
中国水稻科学 ›› 2020, Vol. 34 ›› Issue (4): 368-382.DOI: 10.16819/j.1001-7216.2020.9108
• 研究报告 • 上一篇
李丽1, 莫旭艳2, 李甜甜1, 张丽媛1,3,*( ), 董汉松1,2,3
), 董汉松1,2,3
收稿日期:2019-10-08
修回日期:2020-03-06
出版日期:2020-07-10
发布日期:2020-07-10
通讯作者:
张丽媛
基金资助:
Li LI1, Xuyan MO2, Tiantian LI1, Liyuan ZHANG1,3,*( ), Hansong DONG1,2,3
), Hansong DONG1,2,3
Received:2019-10-08
Revised:2020-03-06
Online:2020-07-10
Published:2020-07-10
Contact:
Liyuan ZHANG
摘要:
【目的】黄单胞菌细胞外泌蛋白质(Xanthomonas outer proteins, Xops)是植物病原黄单胞菌高度保守的毒性效应子或毒性辅助组分,通过细菌Ⅲ型分泌系统分泌到细菌细胞外部,然后转入植物细胞而发挥病理作用。水稻黄单胞菌水稻致病变种即水稻白叶枯病菌的标准菌株PXO99A分泌的XopN是一种毒性效应子,通过影响寄主免疫反应而使水稻发病。但是,XopN对病菌毒性的影响是否因水稻品种的不同而异,还有待研究。【方法】利用同源双交换技术,先敲除了PXO99A的XopN基因,获得了∆XopN突变体,又经过遗传互补,得到了回补菌株∆XopN/XopN。通过营养肉汤液体培养,测定了XopN对病菌繁殖能力的影响;根据文献选用14个水稻品种,通过接种实验,测定了PXO99A对这些品种的毒性与XopN敲除或回补的影响;在XopN发挥毒性作用的水稻品种上,测定了隐性抗病基因OsSWEET11/xa13与显性感病基因OsSWEET11/Xa13受病菌侵染而表达的情况,分析了XopN敲除或回补的影响。【结果】在营养肉汤液体培养过程中,病菌突变体菌株∆XopN的繁殖速度明显低于野生型PXO99A。PXO99A、∆XopN和∆XopN/XopN接种水稻后,根据其在水稻叶片组织内的繁殖量及随后产生的白叶枯病症状的严重程度,将供试的14个水稻品种分为两种情况。一是感病程度与病菌XopN是否敲除或回补无关,这有10个水稻品种(IRBB1、IRBB3、IRBB8、IRBB10、IRBB14、IR24、IRBB203、IRBB204、IRBB205和IRBB211),PXO99A、∆XopN和∆XopN/XopN对它们的毒性无明显差别。二是XopN对病菌毒性发挥作用的水稻品种,包括高度感病的3个品种(IRBB208、Asominori和日本晴)和低感品种IRBB13。与PXO99A或∆XopN/XopN相比,∆XopN对这4个水稻品种的毒性大为降低。在IRBB13上,病菌侵染对隐性抗病基因OsSWEET11/xa13在叶片内的表达发生抑制作用,这一效应与XopN的毒性功能相关。相反,日本晴显性感病基因OsSWEET11/Xa13却受病菌侵染的诱导,在叶片内的表达水平大幅度提高。IRBB208和Asominori携带OsSWEET11/Xa13同源基因,该同源基因在叶片内的表达水平也因病菌侵染而大幅提高。在这4个水稻品种上,PXO99A和∆XopN/XopN能够诱导OsSWEET11/Xa13或其同源基因表达,但∆XopN无此作用。另外,XopN对病菌在非寄主植物烟草上诱发过敏反应有量变贡献,相比PXO99A或∆XopN/XopN,∆XopN引起过敏反应的程度有所降低。【结论】XopN是一个有限广谱性效应子,在拥有OsSWEET11同源基因的水稻品种上发挥毒性作用。XopN也是病菌繁殖所需要的,对病菌在非寄主植物上诱导过敏反应有一定贡献。
中图分类号:
李丽, 莫旭艳, 李甜甜, 张丽媛, 董汉松. 白叶枯病菌效应子XopN在拥有OsSWEET11同源基因的水稻品种上发挥毒性作用[J]. 中国水稻科学, 2020, 34(4): 368-382.
Li LI, Xuyan MO, Tiantian LI, Liyuan ZHANG, Hansong DONG. Effector XopN of Xanthomonas oryzae pv. oryzae Plays a Virulent Role in Rice Varieties Harboring OsSWEET11 Homologs[J]. Chinese Journal OF Rice Science, 2020, 34(4): 368-382.
| 水稻品种 Rice variety | 抗病基因 Resistance gene | 基因产物 Encoding protein | 文献 Reference |
|---|---|---|---|
| IRBB1 | Xa1 | NLR (leucine-rich repeat receptor) | 31-33 |
| IRBB3 | Xa3 | LRR-RLK (leucine-rich repeat-receptor-like kinase) | 31-33 |
| IRBB8 | xa8 | Unknown | 31, 34 |
| IRBB10 | Xa10 | Executor | 31, 33 |
| IRBB13 | xa13 | OsSWEET11 (synonym Xa13) | 31, 33 |
| IRBB14 | Xa14 | Unknown | 31 |
| IRBB203 | Xa3 | LRR-RLK | 31, 33 |
| IRBB204 | Xa4 | RLK | 31, 33 |
| IRBB205 | Xa5 | TFIIA TF | 31, 33 |
| IRBB208 | Xa8 | Putative OsSWEET11 | 本研究This study |
| IRBB211 | Xa11 | RLK | AAY32487.1 |
| IR24 | Xa18 | RLK | AAY32494 |
| Asominori | Putative Xa13 | Putative OsSWEET11 | 本研究This study |
| 日本晴 Nipponbare | Xa13 (synonym Os8N3) | OsSWEET11 (synonym XA13) | 33-36 |
表1 供试水稻品种中白叶枯病的抗病基因及基因表达产物
Table 1 Xoo-relevant resistance gene and encoding proteins in the tested rice varieties.
| 水稻品种 Rice variety | 抗病基因 Resistance gene | 基因产物 Encoding protein | 文献 Reference |
|---|---|---|---|
| IRBB1 | Xa1 | NLR (leucine-rich repeat receptor) | 31-33 |
| IRBB3 | Xa3 | LRR-RLK (leucine-rich repeat-receptor-like kinase) | 31-33 |
| IRBB8 | xa8 | Unknown | 31, 34 |
| IRBB10 | Xa10 | Executor | 31, 33 |
| IRBB13 | xa13 | OsSWEET11 (synonym Xa13) | 31, 33 |
| IRBB14 | Xa14 | Unknown | 31 |
| IRBB203 | Xa3 | LRR-RLK | 31, 33 |
| IRBB204 | Xa4 | RLK | 31, 33 |
| IRBB205 | Xa5 | TFIIA TF | 31, 33 |
| IRBB208 | Xa8 | Putative OsSWEET11 | 本研究This study |
| IRBB211 | Xa11 | RLK | AAY32487.1 |
| IR24 | Xa18 | RLK | AAY32494 |
| Asominori | Putative Xa13 | Putative OsSWEET11 | 本研究This study |
| 日本晴 Nipponbare | Xa13 (synonym Os8N3) | OsSWEET11 (synonym XA13) | 33-36 |
| 菌株及质粒 Strains and plasmids | 相关性状 The study-relevant characteristics | 来源 Resource |
|---|---|---|
| 菌株Bacterial strains 黄单胞菌Xanthomonas oryzae pv. oryzae | ||
| PXO99A | PXO99的氮杂胞苷抗性克隆,也叫菲律宾小种6号 The azacytidine-resistant clone of PXO99, synonym Philippine race 6 | 本实验室This lab |
| ∆XopN | PXO99A XopN敲除突变体 PXO99A XopN-knockout unmarked mutant | 本研究This study |
| ∆XopN/XopN | 用质粒pHMXopN回补PXO99A XopN敲除突变体 PXO99A XopN mutant complemented with pHMXopN | 本研究This study |
| ∆XopQ | PXO99A XopQ敲除突变体 PXO99A XopQ-knockout unmarked mutant | 本实验室This lab |
| 质粒Plasmids | ||
| pMD19-Ts | 基因克隆T载体,含Apr, Mob+, Mob(P), LacZa+, Ampr Gene-cloning T-simple vector, Apr, Mob+, Mob(P), LacZa+, Ampr | TaKaRa |
| pMD20-T | 基因重组T载体,含Kanr, Ampr Gene-recombination T-simple vector, Kanr, Ampr | TaKaRa |
| pMD20-T::F1::Km::F2 | 重组的基因突变载体 Recombinant gene-mutagenicity vector | 本研究This study |
| pHMI | 多个pUC19重组的寄主更广的载体,含SpR Broad-host range vector with pUC19 polylinker, SpR | 本实验室This lab |
| pHMI-XopN | XopN整合到pHMI载体lacz启动子上的重组基因表达载体 Recombinant gene-expression vector containing XopN fused to lacZ promoter of pHMI | 本研究This study |
表2 供试菌株与质粒
Table 2 Bacterial strains and plasmid vectors used and created in this study.
| 菌株及质粒 Strains and plasmids | 相关性状 The study-relevant characteristics | 来源 Resource |
|---|---|---|
| 菌株Bacterial strains 黄单胞菌Xanthomonas oryzae pv. oryzae | ||
| PXO99A | PXO99的氮杂胞苷抗性克隆,也叫菲律宾小种6号 The azacytidine-resistant clone of PXO99, synonym Philippine race 6 | 本实验室This lab |
| ∆XopN | PXO99A XopN敲除突变体 PXO99A XopN-knockout unmarked mutant | 本研究This study |
| ∆XopN/XopN | 用质粒pHMXopN回补PXO99A XopN敲除突变体 PXO99A XopN mutant complemented with pHMXopN | 本研究This study |
| ∆XopQ | PXO99A XopQ敲除突变体 PXO99A XopQ-knockout unmarked mutant | 本实验室This lab |
| 质粒Plasmids | ||
| pMD19-Ts | 基因克隆T载体,含Apr, Mob+, Mob(P), LacZa+, Ampr Gene-cloning T-simple vector, Apr, Mob+, Mob(P), LacZa+, Ampr | TaKaRa |
| pMD20-T | 基因重组T载体,含Kanr, Ampr Gene-recombination T-simple vector, Kanr, Ampr | TaKaRa |
| pMD20-T::F1::Km::F2 | 重组的基因突变载体 Recombinant gene-mutagenicity vector | 本研究This study |
| pHMI | 多个pUC19重组的寄主更广的载体,含SpR Broad-host range vector with pUC19 polylinker, SpR | 本实验室This lab |
| pHMI-XopN | XopN整合到pHMI载体lacz启动子上的重组基因表达载体 Recombinant gene-expression vector containing XopN fused to lacZ promoter of pHMI | 本研究This study |
| 基因或基因片段 a Gene or partial sequence a | 引物 Primer | 酶切位点 Restriction site |
|---|---|---|
| F1 | 5'-AAGCTTGCCGACATGAAGGTGTATGAGGTG-3' | Hind Ⅲ |
| 5'-ACTAGTTGCGGCACCCGATGCTGCTGC-3' | SpeⅠ | |
| F2 | 5'-GGTACCTGGCCGCTGCCGGGTAATGC-3' | KpnⅠ |
| 5'-GAATTCGCGTGCTAATCAGGTTTGCCTTTG-3' | EcoRⅠ | |
| Kan | 5'-AGTGATTGCGCCTACCCGG-3' | 无None |
| 5'-ACGTCTTGAGCGATTGTGTAGG-3' | 无None | |
| XopN | 5'-GACCATGATTACGCCAAGCTTCTGGCAACAGCATCCCTCC-3' | Hind Ⅲ |
| 5'-GACCTGCAGGCATGCAAGCTTTTACGCCGGCGGCAGTGCCCGATC-3' | Hind Ⅲ |
表3 用于病菌XopN基因敲除和回补的PCR引物相关信息
Table 3 Information of primers used in PCR protocols for Xoo XopN knockout and complementation.
| 基因或基因片段 a Gene or partial sequence a | 引物 Primer | 酶切位点 Restriction site |
|---|---|---|
| F1 | 5'-AAGCTTGCCGACATGAAGGTGTATGAGGTG-3' | Hind Ⅲ |
| 5'-ACTAGTTGCGGCACCCGATGCTGCTGC-3' | SpeⅠ | |
| F2 | 5'-GGTACCTGGCCGCTGCCGGGTAATGC-3' | KpnⅠ |
| 5'-GAATTCGCGTGCTAATCAGGTTTGCCTTTG-3' | EcoRⅠ | |
| Kan | 5'-AGTGATTGCGCCTACCCGG-3' | 无None |
| 5'-ACGTCTTGAGCGATTGTGTAGG-3' | 无None | |
| XopN | 5'-GACCATGATTACGCCAAGCTTCTGGCAACAGCATCCCTCC-3' | Hind Ⅲ |
| 5'-GACCTGCAGGCATGCAAGCTTTTACGCCGGCGGCAGTGCCCGATC-3' | Hind Ⅲ |
| 水稻品种 Rice variety | 基因 Gene | 登录号 Locus number | 文献 Reference | 引物 Primer | PCR产物长度 Product size / bp | 用途 Subject | ||
|---|---|---|---|---|---|---|---|---|
| 所有品种 All varieties | OsEF1α | AF030517 | 9-13 | 5'-CGTGCCTGTGGGTCGTGTTG-3' 5'-TCCTGGAGAGCCTCGTGGTG-3' | 120 | 通过RT-qPCR 进行基因表达量分析 Gene expression analysis by RT-qPCR | ||
| 日本晴Nipponbare | Xa13 (OsSWEET11) | AK070510 | 10 | 5'-TGGTTCTGCTACGGCCTCTT-3' 5'-GGTACCAGAAGTAGAGCCCCATCT-3' | 103 | 通过RT-qPCR 进行基因表达量分析 Gene expression analysis by RT-qPCR | ||
| IRBB208, Asominori | 假定的OsSWEET11同源物 Hypothetic OsSWEET11 homologs | AK070510 | 本研究 This study | 5'-TGGTTCTGCTACGGCCTCTT-3' 5'-GGTACCAGAAGTAGAGCCCCATCT-3' | 103 | 通过RT-qPCR 进行基因表达量分析 Gene expression analysis by RT-qPCR | ||
| 日本晴, IRBB208 Asomimori Nipponbare, IRBB208 and Asominori | SWEET11与其假定同源物 OsSWEET11 and its homologs | AK070510 | 本研究 This study | 5'-CCAAGGCCAAACCACACATG-3' 5'-TAACTACTAATAATACCTG-3' | 1517 (Full length) | 通过RT-PCR 进行基因克隆,验证产物序列 Gene cloning by RT-PCR and confir mation of the product sequences | ||
| IRBB13 | xa13 (OsSWEET11) | DQ421394.1 | 33 | 5'-GGCGTGCACAAGGTCGAGGT-3' 5'-CTGGCCGTCATCGATCCGGC-3' | 205(3911-4116 in DQ421394.1) | 通过RT-qPCR进行基因表达量分析 Gene expression analysis by RT-qPCR | ||
| 5'-TACCCGAACGTGGGCGGCTT-3' 5'-AGTAACTACTAATAATACCTGTC-3' | 752(3701-4453 in DQ421394.1) | 通过RT-PCR进行基因克隆,验证产物序列Gene cloning by RT-PCR and confirmation of the product sequence | ||||||
表4 水稻基因及其RT-PCR与RT-qPCR分析所用引物信息
Table 4 Information on rice genes and their primers used for RT-PCR and RT-qPCR analyses.
| 水稻品种 Rice variety | 基因 Gene | 登录号 Locus number | 文献 Reference | 引物 Primer | PCR产物长度 Product size / bp | 用途 Subject | ||
|---|---|---|---|---|---|---|---|---|
| 所有品种 All varieties | OsEF1α | AF030517 | 9-13 | 5'-CGTGCCTGTGGGTCGTGTTG-3' 5'-TCCTGGAGAGCCTCGTGGTG-3' | 120 | 通过RT-qPCR 进行基因表达量分析 Gene expression analysis by RT-qPCR | ||
| 日本晴Nipponbare | Xa13 (OsSWEET11) | AK070510 | 10 | 5'-TGGTTCTGCTACGGCCTCTT-3' 5'-GGTACCAGAAGTAGAGCCCCATCT-3' | 103 | 通过RT-qPCR 进行基因表达量分析 Gene expression analysis by RT-qPCR | ||
| IRBB208, Asominori | 假定的OsSWEET11同源物 Hypothetic OsSWEET11 homologs | AK070510 | 本研究 This study | 5'-TGGTTCTGCTACGGCCTCTT-3' 5'-GGTACCAGAAGTAGAGCCCCATCT-3' | 103 | 通过RT-qPCR 进行基因表达量分析 Gene expression analysis by RT-qPCR | ||
| 日本晴, IRBB208 Asomimori Nipponbare, IRBB208 and Asominori | SWEET11与其假定同源物 OsSWEET11 and its homologs | AK070510 | 本研究 This study | 5'-CCAAGGCCAAACCACACATG-3' 5'-TAACTACTAATAATACCTG-3' | 1517 (Full length) | 通过RT-PCR 进行基因克隆,验证产物序列 Gene cloning by RT-PCR and confir mation of the product sequences | ||
| IRBB13 | xa13 (OsSWEET11) | DQ421394.1 | 33 | 5'-GGCGTGCACAAGGTCGAGGT-3' 5'-CTGGCCGTCATCGATCCGGC-3' | 205(3911-4116 in DQ421394.1) | 通过RT-qPCR进行基因表达量分析 Gene expression analysis by RT-qPCR | ||
| 5'-TACCCGAACGTGGGCGGCTT-3' 5'-AGTAACTACTAATAATACCTGTC-3' | 752(3701-4453 in DQ421394.1) | 通过RT-PCR进行基因克隆,验证产物序列Gene cloning by RT-PCR and confirmation of the product sequence | ||||||
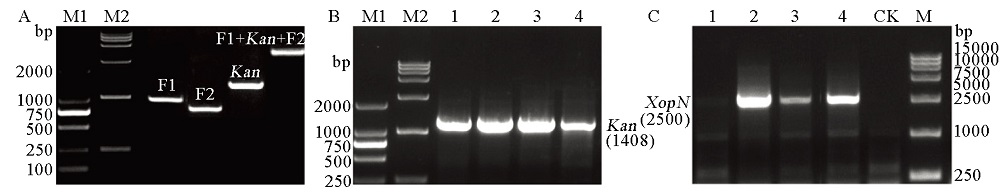
图1 PXO99A的XopN基因敲除 A–通过PCR扩增指定的基因片段来验证基因敲除重组载体pMD20-T::F1::Kan::F2; M1、M2为DNA标记。B–提取1~4号黄单胞菌转化子的基因组DNA对Kan基因进行PCR扩增,验证XopN基因敲除(∆XopN)突变体; M1、M2为DNA标记。C–提取1~4号黄单胞菌转化子的基因组DNA对XopN基因进行PCR扩增,验证∆XopN突变体。对照为以提取的转入空载体pMD20-T的黄单胞菌基因组DNA为模板进行的PCR扩增。CK为空白对照; M为DNA标记。
Fig. 1. XopN gene knockout from PXO99A. A, Verification of the gene-knockout recombinant vector pMD20-T::F1::Kan::F2 by PCR amplification of the indicated specific recombinant elements; M1, M2, DNA markers. B, Screening of the XopN-knockout mutant ∆XopN based on PCR amplification of Kan from genomic DNAs of the conjugate clones #1 to #4. M1, M2, DNA markers. C, Confirmation of the ∆XopN mutant by PCR amplification of the XopN from genomic DNAs of the conjugate clones #1 to #4 analyzed together with a control, which was the clone of bacteria transformed with the empty pMD20-T vector. CK, Blank control; M, DNA marker.
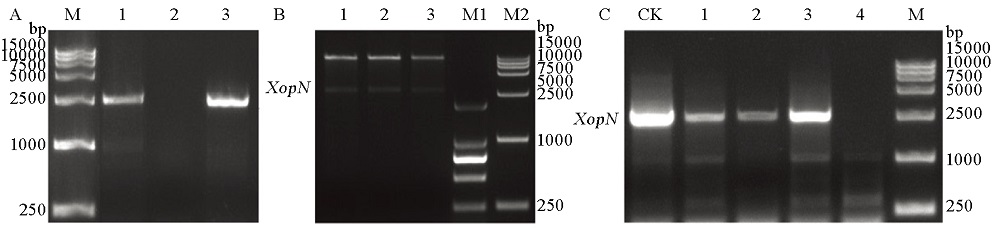
图2 突变体∆XopN遗传回补效果验证 A–对转化pHMI::XopN载体的1~3号大肠杆菌转化子提取基因组DNA进行的菌落PCR验证; M为DNA标记。B–对转化pHMI::XopN载体的1~3号大肠杆菌转化子提取质粒经Hind Ⅲ酶切验证。M1, M2为DNA标记。C–对转化pHMI::XopN载体的1~4号黄单胞菌转化子提取基因组DNA 进行PCR验证。M为DNA标记, CK为对照。在A~C中,均使用XopN特异性引物进行PCR扩增。
Fig. 2. Genetic complementation of the ∆XopN mutant. A, Escherichia coli colonial PCR analysis by using genomic DNA samples from the pHMI::XopN conjugate colonies #1 to #3; M, DNA marker. B, Restriction PCR analysis of the PCR products that were collected from A and digested by Hind Ⅲ. M1, M2, DNA markers. C, PCR analysis of genomic DNA samples from the pHMI::XopN conjugate colonies #1 to #3. CK, Control; M, DNA marker. In A to C, PCR was performed with the XopN-specific primers.
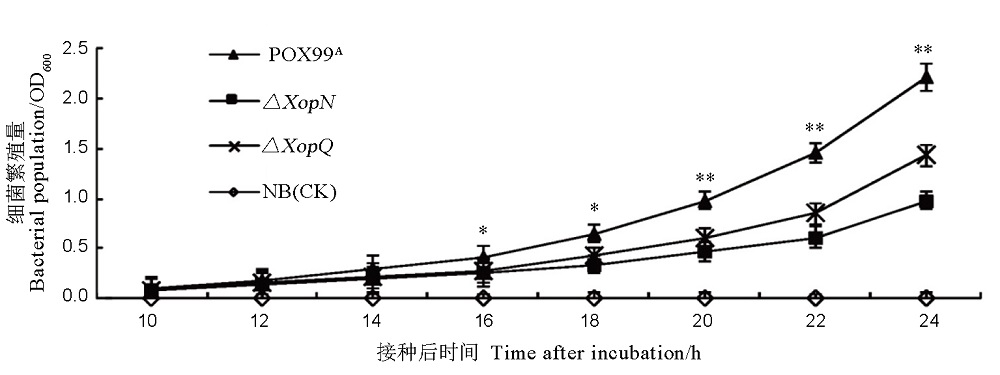
图3 Xoo不同菌株在NB液体培养基中的生长曲线所示数据为平均值 ± 标准差(standard deviation, SD); 对每个时间点获得的数据,都通过ANOVA和邓肯新复极差测验进行了多重比较。多重比较结果表明,在接种后16 h、18 h、20 h、22 h、24 h,PXO99A与两个突变体的生长量均存在显著差异(n = 6,P < 0.05或0.01)。
Fig. 3. Bacterial population of the indicated Xoo strains in NB medium. Data are shown as mean values ± SD. Significant differences exist between PXO99A and both mutants 16, 18, 20, 22 and 24 hours after inoculation based on ANOVA and Duncan’s test (n = 6, P < 0.05 or 0.01).
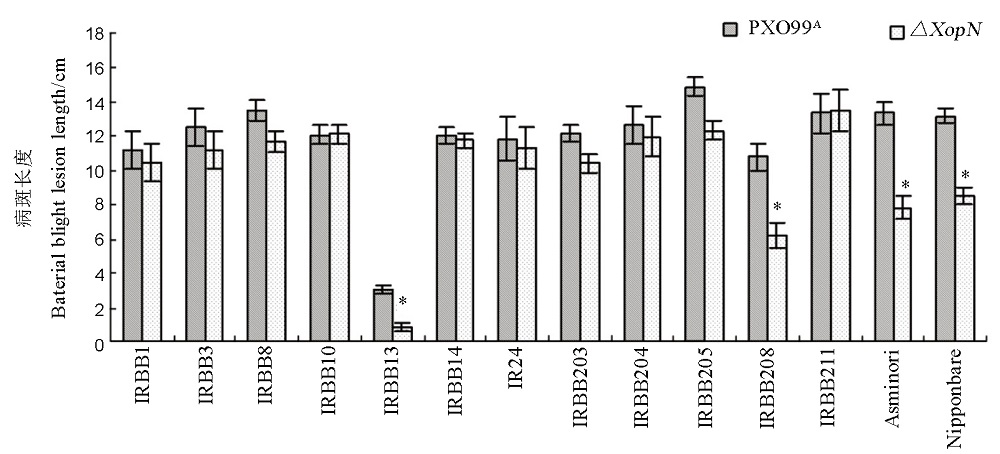
图4 用XopN与PXO99A剪叶接种水稻不同品种引起的白叶枯病斑长度横坐标下面所列的水稻品种幼苗的叶片分别用∆XopN和PXO99A的菌悬液进行剪叶接种,两周后测量叶片白叶枯病的病斑长度。图上所示数据是3次独立实验结果的平均值 ± 标准差,每次实验使用6株幼苗,每株幼苗接种12张叶片。数据通过方差分析(ANOVA)和F测验进行了成对数据比较,图上星号(*)表示Xoo这两个菌株引起的病斑在长度上存在显著差异(P < 0.05,n = 12)。
Fig. 4. Lesion Length of leaf blight caused by the Xoo strains in different rice varieties. Plants of various rice varieties at the bottom were inoculated by leaf-top clipping with bacterial suspensions of ∆XopN and PXO99A, respectively. Lesion length of leaf blight was measured two weeks later. Values are shown as mean ± standard deviation (SD) estimates of 3 independent experiments each involving 12 leaves of 6 plants. Asterisks indicate significant differences in the corresponding pair comparisons based on analysis of variance (ANOVA) and Fisher’s test at P < 0.05 (n = 12).
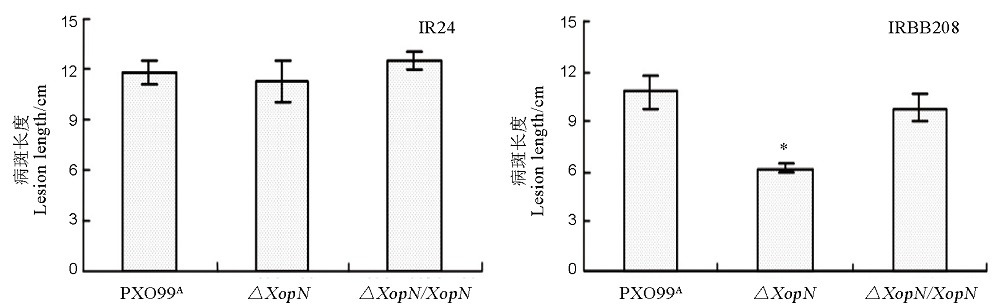
图5 PXO99A、∆XopN、∆XopN/XopN在水稻品种IR24和IRBB208上的白叶枯病病斑长度水稻品种IR14和IRBB208幼苗分别用PXO99A、∆XopN和∆XopN/XopN的菌悬液进行剪叶接种,两周后测量叶片白叶枯病的病斑长度。图上所示数据为平均值 ± 标准差; 通过ANOVA和F测验做了多重比较,星号(*)表示Xoo这三个菌株在IRBB208叶片内的繁殖量存在显著性差异(P < 0.05,n = 12)。
Fig. 5. Lesion length of leaf blight caused by the three Xoo strains in the two rice varieties. Plants of the rice varieties were inoculated by leaf-top clipping with bacterial suspensions of PXO99A, ∆XopN and ∆XopN/XopN, respectively. Lesion length of leaf blight was measured two weeks later. Data are mean values ± SD. Asterisks indicate significant differences in the multiple comparisons by ANOVA and Fisher’s test (P < 0.05, n = 12).
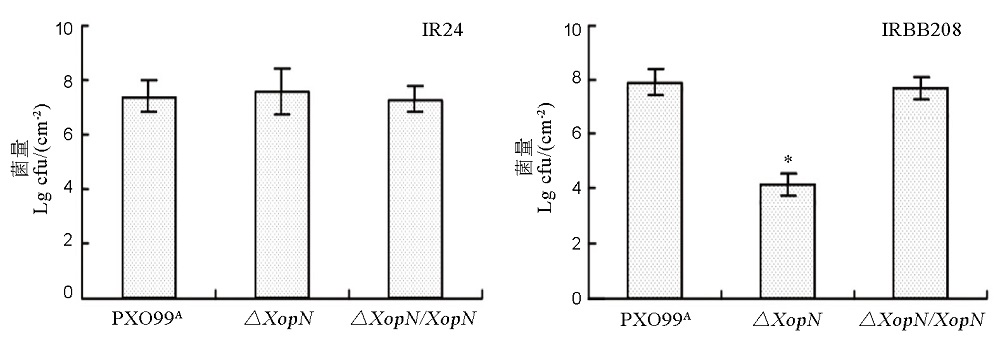
图6 PXO99A、∆XopN和∆XopN/XopN在水稻品种IR24和IRBB208叶片内的繁殖量采取叶片注射接种的方法,用柱状图横坐标下面所列Xoo三个菌株的细菌悬浮液分别接种水稻品种IR24和IRBB208幼苗。3 d后,从接种叶片回收细菌,经过平板培养长出菌落后,计数菌落形成单位(colony formation unit, cfu),并换算成单位叶面积cfu数值。所示数据为平均值 ± 标准差。图上的星号(*)表示通过ANOVA和邓肯新复极差测验进行多重比较的结果,表明来自不同细菌菌株的数据之间存在显著性差异(P < 0.01,n = 6)。
Fig. 6. Bacterial population of PXO99A, ∆XopN and ∆XopN/XopN in leaves of IR24 and IRBB208. Plants of rice varieties IR24 and IRBB208 were inoculated by leaf infiltration with bacterial suspensions prepared from each of the bacterial strains indicated at the bottom. Three days later, bacteria were recovered from leaves and scored as colony formation unit (cfu) in the given leaf size. Data are mean values±SD. Different letters indicate significant differences of multiple comparisons of data from different bacterial strains based on ANOVA and Duncan’s multiple-range test(P<0.01, n= 6).
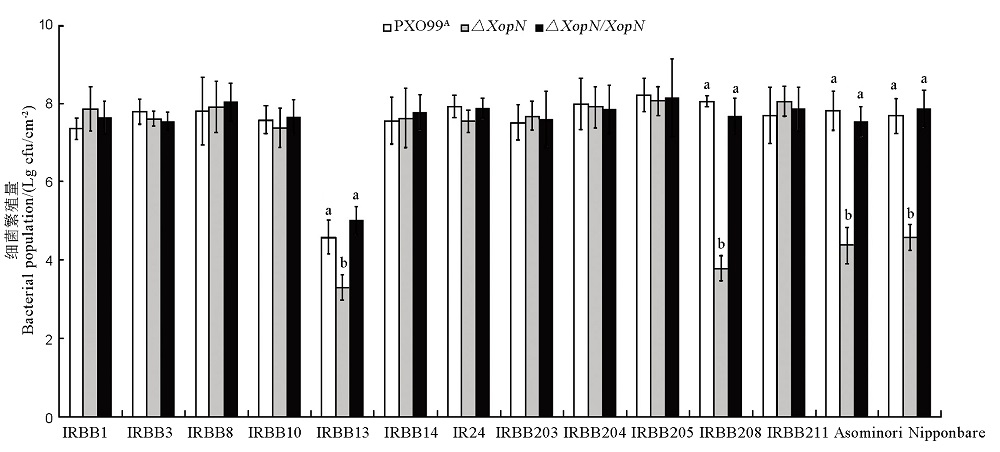
图7 PXO99A、∆XopN和∆XopN/XopN在14个水稻品种体内的繁殖量使用Xoo三个菌株的细菌悬浮液,采取叶片注射接种的方法,分别接种14个水稻品种的幼苗。3 d后,从接种叶片回收细菌,经过培养长出菌落,计数cfu,并换算成单位叶面积cfu数值。所示数据为平均值 ± 标准差,通过ANOVA和邓肯新复极差测验做了多重比较,图上标注的不同字母表示Xoo不同菌株在相应水稻品种幼苗叶片内的繁殖量存在显著性差异(P < 0.01,n = 3)。
Fig. 7. Populations of PXO99A, ∆XopN and ∆XopN/XopN bacteria multiplied in leaves of the 14 rice varieties. Plants of different rice varieties at the bottom were inoculated by leaf infiltration with bacterial suspensions prepared from each of the indicated bacterial strains on the top. Three days later, bacteria were recovered from leaves and scored as cfu in the given leaf size. Data shown are mean values ± SD. Different letters on the graph indicate that ANOVA and Duncan’s multiple-range test disclose significant differences of multiple comparisons of data from the corresponding rice varieties inoculated differently (P < 0.01, n = 3).
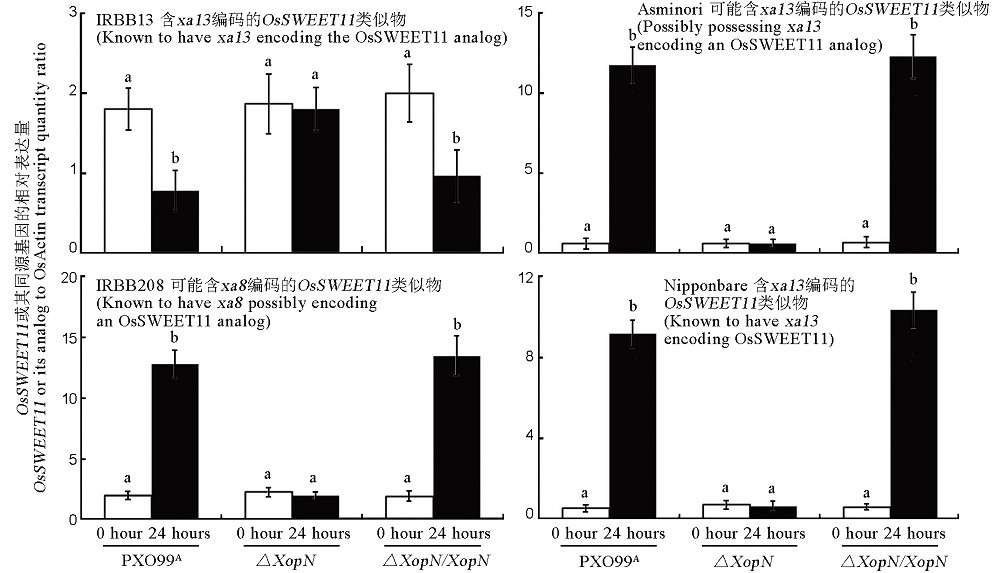
图8 XopN对OsSWEET11或其同源基因在水稻4个品种叶片内表达水平的影响每个水稻品种的名称,连同所含的抗病基因信息,标注在相应的柱状图上方。这4个水稻品种都在14日龄进行接种,用标注于柱状图下方的Xoo菌株的细菌悬浮液,分别注射幼苗叶片。接种前(0h)和接种后24h取样,即切取接种的叶片,用来提取RNA,随后进行RT-qPCR分析。使用IRBB13隐性基因OsSWEET11/xa13的特异性引物,对IRBB13的RNA进行反转录,而另外3个水稻品种的RNA反转录,则使用日本晴显性基因Xa13/OsSWEET11的特异性引物。所有RT-qPCR实验均包括组成型表达的OsEF1α基因,用作参比。所示数据为平均值 ± 标准差,图上的不同字母指出了ANOVA和邓肯新复极差测验结果,表明多重比较的数据之间存在显著性差异(P < 0.05,n = 6)。
Fig. 8. Effects of XopN on expression levels of OsSWEET11 or its analogs in leaves of the 4 rice varieties. Plants were inoculated by leaf infiltration with bacterial suspensions prepared from each of the indicated bacterial strains. Immediately before inoculation (0 hour) and 24 hours after inoculation, inoculated leaves were excised and used to isolate RNAs. RNA samples were subjected to RT-qPCR in two combinations. One was IRBB13 RNA samples and a pair of primers specific to the recessive resistance gene OsSWEET11/xa13 of IRBB13. The other was RNA samples from additional 3 rice varieties and a pair of primers specific to the dominant disease-susceptibility gene OsSWEET11/Xa13 of Nipponbare. Relative levels of the genes were quantified as ratios of their transcript amounts to the transcript amount of OsEF1α, a constitutively expressed gene used as a reference. Data are mean values ± SD estimates. Different letters on graphs indicate significant differences of multiple comparisons of data from differently inoculated plants based on ANOVA and Duncan’s test (P < 0.05, n = 6).
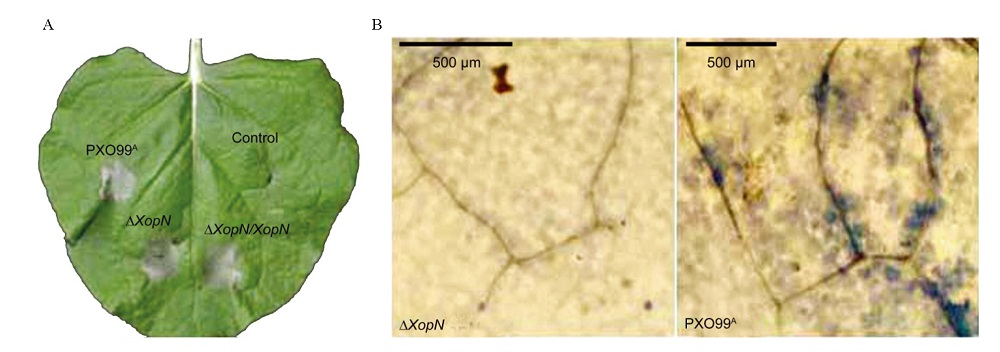
图9 XopN对Xoo诱发烟草叶片过敏反应的影响 A–本氏烟叶片HR观察。烟草叶片分别用纯水(对照)或所指Xoo菌株的菌悬液注射,36 h后拍照。这张照片代表经过类似处理的3株烟草的6张叶片。B–本氏烟叶片微敏反应(micro-HR)观察。烟草叶片用∆XopN或PXO99A的菌悬液进行注射后24 h,切取下来并用台盼蓝染色,然后观察微敏反应。微敏反应通过显微镜观察,可以在右侧图像中观察到,而在左侧图像中观察不到,两张显微图像各代表3株烟草的6张注射过的叶片。
Fig. 9. Effect of XopN on the ability of Xoo to induce hypersensitive response in tobacco leaves. A, A tobacco (Nicotiana benthamiana) leaf photographed at 36 h after infiltration with pure water (control) or with a bacterial suspension of every Xoo strain shown on the leaf. This photo represents 6 leaves of 3 plants that were treated similarly. B, Parts of leaves 24 h after infiltration with the indicated bacterial suspensions and stained with trypan blue to show micro-HR. Micro-HR was visualized with a microscopy and is present in the right image but absent in the left one. Both microscopic images represent 6 infiltrated leaves excised from 3 plants.
| [1] | Mew T W.Current status and future prospects of research on bacterial blight of rice[J]. Annual Review of Phytopathology, 1987, 25: 359-382. |
| [2] | Ou S H.Rice Diseases[M]. Kew, Surrey: Commonwealth Mycological Institute, 1972: 368. |
| [3] | Buttner D, Bonas U.Regulation and secretion of Xanthomonas virulence factors[J]. FEMS Microbiology Reviews, 2010, 34(2): 107-133. |
| [4] | Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer S V, Machado M A, Toth I, Salmond G, Foster G D.Top 10 plant pathogenic bacteria in molecular plant pathology[J]. Molecular Plant Pathology, 2012, 13(6): 614-629. |
| [5] | Niño-Liu D O, Ronald P C, Bogdanove A J. Xanthomonas oryzae pathovars: Model pathogens of a model crop[J]. Molecular Plant Pathology, 2006, 7: 303-324. |
| [6] | Noda T, Kaku H.Growth of Xanthomonas oryzae pv. oryzae in planta and in guttation fluid of rice[J]. Annals of the Phytopathological Society of Japan, 1999, 65: 9-14. |
| [7] | Ou S H.Rice Diseases[M]. Kew, Surrey: Commonwealth Agricultural Bureaux, 1985: 109-201. |
| [8] | Park C J, Kazunari N, Ronald P C.Quantitative measurements of Xanthomonas oryzae pv. oryzae distribution in rice using fluorescent-labelling[J]. Journal of Plant Biology, 2010, 15: 595-599. |
| [9] | Bian H J, Zhang L Y, Chen L, Wang W Z, Ji H T, Dong H S.Real-time monitoring of translocation of selected type-III effectors from Xanthomonas oryzae pv. oryzae into rice cells[J]. Journal of Biosciences, 2019, 44: 4. |
| [10] | Li P, Zhang L Y, Mo X Y, Ji H T, Bian H J, Hu Y Q, Majid T, Long J Y, Pang H, Tao Y, Ma J B, Dong H S.Rice aquaporin PIP1;3 and harpin Hpa1 of bacterial blight pathogen cooperate in a type III effector translocation[J]. Journal of Experimental Botany, 2019, 70(12): 3057-3073. |
| [11] | Zhang L Y, Chen L, Dong H S.Plant aquaporins in infection by and immunity against pathogens: A critical review[J]. Frontiers in Plant Science, 2019, 10: 632. |
| [12] | Wang X, Zhang L Y, Ji H T, Mo X Y, Li P, Wang J Z, Dong H S.Hpa1 is a type III translocator in Xanthomonas oryzae pv. oryzae[J]. BMC Microbiology, 2018, 18: 105. |
| [13] | Zhang L Y, Hu Y Q, Li P, Wang X B, Dong H S.Silencing of an aquaporin gene diminishes bacterial blight disease in rice[J]. Australasian Plant Pathology, 2019, 48(2): 143-158. |
| [14] | Zhu P L, Zhao S, Tang J L, Feng J X.The rsmA-like gene rsmAxoo of Xanthomonas oryzae pv. oryzae regulates bacterial virulence and production of diffusible signal factor[J]. Molecular Plant Pathology, 2011, 12(3): 227-237. |
| [15] | White F F, Potnis N, Jones J B, Koebnik R.The type III effectors of Xanthomonas[J]. Molecular Plant Pathology, 2009, 10(6): 749-766. |
| [16] | Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U.Breaking the code of DNA binding specificity of TAL-type III effectors[J]. Science, 2009, 326: 1509-1512. |
| [17] | Moscou M J, Bogdanove A J.A simple cipher governs DNA recognition by TAL effectors[J]. Science, 2009, 326: 1501. |
| [18] | Oliva R, Ji C H, Atienza-Grande1 G, Huguet-Tapia J C, Perez-Quintero A, Li T, Eom J S, Li C H, Nguyen H, Liu B, Auguy F, Sciallano C, Luu V T, Dossa G S, Cunnac S, Schmidt S M, Slamet-Loedin I H, Cruz C V, Szurek B, Frommer W B, White F F, Yang B. Broad-spectrum resistance to bacterial blight in rice using genome editing[J]. Nature Biotechnology, 2019, 37: 1344-1350. |
| [19] | Richter A, Streubel J, Blücher C, Szurek B, Reschke M, Grau J, Boch J.A TAL effector repeat architecture for frameshift binding[J]. Nature Communcations, 2014, 5: 3447. |
| [20] | Kim J G, Stork W, Mudgett M B.Xanthomonas type Ⅲ effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth[J]. Cell Host Microbe, 2013, 13(2): 143-154. |
| [21] | Kim J G, Taylor K W, Hotson A, Keegan M, Schmelz E A, Mudgett M B.XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves[J]. Plant Cell, 2008, 20: 1915-1929. |
| [22] | Long J, Song C, Yan F, Zhou J, Zhou H, Yang B.Non-TAL effectors from Xanthomonas oryzae pv. oryzae suppress peptidoglycan-triggered MAPK activation in rice[J]. Frontiers in Plant Science, 2018, 12(9): 1857. |
| [23] | Medina C A, Reyes P A, Trujillo C A, Gonzalez J L, Bejarano D A, Montenegro N A, Jacobs J M, Joe A, Restrepo S, Alfano J R, Bernal A.The role of type Ⅲ effectors from Xanthomonas axonopodis pv. manihotis in virulence and suppression of plant immunity[J]. Molecular Plant Pathology, 2018, 19(3): 593-606. |
| [24] | Sinha D, Gupta M K, Patel H K, Ranjan A, Sonti R V.Cell wall degrading enzyme induced rice innate immune responses are suppressed by the type 3 secretion system effectors XopN, XopQ, XopX and XopZ of Xanthomonas oryzae pv. oryzae[J/OL]. PLoS ONE, 2013, 8(9): e75867. |
| [25] | Taylor K W, Kim J G, Su X B, Aakre C D, Roden J A, Adams C M, Mudgett M B.Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector XopN from binding to TFT1 attenuate Xanthomonas virulence[J/OL]. PLoS Pathogens, 2012, 8(6): e1002768. |
| [26] | Li N, Han X, Feng D, Yuan D, Huang L J.Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering[J]? International Journal of Molecular Sciences, 2019, 20(3): 671. |
| [27] | Roden J A, Belt B, Ross J B, Tachibana T, Vargas J, Mudgett M B.A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(47): 16 624-16 629. |
| [28] | Jiang B L, He Y Q, Cen W J, Wei H Y, Jiang G F, Jiang W, Hang X H, Feng J X, Lu G T, Tang D J, Tang J L.The type III secretion effector XopXccN of Xanthomonas campestris pv. campestris is requried for full virulence[J]. Research in Microbiology, 2008, 159(3): 216-220. |
| [29] | Liao Z X, Li J Y, Mo X Y, Ni Z, Jiang W, He Y Q, Huang S.Type III effectors xopN and avrBS2 contribute to the virulence of Xanthomonas oryzae pv. oryzicola strain GX01[J]. Research in Microbiology, 2019: 10.002. |
| [30] | Cheong H, Kim C Y, Jeon J S, Lee B M, Sun Moon J, Hwang I.Xanthomonas oryzae pv. oryzae type III effector XopN targets OsVOZ2 and a putative thiamine synthase as a virulence factor in rice[J/OL]. PLoS One, 2013, 8(9): e73346. |
| [31] | 孙荣华. 水稻白叶枯菌JXOV中tal17.5基因的克隆与功能研究[D]. 南京:南京农业大学, 2016. |
| Sun R H.Cloning and functional characterization of tal 17.5 gene from Xanthomonas oryzae pv. oryzae JXOV strain[D]. Nanjing: Nanjing Agricultural University, 2016. | |
| [32] | Jiang N, Yan J, Liang Y, Shi Y, He Z, Wu Y, Zeng Q, Liu X, Peng J.Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.): An updated review[J]. Rice (NY), 2020, 13(1): 3. |
| [33] | Ji Z, Wang C, Zhao K.Rice routes of counting Xanthomonas oryzae[J]. International Journal of Molecular Sciences, 2018, 19: 3008. |
| [34] | Yang S Q, Liu S Y, Zhao S, Yu Y H, Li R B, Duan C J, Tang J L, Feng J X.Molecular and pathogenic characterization of new Xanthomonas oryzae pv. oryzae strains from the coastline region of Fangchenggang city in China[J]. World Journal of Microbiology & Biotechnology, 2013, 29(4): 713-720. |
| [35] | Yang B, Sugio A, White F F.Os8N3 is a host disease-susceptibility gene for bacterial blight of rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103: 10 503-10 508. |
| [36] | 胡立. 水稻白叶枯病菌效应蛋白XopN病理功能与核黄素对拟南芥开花调控的影响[D]. 南京:南京农业大学, 2016. |
| Hu L.Pathological roles of Xanthomonas oryzae pv. oryzae effector XopN and Arabidopsis flowering regulation by Riboflavin content[D]. Nanjing: Nanjing Agricultural University, 2016. | |
| [37] | Chen L, Zhang S J, Zhang S S, Qu S P, Ren X Y, Long J Y, Yin Q, Qian J, Sun F, Zhang C L, Wang L X, Wu X L, Wu T Q, Zhang Z K, Chen Z Q, Hayes M, Beer S V, Dong H S.A fragment of the Xanthomonas oryzae pv. oryzicola harpin HpaGXooc reduces disease and increases yield of rice in extensive grower plantings[J]. Phytopathology, 2008, 98(7): 792-802. |
| [38] | 时立文. SPSS19.0统计分析从入门到精通[M]. 北京: 清华大学出版社, 2012. |
| Shi L W.SPSS19.0 statistical analysis from entry to the master[M]. Beijing: Tsinghua University Press, 2012. | |
| [39] | Zhu Y, Chen H, Fan J, Wang Y, Li Y, Chen J, Fan J, Yang S, Hu L, Leung H, Mew T W, Teng P S, Wang Z, Mundt C C.Genetic diversity and disease control in rice[J]. Nature, 2000, 406(6797): 718-722. |
| [40] | Li C, Li W, Zhou Z, Vhen H, Xie C, Lin Y.A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice[J]. Plant Biotechnology Journal, 2020, 18: 313-315. |
| [41] | Yuan M, Chu Z, Li X, Xu, C, Wang S. Pathogen-induced expression loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice[J]. Plant & Cell Physiology, 2009, 50(5): 947-955. |
| [42] | Song C, Yang B.Mutagenesis of 18 type III effectors reveals virulence function of XopZPXO99 in Xanthomonas oryzae pv. oryzae[J]. Molecular Plant-Microbe Interactions, 2010, 23: 893-902. |
| [43] | Zhao S, Mo W L, Wu F, Tang W, Tang J L, Szurek B, Verdier V, Koebnik R, Feng J X.Identification of non-TAL effectors in Xanthomonas oryzae pv. oryzae Chinese strain 13751 and analysis of their role in the bacterial virulence[J]. World Journal of Microbiology & Biotechnology, 2013, 29: 733-744. |
| [44] | 董汉松, 陈蕾, 邹珅珅. 植物分子免疫学[M]. 北京: 科学出版社, 2020 (待印). |
| Dong H S, Chen L, Zhou S S.Plant molecular immunology[M]. Beijing: Science Press, 2020 | |
| (To be printed). | |
| [45] | Akimoto-Tomiyama C, Furutani A, Tsuge S, Washington E J, Nishizawa Y, Minami E, Ochiai H.XopR, a type III effector secreted by Xanthomonas oryzae pv. oryzae, suppresses microbe-associated molecular pattern triggered immunity in Arabidopsis thaliana[J]. Molecular Plant-Microbe Interactions, 2012, 25: 505-514. |
| [46] | Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen J L, Zhang Q, Wang S.Promoter mutations of an essential gene for pollen development result in disease resistance in rice[J]. Genes & Development, 2006, 20(10): 1250-1255. |
| [47] | Sobhani A, Khanlarkhani N, Baazm M, Mohammadzadeh F, Najafi A, Mehdinejadiani S, Sargolzaei Aval F.Multipotent stem cell and current application[J]. Acta Medica Iranica, 2017, 55(1): 6-23. |
| [48] | Kim J G, Li X, Roden J A, Taylor K W, Aakre C D, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, McLane H, Martin G B, Mudgett M B. Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1[J]. Plant Cell, 2009, 21: 1305-1323. |
| [49] | Ji H, Dong H.Key steps in type III secretion system (T3SS) towards translocon assembly with potential sensor at plant plasma membrane[J]. Molecular Plant Pathology, 2015, 16(7): 762-773. |
| [50] | Kvitko B H, Ramos A R, Morello J E, Oh H S, Collmer A.Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors[J]. Journal of Bacteriology, 2007, 189: 8059-8072. |
| [51] | Römer P, Recht S, Strauß T, Elsaesser J, Schornack S, Boch J, Wang S, Lahaye T.Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae[J]. New Phytology, 2010, 187: 1048-1057. |
| [1] | 马静静, 潘妍妍, 杨孙玉悦, 王嘉琦, 蒋冬花. 硫藤黄链霉菌St-79对水稻白叶枯病的防效和促生作用[J]. 中国水稻科学, 2022, 36(6): 623-638. |
| [2] | 朱引引,刘永庭,李士河,宋从凤*. 水稻白叶枯菌OS198中talR26.5基因的克隆及功能分析[J]. 中国水稻科学, 2014, 28(4): 343-350. |
| [3] | 刘希玲,邹华松,邹丽芳,陈功友 . 水稻白叶枯病菌hrcU基因缺失突变体构建及功能研究[J]. 中国水稻科学, 2010, 24(4): 348-352 . |
| [4] | 胡泽友邓小波,彭喜旭,何艳,刘文海,戴光宇,王海华,. 外源钙对镍胁迫下水稻幼苗抗氧化酶活性及膜脂过氧化的影响[J]. 中国水稻科学, 2007, 21(4): 367-371 . |
| [5] | 陈河王慧中赵文华,蔡玲斐,王灵均,陈小囡,黄大年. 转mtlD/gutD基因稻米对大鼠性腺毒性的实验研究[J]. 中国水稻科学, 2007, 21(4): 341-344 . |
| [6] | 陈功友,邹丽芳,武晓敏,李玉蓉,王金生. 水稻条斑病细菌中新发现的avrBs3/PthA家族新成员avr/pth13基因[J]. 中国水稻科学, 2005, 19(4): 291-296 . |
| [7] | 王慧中, 应奇才, 钟国庆, 田菊霞, 陈 河, 黄大年, 钱 前, 张红生. 转mtlD/gutD双价基因稻米对大鼠的毒性试验[J]. 中国水稻科学, 2004, 18(3): 208-212 . |
| [8] | 陈忠孝, 胡国文,凌逸军. 噻嗪酮对稻纵卷叶螟F1代生存的影响[J]. 中国水稻科学, 1994, 8(1): 57-60 . |
| [9] | 谢关林, 苗东华. 稻白叶枯病菌在人工培养基上毒性降低因素研究(英文)[J]. 中国水稻科学, 1987, 1(2): 118-126 . |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||