
中国水稻科学 ›› 2024, Vol. 38 ›› Issue (6): 638-652.DOI: 10.16819/j.1001-7216.2024.240101
收稿日期:2024-01-01
修回日期:2024-03-18
出版日期:2024-11-10
发布日期:2024-11-15
通讯作者:
*email: caiyicongcyc@163.com
基金资助:
ZHONG Zhihu, QIN Lu, LI Zhili, YANG Zhen, HE Xiaopeng, CAI Yicong*( )
)
Received:2024-01-01
Revised:2024-03-18
Online:2024-11-10
Published:2024-11-15
Contact:
*email: caiyicongcyc@163.com
摘要:
【目的】明确植物特异转录因子OsIDD基因家族成员特征特性,验证其表达模式,为深入研究水稻IDD基因家族生物学功能奠定基础。【方法】通过Pfam鉴定与明确水稻IDD家族成员,对其核酸与蛋白序列特征、蛋白结构、系统发育与共线性、启动子元件进行综合分析;通过RT-PCR技术验证OsIDD基因在各组织、不同激素处理及不同非生物胁迫下的表达模式。【结果】发现OsIDD家族具有15个成员,为3个亚家族,在染色体上呈不均匀分布,N端均具有核定位信号及高度保守的ID结构域。多重序列比对及motif分析表明其在各个类型中相对保守,与玉米IDD基因家族共线性分析表明两个物种间具有高度同源性。芯片及RT-PCR分析发现OsIDD主要在穗及发育胚乳中表达量较高,其启动子序列均包含激素/胁迫响应顺式元件。【结论】本研究对15个OsIDD成员进行了综合分析,OsIDD结构保守,与ZmIDD共线性强,主要在穗及发育胚乳组织中高表达,可能参与不同激素及多种非生物逆境胁迫响应过程。
钟智慧, 秦璐, 黎志力, 杨珍, 贺晓鹏, 蔡怡聪. 水稻IDD基因家族的全基因组鉴定及综合分析[J]. 中国水稻科学, 2024, 38(6): 638-652.
ZHONG Zhihu, QIN Lu, LI Zhili, YANG Zhen, HE Xiaopeng, CAI Yicong. Genome-wide Identification and Comprehensive Analysis of IDD Gene Family in Rice[J]. Chinese Journal OF Rice Science, 2024, 38(6): 638-652.
| 引物名称 Primer name | 正向引物序列 Forward sequence (5’-3’) | 反向引物序列 Revers sequence(5’-3’) |
|---|---|---|
| Actin-RT | CGGGAAATTGTGAGGGACAT | AGGAAGGCTGGAAGAGGACC |
| OsID1-RT | CTCTTCTCCAGGAAGGACAGCC | GTAGTAGTGATGCTGCTGCTGTTG |
| OsIDD1-RT | GTTCTGGTCGCGCTGGAAC | GACACAATCATTAGGAGCGGCA |
| OsIDD2-RT | CGGCAACCCAGATCCTGAG | CTGGTCTCGCTGGAACCC |
| OsIDD3-RT | GGAAACCCAAATCCAGATGCG | CCGCTGGAACCCCTTGTTG |
| OsIDD4-RT | CCTTACCAGACCCGGACGC | GTTCTGCTCCCGCTGGAAC |
| OsIDD5-RT | GGATGAGCAAGAGGTGGTTG | ATCCAGCAGGTTTGAGTCCT |
| OsIDD6-RT | CCCGGGAATCCAAATCCTGATGC | CTGCTCCCTCTGGAACCCC |
| OsIDD7-RT | GGAAACCCAGATCCAGATGTTGAG | GGAGGTTCTGGTCTCTCTGGAAG |
| OsIDD8-RT | CGGCACACCAGATCCGG | CAGGTTCTGGTCTCGCTGG |
| OsIDD9-RT | GTCTTCTCCAGGCGCGAC | CAGAGCGGAGGTGACCG |
| OsIDD10-RT | CAACCAACCCAAACCCGGAC | GGTTCTGCTCCCGCTGG |
| OsIDD11-RT | GCAACCCAGACCCGGAAG | CTGCAGGTTCTGGTCCCTC |
| OsIDD12-RT | CAGATCACCTGCTACAGCTG | TGACGACGGACGAGACGTA |
| OsIDD13-RT | GCACGCCAGACCCGGAC | CTGCAGGTTCTGGTCTCGC |
| OsIDD14-RT | GCGTCTTCTCCCGAGTGG | GGATGACCGGCAGCGTC |
表1 本研究用到的引物信息
Table 1. Primers used in the study.
| 引物名称 Primer name | 正向引物序列 Forward sequence (5’-3’) | 反向引物序列 Revers sequence(5’-3’) |
|---|---|---|
| Actin-RT | CGGGAAATTGTGAGGGACAT | AGGAAGGCTGGAAGAGGACC |
| OsID1-RT | CTCTTCTCCAGGAAGGACAGCC | GTAGTAGTGATGCTGCTGCTGTTG |
| OsIDD1-RT | GTTCTGGTCGCGCTGGAAC | GACACAATCATTAGGAGCGGCA |
| OsIDD2-RT | CGGCAACCCAGATCCTGAG | CTGGTCTCGCTGGAACCC |
| OsIDD3-RT | GGAAACCCAAATCCAGATGCG | CCGCTGGAACCCCTTGTTG |
| OsIDD4-RT | CCTTACCAGACCCGGACGC | GTTCTGCTCCCGCTGGAAC |
| OsIDD5-RT | GGATGAGCAAGAGGTGGTTG | ATCCAGCAGGTTTGAGTCCT |
| OsIDD6-RT | CCCGGGAATCCAAATCCTGATGC | CTGCTCCCTCTGGAACCCC |
| OsIDD7-RT | GGAAACCCAGATCCAGATGTTGAG | GGAGGTTCTGGTCTCTCTGGAAG |
| OsIDD8-RT | CGGCACACCAGATCCGG | CAGGTTCTGGTCTCGCTGG |
| OsIDD9-RT | GTCTTCTCCAGGCGCGAC | CAGAGCGGAGGTGACCG |
| OsIDD10-RT | CAACCAACCCAAACCCGGAC | GGTTCTGCTCCCGCTGG |
| OsIDD11-RT | GCAACCCAGACCCGGAAG | CTGCAGGTTCTGGTCCCTC |
| OsIDD12-RT | CAGATCACCTGCTACAGCTG | TGACGACGGACGAGACGTA |
| OsIDD13-RT | GCACGCCAGACCCGGAC | CTGCAGGTTCTGGTCTCGC |
| OsIDD14-RT | GCGTCTTCTCCCGAGTGG | GGATGACCGGCAGCGTC |
| 名称 Name | 染色体 Chr. | 基因长度 Gene length | 氨基酸数目 No. of amino acids | 分子量 Molecular weight | 理论等电点 Theoretical pI | 蛋白质不稳定 指数 Instability index | 脂溶指数 Aliphatic index | 亲水性平均系数 Grand average of hydropathicity | 亚细胞定位预测 Predicted subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| OsID1 | 10 | 1230 | 409 | 44267.44 | 8.04870033 | 51.19 | 51.19 | −0.467 | 细胞核Nucleus |
| OsIDD1 | 3 | 1659 | 552 | 57540.01 | 8.55090046 | 75.58 | 75.58 | −0.486 | 细胞核Nucleus |
| OsIDD2 | 1 | 1464 | 487 | 51815.90 | 8.60980034 | 48.96 | 48.96 | −0.527 | 细胞核Nucleus |
| OsIDD3 | 9 | 1608 | 535 | 54893.05 | 9.41469955 | 46.65 | 46.65 | −0.460 | 细胞核Nucleus |
| OsIDD4 | 2 | 1848 | 615 | 63597.71 | 8.63129997 | 46.29 | 46.29 | −0.497 | 细胞核Nucleus |
| OsIDD5 | 7 | 1902 | 633 | 67636.09 | 6.01849985 | 55.08 | 55.08 | −0.567 | 细胞核Nucleus |
| OsIDD6 | 8 | 1659 | 552 | 57262.74 | 9.38479996 | 49.81 | 49.81 | −0.500 | 细胞核Nucleus |
| OsIDD7 | 2 | 1479 | 492 | 54491.89 | 7.31449986 | 59.23 | 59.23 | −0.697 | 细胞核Nucleus |
| OsIDD8 | 1 | 1488 | 495 | 51394.20 | 8.89400005 | 49.83 | 49.83 | −0.293 | 细胞核Nucleus |
| OsIDD9 | 1 | 1431 | 476 | 50107.14 | 7.95529985 | 52.62 | 52.62 | −0.265 | 细胞核Nucleus |
| OsIDD10 | 4 | 1842 | 613 | 63814.37 | 9.93620014 | 52.40 | 52.4 | −0.476 | 细胞核Nucleus |
| OsIDD11 | 1 | 1614 | 537 | 57410.90 | 7.97130013 | 62.67 | 62.67 | −0.703 | 细胞核Nucleus |
| OsIDD12 | 8 | 1602 | 533 | 56426.28 | 9.51509953 | 62.47 | 62.47 | −0.467 | 细胞核Nucleus |
| OsIDD13 | 9 | 1515 | 504 | 53574.19 | 8.50090027 | 62.67 | 62.67 | −0.459 | 细胞核Nucleus |
| OsIDD14 | 3 | 1317 | 438 | 45959.66 | 10.12440010 | 70.46 | 70.46 | −0.509 | 细胞核Nucleus |
表2 OsIDD蛋白序列理化性质
Table 2. Physical and chemical properties of OsIDD
| 名称 Name | 染色体 Chr. | 基因长度 Gene length | 氨基酸数目 No. of amino acids | 分子量 Molecular weight | 理论等电点 Theoretical pI | 蛋白质不稳定 指数 Instability index | 脂溶指数 Aliphatic index | 亲水性平均系数 Grand average of hydropathicity | 亚细胞定位预测 Predicted subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| OsID1 | 10 | 1230 | 409 | 44267.44 | 8.04870033 | 51.19 | 51.19 | −0.467 | 细胞核Nucleus |
| OsIDD1 | 3 | 1659 | 552 | 57540.01 | 8.55090046 | 75.58 | 75.58 | −0.486 | 细胞核Nucleus |
| OsIDD2 | 1 | 1464 | 487 | 51815.90 | 8.60980034 | 48.96 | 48.96 | −0.527 | 细胞核Nucleus |
| OsIDD3 | 9 | 1608 | 535 | 54893.05 | 9.41469955 | 46.65 | 46.65 | −0.460 | 细胞核Nucleus |
| OsIDD4 | 2 | 1848 | 615 | 63597.71 | 8.63129997 | 46.29 | 46.29 | −0.497 | 细胞核Nucleus |
| OsIDD5 | 7 | 1902 | 633 | 67636.09 | 6.01849985 | 55.08 | 55.08 | −0.567 | 细胞核Nucleus |
| OsIDD6 | 8 | 1659 | 552 | 57262.74 | 9.38479996 | 49.81 | 49.81 | −0.500 | 细胞核Nucleus |
| OsIDD7 | 2 | 1479 | 492 | 54491.89 | 7.31449986 | 59.23 | 59.23 | −0.697 | 细胞核Nucleus |
| OsIDD8 | 1 | 1488 | 495 | 51394.20 | 8.89400005 | 49.83 | 49.83 | −0.293 | 细胞核Nucleus |
| OsIDD9 | 1 | 1431 | 476 | 50107.14 | 7.95529985 | 52.62 | 52.62 | −0.265 | 细胞核Nucleus |
| OsIDD10 | 4 | 1842 | 613 | 63814.37 | 9.93620014 | 52.40 | 52.4 | −0.476 | 细胞核Nucleus |
| OsIDD11 | 1 | 1614 | 537 | 57410.90 | 7.97130013 | 62.67 | 62.67 | −0.703 | 细胞核Nucleus |
| OsIDD12 | 8 | 1602 | 533 | 56426.28 | 9.51509953 | 62.47 | 62.47 | −0.467 | 细胞核Nucleus |
| OsIDD13 | 9 | 1515 | 504 | 53574.19 | 8.50090027 | 62.67 | 62.67 | −0.459 | 细胞核Nucleus |
| OsIDD14 | 3 | 1317 | 438 | 45959.66 | 10.12440010 | 70.46 | 70.46 | −0.509 | 细胞核Nucleus |
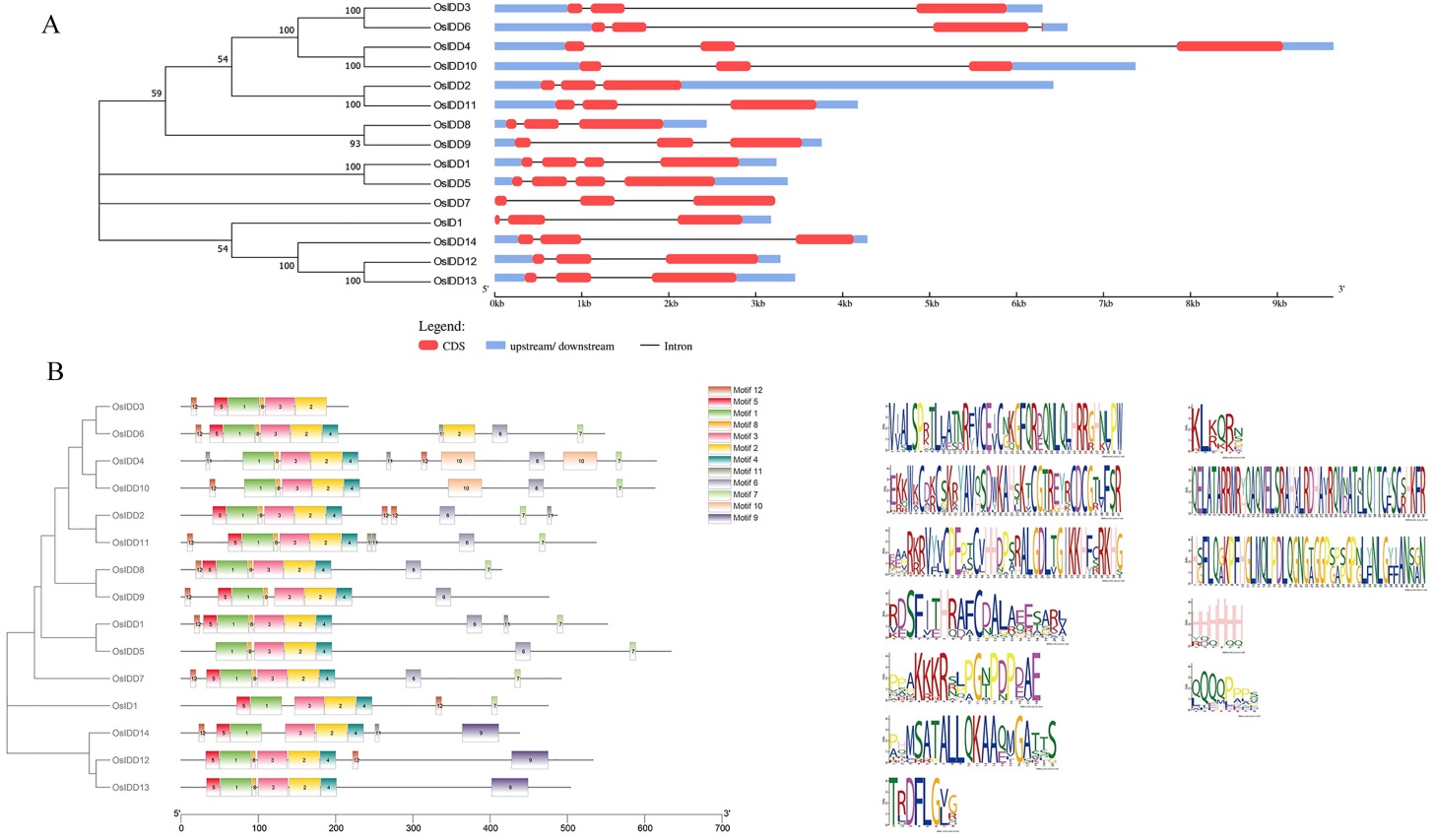
图2 OsIDD基因结构与Motif分析 A:OsIDD基因结构;B:OsIDD蛋白Motif分析。
Fig. 2. Schematic graph of OsIDD gene structure and OsIDD Motif analysis A, Analysis of gene structure for OsIDD family; B, Analysis of OsIDD-protein motif domain.

图3 OsIDD蛋白序列比对 蓝框表明核定位信号区域(NLS);红框表明锌指蛋白结构域(代表C、H残基)。
Fig. 3. Sequence alignment of OsIDD proteins The blue boxes indicate nuclear localization signal region (NLS); the red boxes indicate zinc finger domains (C, H residues).
| 基因名称 Gene name | α-螺旋 α-helix | β-折叠 β-bridge | 延伸链 Extended strand | β-转角 β-turn | 无规则卷曲 Random coil |
|---|---|---|---|---|---|
| OsID1 | 92 (22.49%) | 0 | 89 (21.76%) | 33 (8.07%) | 195 (47.68%) |
| OsIDD1 | 78 (14.13%) | 0 | 66 (11.96%) | 31 (5.62%) | 377 (68.30%) |
| OsIDD2 | 149 (30.60%) | 0 | 42 (8.62%) | 32 (6.57%) | 264 (54.21%) |
| OsIDD3 | 97 (18.13%) | 0 | 71 (13.27%) | 30 (5.61%) | 337 (62.99%) |
| OsIDD4 | 182 (29.59%) | 0 | 77 (12.52%) | 48 (7.80%) | 308 (50.08%) |
| OsIDD5 | 132 (20.85%) | 0 | 97 (15.32%) | 50 (7.90%) | 354 (55.92%) |
| OsIDD6 | 150 (27.17%) | 0 | 87 (15.76%) | 48 (8.70%) | 267 (48.37%) |
| OsIDD7 | 99 (20.12%) | 0 | 48 (9.76%) | 16 (3.25%) | 329 (66.87%) |
| OsIDD8 | 147 (29.70%) | 0 | 64 (12.93%) | 42 (8.48%) | 242 (48.89%) |
| OsIDD9 | 112 (23.53%) | 0 | 68 (14.29%) | 45 (9.45%) | 251 (52.73%) |
| OsIDD10 | 172 (28.06%) | 0 | 82 (13.38%) | 37 (6.04%) | 322 (52.53%) |
| OsIDD11 | 195 (36.31%) | 0 | 58 (10.80%) | 46 (8.57%) | 238 (44.32%) |
| OsIDD12 | 200 (37.52%) | 0 | 69 (12.95%) | 33 (6.19%) | 231 (43.34%) |
| OsIDD13 | 225 (44.64%) | 0 | 40 (7.94%) | 13 (2.58%) | 226 (44.84%) |
| OsIDD14 | 190 (43.38%) | 0 | 41 (9.36%) | 35 (7.99%) | 172 (39.27%) |
表3 OsIDD蛋白的二级结构预测分析
Table 3. Secondary structure prediction analysis of the OsIDD proteins
| 基因名称 Gene name | α-螺旋 α-helix | β-折叠 β-bridge | 延伸链 Extended strand | β-转角 β-turn | 无规则卷曲 Random coil |
|---|---|---|---|---|---|
| OsID1 | 92 (22.49%) | 0 | 89 (21.76%) | 33 (8.07%) | 195 (47.68%) |
| OsIDD1 | 78 (14.13%) | 0 | 66 (11.96%) | 31 (5.62%) | 377 (68.30%) |
| OsIDD2 | 149 (30.60%) | 0 | 42 (8.62%) | 32 (6.57%) | 264 (54.21%) |
| OsIDD3 | 97 (18.13%) | 0 | 71 (13.27%) | 30 (5.61%) | 337 (62.99%) |
| OsIDD4 | 182 (29.59%) | 0 | 77 (12.52%) | 48 (7.80%) | 308 (50.08%) |
| OsIDD5 | 132 (20.85%) | 0 | 97 (15.32%) | 50 (7.90%) | 354 (55.92%) |
| OsIDD6 | 150 (27.17%) | 0 | 87 (15.76%) | 48 (8.70%) | 267 (48.37%) |
| OsIDD7 | 99 (20.12%) | 0 | 48 (9.76%) | 16 (3.25%) | 329 (66.87%) |
| OsIDD8 | 147 (29.70%) | 0 | 64 (12.93%) | 42 (8.48%) | 242 (48.89%) |
| OsIDD9 | 112 (23.53%) | 0 | 68 (14.29%) | 45 (9.45%) | 251 (52.73%) |
| OsIDD10 | 172 (28.06%) | 0 | 82 (13.38%) | 37 (6.04%) | 322 (52.53%) |
| OsIDD11 | 195 (36.31%) | 0 | 58 (10.80%) | 46 (8.57%) | 238 (44.32%) |
| OsIDD12 | 200 (37.52%) | 0 | 69 (12.95%) | 33 (6.19%) | 231 (43.34%) |
| OsIDD13 | 225 (44.64%) | 0 | 40 (7.94%) | 13 (2.58%) | 226 (44.84%) |
| OsIDD14 | 190 (43.38%) | 0 | 41 (9.36%) | 35 (7.99%) | 172 (39.27%) |
| 基因名称Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number |
|---|---|---|---|---|---|---|---|
| OsID1 | LOC_Os10g28330 | AtIDD1 | At5G66730 | AtIDD16 | AT1G25250 | ZmIDD14 | GRMZM2G141031 |
| OsIDD1 | LOC_Os03g10140 | AtIDD2 | At3G50700 | ZmID1 | GRMZM2G011357 | ZmIDD15 | GRMZM2G123094 |
| OsIDD2 | LOC_Os01g09850 | AtIDD3 | At1G03840 | ZmIDD1 | GRMZM2G171073 | ZmIDD16 | GRMZM2G074032 |
| OsIDD3 | LOC_Os09g38340 | AtIDD4 | AT2G02080 | ZmIDD2 | GRMZM2G143723 | ZmIDD17 | GRMZM2G110107 |
| OsIDD4 | LOC_Os02g45054 | AtIDD5 | AT2G02070 | ZmIDD3 | GRMZM5G828179 | ZmIDD18 | GRMZM2G465595 |
| OsIDD5 | LOC_Os07g39310 | AtIDD6 | AT1G14580 | ZmIDD4 | GRMZM2G151309 | ZmIDDp1 | GRMZM2G179677 |
| OsIDD6 | LOC_Os08g44050 | AtIDD7 | At1G55110 | ZmIDD5 | GRMZM2G046290 | ZmIDDveg7 | GRMZM2G042666 |
| OsIDD7 | LOC_Os02g31890 | AtIDD8 | At5G44160 | ZmIDD6 | GRMZM2G035625 | ZmIDDveg9 | GRMZM2G129261 |
| OsIDD8 | LOC_Os01g14010 | AtIDD9 | At3G45260 | ZmIDD7 | GRMZM2G320287 | ZmIDDp10 | GRMZM2G090595 |
| OsIDD9 | LOC_Os01g70870 | AtIDD10 | At5G03150 | ZmIDD8 | GRMZM2G022213 | ||
| OsIDD10 | LOC_Os04g47860 | AtIDD11 | At3G13810 | ZmIDD9 | GRMZM5G884137 | ||
| OsIDD11 | LOC_Os01g39110 | AtIDD12 | At4G02670 | ZmIDD10 | GRMZM2G058197 | ||
| OsIDD12 | LOC_Os08g36390 | AtIDD13 | At5G60470 | ZmIDD11 | GRMZM2G177693 | ||
| OsIDD13 | LOC_Os09g27650 | AtIDD14 | AT1G68130 | ZmIDD12 | GRMZM2G027333 | ||
| OsIDD14 | LOC_Os03g13400 | AtIDD15 | AT2G01940 | ZmIDD13 | GRMZM2G021587 |
表4 水稻、拟南芥和玉米IDD基因成员汇总
Table 4. Summary of the IDD gene members in Oryza sativa, Arabidopsis, and Zea mays
| 基因名称Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number |
|---|---|---|---|---|---|---|---|
| OsID1 | LOC_Os10g28330 | AtIDD1 | At5G66730 | AtIDD16 | AT1G25250 | ZmIDD14 | GRMZM2G141031 |
| OsIDD1 | LOC_Os03g10140 | AtIDD2 | At3G50700 | ZmID1 | GRMZM2G011357 | ZmIDD15 | GRMZM2G123094 |
| OsIDD2 | LOC_Os01g09850 | AtIDD3 | At1G03840 | ZmIDD1 | GRMZM2G171073 | ZmIDD16 | GRMZM2G074032 |
| OsIDD3 | LOC_Os09g38340 | AtIDD4 | AT2G02080 | ZmIDD2 | GRMZM2G143723 | ZmIDD17 | GRMZM2G110107 |
| OsIDD4 | LOC_Os02g45054 | AtIDD5 | AT2G02070 | ZmIDD3 | GRMZM5G828179 | ZmIDD18 | GRMZM2G465595 |
| OsIDD5 | LOC_Os07g39310 | AtIDD6 | AT1G14580 | ZmIDD4 | GRMZM2G151309 | ZmIDDp1 | GRMZM2G179677 |
| OsIDD6 | LOC_Os08g44050 | AtIDD7 | At1G55110 | ZmIDD5 | GRMZM2G046290 | ZmIDDveg7 | GRMZM2G042666 |
| OsIDD7 | LOC_Os02g31890 | AtIDD8 | At5G44160 | ZmIDD6 | GRMZM2G035625 | ZmIDDveg9 | GRMZM2G129261 |
| OsIDD8 | LOC_Os01g14010 | AtIDD9 | At3G45260 | ZmIDD7 | GRMZM2G320287 | ZmIDDp10 | GRMZM2G090595 |
| OsIDD9 | LOC_Os01g70870 | AtIDD10 | At5G03150 | ZmIDD8 | GRMZM2G022213 | ||
| OsIDD10 | LOC_Os04g47860 | AtIDD11 | At3G13810 | ZmIDD9 | GRMZM5G884137 | ||
| OsIDD11 | LOC_Os01g39110 | AtIDD12 | At4G02670 | ZmIDD10 | GRMZM2G058197 | ||
| OsIDD12 | LOC_Os08g36390 | AtIDD13 | At5G60470 | ZmIDD11 | GRMZM2G177693 | ||
| OsIDD13 | LOC_Os09g27650 | AtIDD14 | AT1G68130 | ZmIDD12 | GRMZM2G027333 | ||
| OsIDD14 | LOC_Os03g13400 | AtIDD15 | AT2G01940 | ZmIDD13 | GRMZM2G021587 |
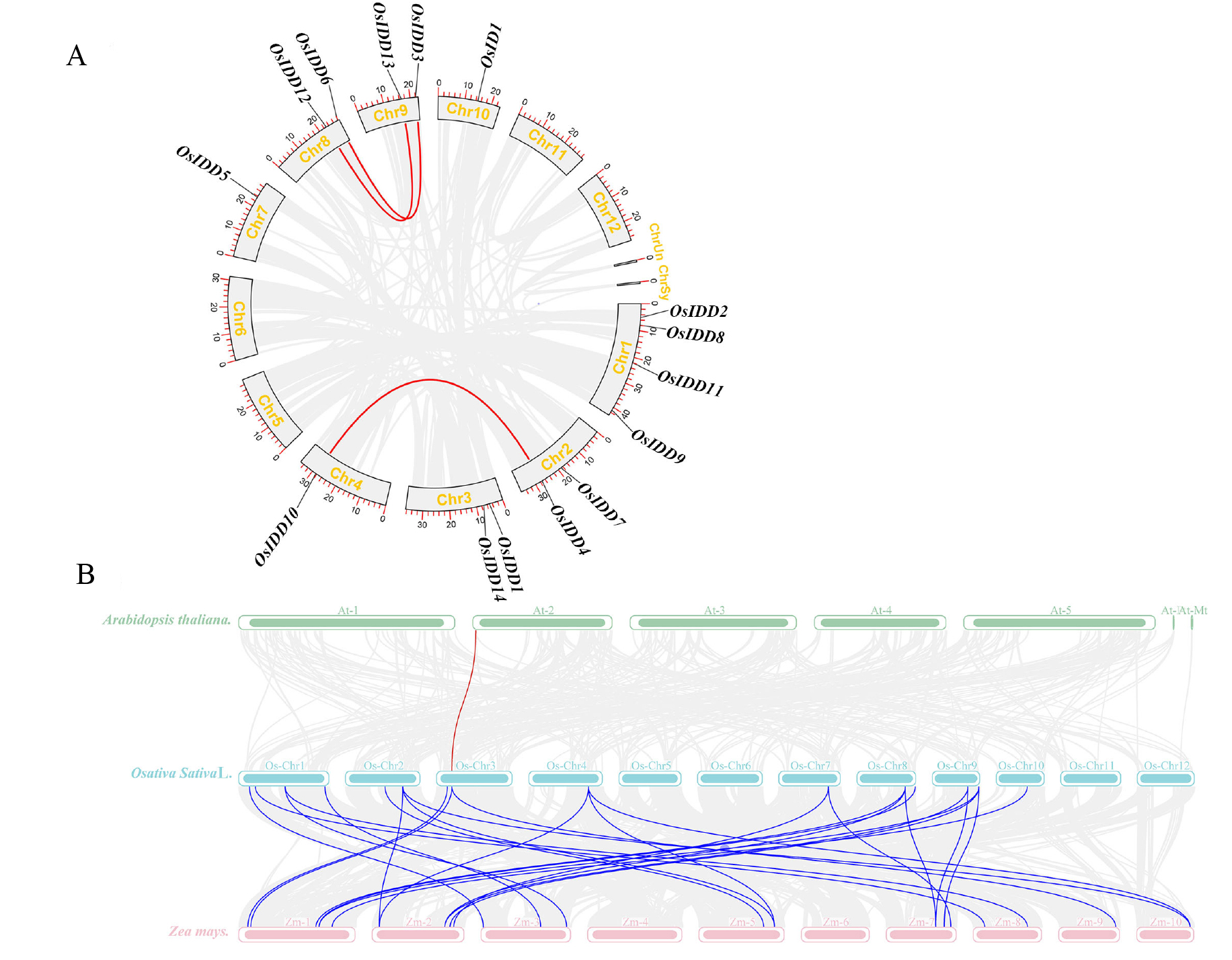
图6 水稻IDD基因的复制关系及其与拟南芥、玉米IDD基因的共线性分析 A:水稻IDDs基因的复制关系;B:水稻IDDs与拟南芥、玉米IDDs基因的共线性分析;线段代表了片段复制基因对。
Fig. 6. Segmental gene pairs of OsIDD genes and collinearity analysis of IDD gene with Arabidopsis and Zea mays A, Segmental gene pairs of OsIDDs genes; B, Collinearity analysis of OsIDDs and AtIDDs, ZmIDDs genes; Lines represents segmental gene pairs.
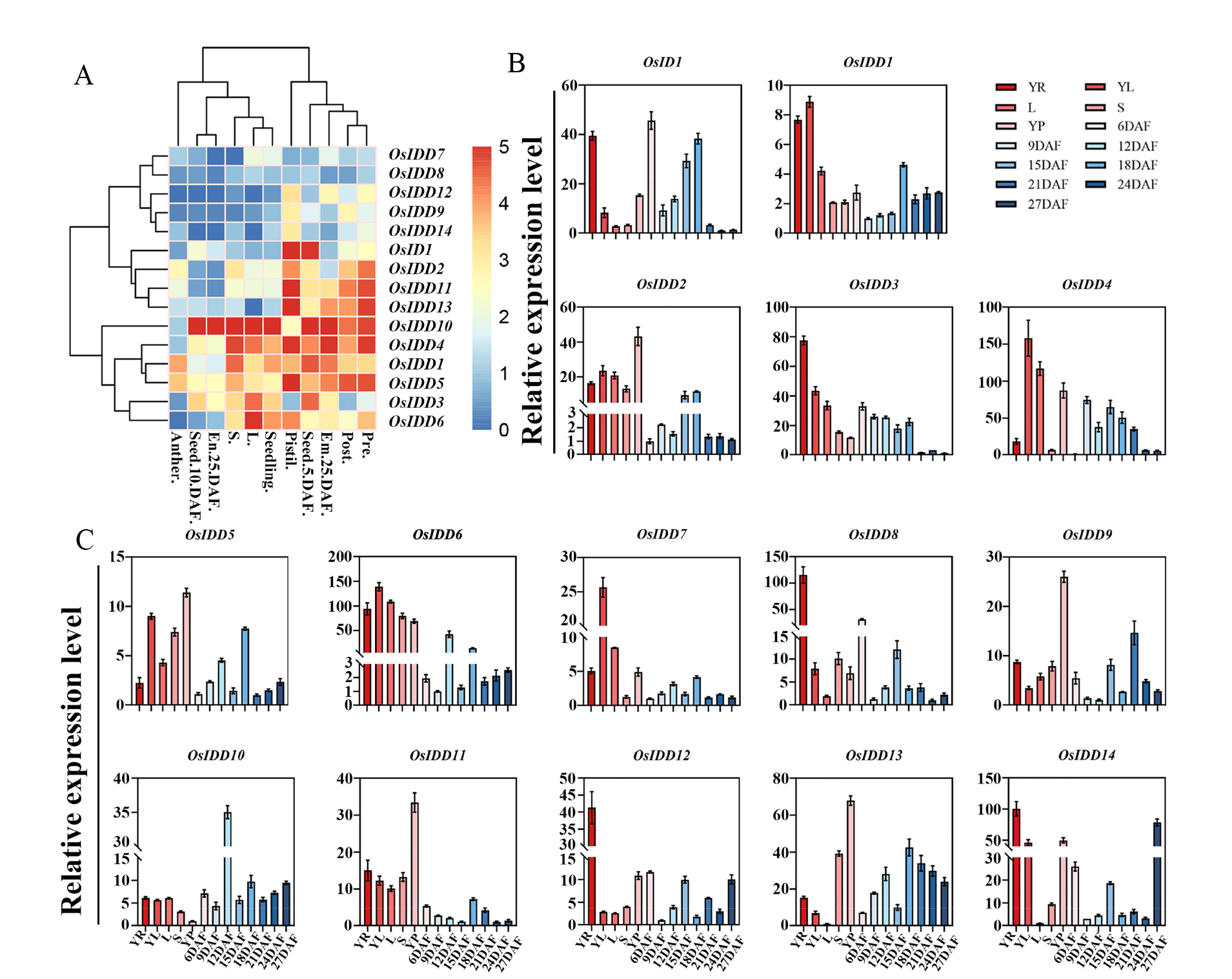
图7 OsIDD基因的表达模式 A:OsIDDs芯片数据表达量热图;B~C:OsIDDs在水稻各组织中的表达量;YR: 三叶期的根;YL:三叶期的叶;L:孕穗期的叶;S:孕穗期的茎;YP:幼穗;6DAF、9DAF、12DAF、15DAF、18DAF、21DAF和24DAF分别代表发育6 d, 9 d, 12 d, 15 d, 18 d, 21 d和24 d的籽粒;27DAF:发育27 d的籽粒。数据为3次生物学重复的平均值±标准差。
Fig. 7. Expression pattern of OsIDD A, OsIDDs expression level thermogram; B~C, OsIDDs expression level in rice tissues; YR, Root at seedling stage with three fully expanded leaves; YL; Leaves at seedling stage with three fully expanded leaves; L, Leaf at booting stage; S, Stem at booting stage; YP, Young spikelet; 6DAF, Developed endosperm for 6 days; 9DAF, Developed endosperm for 9 days; 12DAF, Developed endosperm for 12 days; 15DAF, Developed endosperm for 15 days; 18DAF, Developed endosperm for 18 days; 21DAF, Developed endosperm for 21 days; 24DAF, Developed endosperm for 24 days; 27DAF, Developed endosperm for 27 days. Data are mean ± SD from three biological replicates.
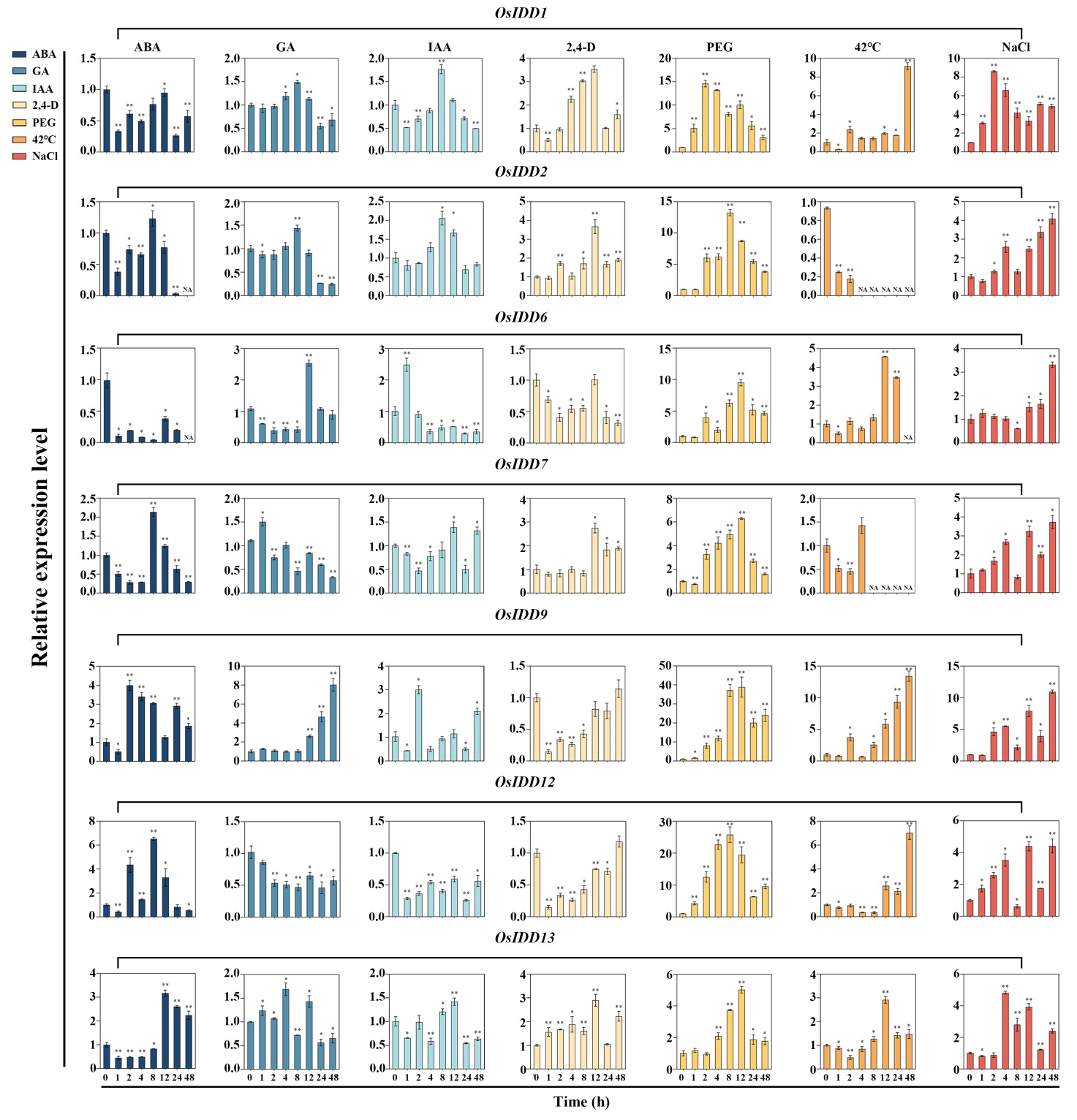
图9 OsIDD基因(部分)在不同激素处理及不同胁迫条件下的表达分析 ABA、GA、IAA、2, 4-D为不同激素处理;PEG、42℃、NaCl为不同胁迫处理;数据是来自三组生物学重复的平均值±标准差(n=3);顶部的星号表示与 0 h 相比差异显著(*,P<0.05;**,P<0.01)。
Fig. 9. Expression analysis of OsIDD genes (part) under different hormone treatments and different stress conditions ABA, GA, IAA, 2, 4-D were different hormones treatments; PEG, 42℃, and NaCl were different stress conditions. Mean ± SD (n=3); The asterisks on the top of the columns indicate significant differences from the value at 0 h (*, P < 0.05; **, P < 0.01).
| [1] | 孙燕, 苟德明, 李文鑫. C2H2型锌指蛋白研究进展[J]. 生命的化学, 2001 (6): 473-475. |
| Sun Y, Gou D M, Li W X. Advances in the study of C2H2 type zinc finger proteins[J]. Chemistry of Life, 2001(6): 473-475. (in Chinese with English abstract) | |
| [2] | Macpherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins[J]. Microbiology and Molecular Biology Reviews, 2006, 70(3): 583-604. |
| [3] | Lee M S, Gippert G P, Soman K V, Case D A, Wright P E. Three-dimensional solution structure of a single zinc finger DNA-binding domain[J]. Science, 1989, 245(4918): 635-637. |
| [4] | Párraga G, Horvath S J, Eisen A, Taylor W E, Hood L, Young E T, Klevit R E. Zinc-dependent structure of a single-finger domain of yeast ADR1[J]. Science, 1988, 241(4872): 1489-1492. |
| [5] | Xu Q K, Yu H P, Xia S S, Cui Y J, Yu X Q, Liu H, Zeng D L, Hu J, Zhang Q, Gao Z Y, Zhang G H, Zhu L, Shen L, Guo L B, Rao Y C, Qian Q, Ren D Y. The C2H2 zinc-finger protein LACKING RUDIMENTARY GLUME 1 regulates spikelet development in rice[J]. Science Bulletin, 2020, 65(9): 753-764. |
| [6] | 侯思宇, 孙朝霞, 郭彬, 王玉国, 李贵全, 韩渊怀. 大豆两个C2H2型转录因子基因序列特征及表达分析[J]. 植物生理学报, 2014, 50(5): 665-674. |
| Hou S Y, Sun Z X, Guo B, Wang Y G, Li G Q, Han Y H. Cloning and expression analysis of two C2H2 transcription factors in soybean[J]. Plant Physiology Journal, 2014, 50(5): 665-674. (in Chinese with English abstract) | |
| [7] | 孟繁君, 陈明, 徐长营, 朴秀吉, 汪可心, 葛善欣, 金玄吉. 玉米C2H2型锌指蛋白基因ZFP225的鉴定、生物信息学分析与克隆[J]. 作物杂志, 2014 (1): 49-53. |
| Meng F J, Chen M, Xu C Y, Piao X J, Wang K X, Ge S X, Jin X J. Identification, bioinformatic analysis and cloning of type C2H2 zinc finger protein gene ZFP225 in maize[J]. Crops, 2014(1): 49-53. (in Chinese with English abstract) | |
| [8] | Sakamoto H, Araki T, Meshi T, Iwabuchi M. Expression of a subset of the Arabidopsis Cys(2)/His(2)-type zinc-finger protein gene family under water stress[J]. Gene, 2000, 248(1-2): 23-32. |
| [9] | Park S J, Kim S L, Lee S, Je B I, Piao H L, Park S H, Kim C M, Ryu C H, Park S H, Xuan Y H, Colasanti J, An G, Han C D. Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod[J]. The Plant Journal, 2008, 56(6): 1018-1029. |
| [10] | Matsubara K, Yamanouchi U, Wang Z X, Minobe Y, Izawa T, Yano M. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1[J]. Plant Physiology, 2008, 148(3): 1425-1435. |
| [11] | Colasanti J, Tremblay R, Wong A Y, Coneva V, Kozaki A, Mable B K. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants[J]. BMC Genomics, 2006, 7: 158. |
| [12] | Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize[J]. Cell, 1998, 93(4): 593-603. |
| [13] | Wu C Y, You C J, Li C S, Long T, Chen G X, Byrne M E, Zhang Q F. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(35): 12915-12920. |
| [14] | Coneva V, Guevara D, Rothstein S J, Colasanti J. Transcript and metabolite signature of maize source leaves suggests a link between transitory starch to sucrose balance and the autonomous floral transition[J]. Journal of Experimental Botany, 2012, 63(2): 5079-5092. |
| [15] | Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions[J]. Plant Physiology, 2004, 136(1): 2734-2746. |
| [16] | Gontarek B C, Neelakandan A K, Wu H, Becraft P W. NKD transcription factors are central regulators of maize endosperm development[J]. The Plant Cell, 2016, 28(12): 2916-2936. |
| [17] | Coelho C P, Huang P, Lee D Y, Brutnell T P. Making roots, shoots, and seeds: IDD gene family diversification in plants[J]. Trends in Plant Science, 2018, 23(1): 66-78. |
| [18] | Morita M T, Sakaguchi K, Kiyose S I, Taira K, Kato T, Nakamura M, Tasaka M. A C2H2‐type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems[J]. The Plant Journal, 2006, 47(4): 619-628. |
| [19] | Cui D Y, Zhao J B, Jing Y J, Fan M Z, Liu J, Wang Z C, Xin W, Hu Y X. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport[J]. PLoS Genetics, 2013, 9(9): e1003759. |
| [20] | Feurtado J A, Huang D, Wicki-Stordeur L, Hemstock L E, Potentier M S, Tsang E W, Cutler A J. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation[J]. The Plant Cell, 2011, 23(5): 1772-1794. |
| [21] | Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson A G, Escobar M A. Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants[J]. Plant, Cell & Environment, 2010, 33(9): 1486-1501. |
| [22] | Seo P J, Ryu J, Kang S K, Park C M. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis[J]. The Plant Journal, 2011, 65(3): 418-429. |
| [23] | Matsubara K, Yamanouchi U, Wang Z X, Minobe Y, Izawa T, Yano M. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1[J]. Plant Physiology, 2008, 148(3): 1425-1435. |
| [24] | Deng L, Li L M, Zhang S, Shen J Q, Li S B, Hu S F, Peng Q, Xiao J H, Wu C Y. Suppressor of rid1 (SID1) shares common targets with RID1 on florigen genes to initiate floral transition in rice[J]. PLOS Genetics, 2017, 13(2): e1006642. |
| [25] | Dou M Z, Cheng S, Zhao B T, Xuan Y H, Shao M L. The Indeterminate Domain Protein ROC1 regulates chilling tolerance via activation of DREB1B/CBF1 in rice[J]. International Journal of Molecular Sciences, 2016, 17(3): 233. |
| [26] | Liu J M, Park S J, Huang J, Lee E J, Xuan Y H, Je B I, Kumar V, Priatama R A, Raj K V, Kim S H, Min M K, Cho J H, Kim T H, Chandran A K, Jung K H, Takatsuto S, Fujioka S, Han C D. Loose Plant Architecture1 (LPA1) determines lamina joint bending by suppressing auxin signalling that interacts with C-22-hydroxylated and 6-deoxo brassinosteroids in rice[J]. Journal of Experimental Botany, 2016, 67(6): 1883-1895. |
| [27] | Wu X R, Tang D, Li M, Wang K J, Cheng Z K. Loose Plant Architecture1, an INDETERMINATE DOMAIN Protein involved in shoot gravitropism, regulates plant architecture in rice[J]. Plant Physiology, 2013, 161(1): 317-329. |
| [28] | Cui Z B, Xue C Y, Mei Q, Xuan Y H. Malectin Domain Protein Kinase (MDPK) promotes rice resistance to sheath blight via IDD12, IDD13, and IDD14[J]. International Journal of Molecular Sciences, 2022, 23(15): 8214. |
| [29] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using realtime quantitative pcr and the 2-△△CT method[J]. Methods, 2001, 25(4): 402-408. |
| [30] | Malheiros R S P, Costa L C, Ávila R T, Pimenta T M, Teixeira L S, Brito F A L, Zsögön A, Araújo W L, Ribeiro D M. Selenium downregulates auxin and ethylene biosynthesis in rice seedlings to modify primary metabolism and root architecture[J]. Planta, 2019, 250(1): 333-345. |
| [31] | Fu Z Z, Yu J, Cheng X W, Zong X, Xu J, Chen M J, Li Z Y, Zhang D B, Liang W Q. The rice Basic Helix-Loop- Helix Transcription Factor TDR INTERACTING PROTEIN2 is a central switch in early anther development[J]. The Plant Cell, 2014, 26(4): 1512-1524. |
| [32] | Ko S S, Li M J, Sun-Ben Ku M, Ho Y C, Lin Y J, Chuang M H, Hsing H X, Lien Y C, Yang H T, Chang H C, Chan M T. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice[J]. The Plant Cell, 2014, 26(6): 2486-2504. |
| [33] | 杜逸凡, 方希林, 曾红丽, 于玉凤, 张露倩, 王悦. 外源多效唑, ABA, 2, 4-D和NAA对水稻秧苗生长特性的影响[J]. 分子植物育种, 2020, 18(8): 2687-2694. |
| Du Y F, Fang X L, Zeng H L, Yu Y F, Zhang L Q, Wang Y. Effects of exogenous Paclobutrazol, ABA, 2, 4-D, and NAA on growth characteristics of rice seedlings[J]. Molecular Plant Breeding, 18(8): 2687-2694. (in Chinese with English abstract) | |
| [34] | Huang S Y, Liu M M, Chen G L, Si F F, Fan F F, Guo Y, Yuan L, Yang F, Li S Q. Favorable QTLs from Oryza longistaminata improve rice drought resistance[J]. BMC Plant Biology, 2022, 22(1): 136. |
| [35] | Song Y, Zhang C J, Ge W N, Zhang Y F, Burlingame A L, Guo Y. Identification of NaCl stress-responsive apoplastic proteins in rice shoot stems by 2D-DIGE[J]. Journal of Proteomics, 2011, 74(7): 1045-1067. |
| [36] | Kozaki A, Hake S, Colasanti J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties[J]. Nucleic Acids Research, 2004, 32(5): 1710-1720. |
| [37] | Seo P J, Kim M J, Ryu J Y, Jeong E Y, Park C M. Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism[J]. Nature Communications, 2011, 2: 303. |
| [38] | Sun Q, Li T Y, Li D D, Wang Z Y, Li S, Li D P, Han X, Liu J M, Xuan Y H. Overexpression of Loose Plant Architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN‐FORMED 1a in rice[J]. Plant Biotechnology Journal, 2019, 17(5): 855-857. |
| [39] | Lu Y Z, Feng Z, Meng Y L, Bian L Y, Xie H, Mysore K S, Liang J S. SLENDER RICE1 and Oryza sativa INDETERMINATE DOMAIN 2 regulating OsmiR396 are involved in stem elongation[J]. Plant Physiology, 2020, 182(4): 2213-2227. |
| [1] | 随晶晶, 赵桂龙, 金欣, 卜庆云, 唐佳琦. 水稻孕穗期耐冷调控的分子及生理机制研究进展 [J]. 中国水稻科学, 2025, 39(1): 1-10. |
| [2] | 任宁宁, 孙永建, 申聪聪, 朱双兵, 李慧菊, 张志远, 陈凯. 水稻中胚轴研究进展 [J]. 中国水稻科学, 2025, 39(1): 11-23. |
| [3] | 张丰勇, 应晓平, 张健, 杨隆维, 应杰政. 半矮秆基因sd1调控水稻重要农艺性状的研究进展 [J]. 中国水稻科学, 2025, 39(1): 24-32. |
| [4] | 陈智慧, 陶亚军, 范方军, 许扬, 王芳权, 李文奇, 古丽娜尔·巴合提别克, 蒋彦婕, 朱建平, 李霞, 杨杰. 水稻抽穗期调控基因Hd6功能标记的开发及应用 [J]. 中国水稻科学, 2025, 39(1): 47-54. |
| [5] | 胡风越, 王健, 王春, 王克剑, 刘朝雷. 水稻DMP1、DMP2、DMP3基因突变体的创制及其单倍体诱导能力鉴定 [J]. 中国水稻科学, 2025, 39(1): 55-66. |
| [6] | 陈书融, 朱练峰, 秦碧蓉, 王婕, 朱旭华, 田文昊, 朱春权, 曹小闯, 孔亚丽, 张均华, 金千瑜. 增氧灌溉下配施硝化抑制剂对水稻生长、产量和氮肥利用的影响 [J]. 中国水稻科学, 2025, 39(1): 92-100. |
| [7] | 吴猛, 倪川, 康钰莹, 毛雨欣, 叶苗, 张祖建. 水稻分蘖早发特性的品种间差异及其氮素响应 [J]. 中国水稻科学, 2025, 39(1): 101-114. |
| [8] | 王晓茜, 蔡创, 宋练, 周伟, 杨雄, 顾歆悦, 朱春梧. 开放式大气CO2浓度升高和温度升高对扬稻6号稻米品质的影响 [J]. 中国水稻科学, 2025, 39(1): 115-127. |
| [9] | 江敏, 王广伦, 李明璐, 苗波, 李明煊, 石春林. 基于模型的水稻高温热害风险评估与动态预警 [J]. 中国水稻科学, 2025, 39(1): 128-142. |
| [10] | 冯向前, 王爱冬, 洪卫源, 李子秋, 覃金华, 詹丽钏, 陈里鹏, 张运波, 王丹英, 陈松. 基于低空无人机遥感的水稻产量估测方法研究进展[J]. 中国水稻科学, 2024, 38(6): 604-616. |
| [11] | 叶苗, 毛雨欣, 张德海, 康钰莹, 袁榕, 张祖建. 高光效水稻品种的叶片和冠层生理生态特征及其氮素调控机制研究进展[J]. 中国水稻科学, 2024, 38(6): 617-626. |
| [12] | 汪晴, 王艳茹, 张秀丽, 吕启明. 水稻孤雌生殖诱导基因BBM1序列变异分析[J]. 中国水稻科学, 2024, 38(6): 627-637. |
| [13] | 杜彦修, 孙文玉, 袁泽科, 张倩倩, 李富豪, 李俊周, 孙红正. 利用QTL-Seq结合分子标记定位粳稻垩白粒率控制位点qChalk8[J]. 中国水稻科学, 2024, 38(6): 665-671. |
| [14] | 毋翔, 张义凯, 张鹏, 马昕伶, 陈玉林, 陈惠哲, 张玉屏, 向镜, 王亚梁, 王志刚, 李良涛. 2,4-表油菜素内酯对生物炭基质育秧水稻秧苗根系生长及生理特性的影响[J]. 中国水稻科学, 2024, 38(6): 685-694. |
| [15] | 汪邑晨, 朱本顺, 周磊, 朱骏, 杨仲南. 光/温敏核不育系的不育机理及两系杂交稻的发展与展望[J]. 中国水稻科学, 2024, 38(5): 463-474. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||