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou J M, He S Y, Xin X F. Pattern-recognition receptors are required for NLR-mediated plant immunity[J]. Nature, 2021: 1-5. |
| [6] | Umate P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice[J]. Plant Signaling & Behavior, 2011, 6(3): 335-338. |
| [7] | Marla S S, Singh V K. LOX genes in blast fungus (Magnaporthe grisea) resistance in rice[J]. Functional & Integrative Genomics, 2012, 12(2): 265-275. |
| [8] | Mizuno K, Iida T, Takano A, Yokoyama M, Fujimura T. A new 9-lipoxygenase cDNA from developing rice seeds[J]. Plant Cell Physiology, 2003, 44: 1168-1175. |
| [9] | Shirasawa K, Takeuchi Y, Ebitani T, Suzuki Y. Identification of gene for rice (Oryza sativa) seed lipoxygenase-3 involved in the generation of stale flavor and development of SNP markers for lipoxygenase-3 deficiency[J]. Breeding Science, 2008, 58: 169-176. |
| [10] | Gayen D, Ali N, Ganguly M, Paul S, Datta S K. RNAi mediated silencing of lipoxygenase gene to maintain rice grain quality and viability during storage[J]. Plant Cell Tissue and Organ Culture, 2014, 118(2): 229-243. |
| [11] | Gayen D, Ali N, Sarkar S N, Datta S K, Datta K. Down-regulation of lipoxygenase gene reduces degradation of carotenoids of golden rice during storage[J]. Planta, 2014, 242(1): 353-363. |
| [12] | Ma L, Zhu F, Li Z, Zhang J, Li X, Dong J, Wang T. TALEN-based mutagenesis of lipoxygenase LOX3 enhances the storage tolerance of rice (Oryza sativa) seeds[J]. PLoS ONE, 2015, 10(12): e0143877. |
| [13] | Xu H, Wei Y, Zhu Y, Lian L, Xie H, Cai Q, Chen Q, Lin Z, Xie H, Zhang J. Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity[J]. Plant Biotechnology Journal, 2015, 13: 526-539. |
| [14] | Huang J, Cai M, Long Q, Liu L, Lin Q, Jiang L, Chen S, Wan J. OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity[J]. Transgenic Research, 2014, 23(4): 643-655 |
| [15] | Ohta H, Shida K, Peng Y L, Furusawa I, Aibara S, Morita Y. A lipoxygenase pathway is activated in rice after infection with the rice blast fungus Magnaporthe grisea[J]. Plant Physiology, 1991, 97: 94-98. |
| [16] | Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu Y G. A robust CRISPR/Cas9 system for convenient, high- efficiency multiplex genome editing in monocot and dicot plants[J]. Molecular Plant, 2015, 8(8): 1274-1284. |
| [17] | Chen S, Songkumarn P, Liu J, Wang G L. A versatile zero background T-vector system for gene cloning and functional genomics[J]. Plant Physiology, 2009, 150(3): 1111-1121. |
| [18] | Hong Y, Liu Q, Cao Y, Zhang Y, Cheng S, Cao L. The OsMPK15 negatively regulates Magnaporthe oryza and Xoo disease resistance via SA and JA signaling pathway in rice[J]. Frontiers in Plant Science, 2019, 10: 752 |
| [19] | Wang G, Ding X, Yuan M, Qiu D, Li X, Xu C, Wang S. Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation[J]. Plant Molecular Biology, 2006, 60: 437-449. |
| [20] | Qiu D, Xiao J, Ding X, Xiong M, Cai M, Li X, Xu C, Wang S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling[J]. Molecular Plant- Microbe Interactions, 2007, 20(5): 492-499. |
| [21] | Qiu D, Xiao J, Xie W, Liu H, Li X, Wang S. rice gene network inferred from expression profiling of plants overexpressing oswrky13, a positive regulator of disease resistance[J]. Molecular Plant, 2008, 1(3): 538-551. |
| [22] | Tao Z, Liu H, Qiu D, Zhou Y, Xu C, Wang S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions[J]. Plant Physiology, 2009, 151: 936-948. |
| [23] | Harkenrider M, Sharma R, de Vleesschauwer D, Tsao L, Zhang X, Chern M, Canlas P, Zuo S, Ronald P C. Overexpression of rice wall-associated kinase25 (OsWAK25) alters resistance to bacterial and fungal pathogens[J]. PLoS ONE, 2016, 11: e0147310. |
| [24] | Tonnessen B W, Manosalva P, Lang J M, Baraoidan M, Bordeos A, Mauleon R, Oard J, Leung H, Leach J E. Rice phenylalanine ammonialyase gene OsPAL4 is associated with broad spectrum disease resistance[J]. Plant Molecular Biology, 2015, 87: 273-286. |
| [25] | Dubouzet J G, Maeda S, Sugano S, Ohtake M, Hayashi N, Ichikawa T, Kondou Y, Horii Y, Matsui M, Hirochika H, Takatsuji H, Mori M. Screening for resistance against Pseudomonas syringae in rice-foxarabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice[J]. Plant Biotechnology Journal, 2010, 9: 466-485. |
| [26] | Zhao X, Zhang T, Feng H, Qiu T, Zhao W. OsNBL1, a multi-organelle localized protein, plays essential roles in rice senescence, disease resistance, and salt tolerance[J]. Rice, 2021, 14(1): 10. |
| [27] | Narayanan S P, Lung S C, Liao P, Lo C, Chye M L. The Overexpression of Osacbp5 protects transgenic rice against necrotrophic, hemibiotrophic and biotrophic pathogens[J]. Scientific Reports, 2020, 10(1): 14918. |
| [28] | Han B, Zhou Y, Zhou Z H, Sun B, Zhou F, Yin C X, Ma W H, Chen H, Lin Y J. Repressed OSMESL expression-triggers reactive oxygen species mediated broad-spectrum disease resistance in rice[J]. Plant Biotechnology Journal, 2021, 19(8): 1511-1522 |
| [29] | Mei C, Qi M, Sheng G, Yang Y Y. inducible overexpression of a rice allele oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection[J]. Molecular Plant Microbe Interactions, 2006, 19(10): 1127-1137. |
| [30] | Nalam V J, Alam S, Keereetaweep J, Shah J. Facilitation of Fusarium graminearum infection by 9-lipoxygenases in Arabidopsis and wheat[J]. Molecular Plant Microbe Interactions, 2015, 28(10): 1142-1152 |
| [31] | Christensen S A, Nemchenko A, Borrego E, Murray I, Sobhy I S, Bosak L, Deblasio S, Vaughn K A, Herrfurth C, Tumlinson J, Nansen C, Meeley R, Kolomiets M V. The maize lipoxygenase, zmlox10, mediates green leaf volatile, jasmonate and-herbivore-induced plant volatile production for defense against insect attack[J]. Plant Journal, 2013, 74(1): 59-73. |
| [32] | Xing Q J, Liao J J, Cao S X, Li M, Lü T H, Qi H Y. Cmlox10 positively regulates drought tolerance through jasmonic acid-mediated stomatal closure in oriental melon (Cucumis melo var. Makuwa Makino)[J]. Scientific Reports, 2020, 10: 17452 |
| [33] | Torres M A, Jones J D, Dangl J L. Reactive oxygen species signaling in response to pathogens[J]. Plant Physiology, 2006, 141: 373-378. |
| [1] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [2] | 童琪, 王春燕, 阙亚伟, 肖宇, 王政逸. 稻瘟病菌热激蛋白(HSP)40编码基因MoMHF6的鉴定及功能研究[J]. 中国水稻科学, 2023, 37(6): 563-576. |
| [3] | 陈明亮, 熊文涛, 沈雨民, 熊焕金, 罗世友, 吴小燕, 胡兰香, 肖叶青. 广谱抗稻瘟病水稻保持系赣香B的抗性遗传解析[J]. 中国水稻科学, 2023, 37(5): 470-477. |
| [4] | 李刚, 高清松, 李伟, 张雯霞, 王健, 程保山, 王迪, 高浩, 徐卫军, 陈红旗, 纪剑辉. 定向敲除SD1基因提高水稻的抗倒性和稻瘟病抗性[J]. 中国水稻科学, 2023, 37(4): 359-367. |
| [5] | 王雨, 孙全翌, 杜海波, 许志文, 吴科霆, 尹力, 冯志明, 胡珂鸣, 陈宗祥, 左示敏. 利用抗稻瘟病基因Pigm和抗纹枯病数量性状基因qSB-9TQ、qSB-11HJX改良南粳9108的抗性[J]. 中国水稻科学, 2023, 37(2): 125-132. |
| [6] | 马静静, 潘妍妍, 杨孙玉悦, 王嘉琦, 蒋冬花. 硫藤黄链霉菌St-79对水稻白叶枯病的防效和促生作用[J]. 中国水稻科学, 2022, 36(6): 623-638. |
| [7] | 曹煜东, 肖湘谊, 叶乃忠, 丁晓雯, 易晓璇, 刘金灵, 肖应辉. 生长素调控因子OsGRF4协同调控水稻粒形和稻瘟病抗性[J]. 中国水稻科学, 2021, 35(6): 629-638. |
| [8] | 刘树芳, 董丽英, 李迅东, 周伍民, 杨勤忠. 持有Pi9基因的水稻单基因系IRBL9-W对稻瘟病菌苗期和成株期抗性差异[J]. 中国水稻科学, 2021, 35(3): 303-310. |
| [9] | 周天顺, 余东, 刘玲, 欧阳宁, 袁贵龙, 段美娟, 袁定阳. 利用CRISPR/Cas9技术编辑AFP1基因提高水稻耐逆性[J]. 中国水稻科学, 2021, 35(1): 11-18. |
| [10] | 朱华珺, 周瑚, 任佐华, 刘二明. 枯草芽孢杆菌JN005胞外抗菌物质及对水稻叶瘟防治效果[J]. 中国水稻科学, 2020, 34(5): 470-478. |
| [11] | 李丽, 莫旭艳, 李甜甜, 张丽媛, 董汉松. 白叶枯病菌效应子XopN在拥有OsSWEET11同源基因的水稻品种上发挥毒性作用[J]. 中国水稻科学, 2020, 34(4): 368-382. |
| [12] | 许赵蒙, 李利华, 高晓庆, 袁正杰, 李莘, 田旭丹, 王岚岚, 瞿绍洪. 转Pi9抗稻瘟病基因水稻株系的比较转录组分析[J]. 中国水稻科学, 2020, 34(3): 245-255. |
| [13] | 孟峰, 张亚玲, 靳学慧. 黑龙江省稻瘟病菌无毒基因AVR-Pita及其同源基因的检测与分析[J]. 中国水稻科学, 2020, 34(2): 143-149. |
| [14] | 李金璐, 张慧, 焦泽宇, 刘剑宇, 韩光煜, 卓晓轩, 罗琼. 水稻子预44和江南香糯基因组比较鉴定稻瘟病抗性相关基因[J]. 中国水稻科学, 2020, 34(1): 8-16. |
| [15] | 陈涛, 孙旭超, 张善磊, 梁文化, 周丽慧, 赵庆勇, 姚姝, 赵凌, 赵春芳, 朱镇, 张亚东, 王才林. 稻瘟病广谱抗性基因Pigm特异性分子标记的开发和应用[J]. 中国水稻科学, 2020, 34(1): 28-36. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||