
中国水稻科学 ›› 2024, Vol. 38 ›› Issue (6): 627-637.DOI: 10.16819/j.1001-7216.2024.231210
汪晴1,2, 王艳茹2,3, 张秀丽1,2, 吕启明1,2,*( )
)
收稿日期:2023-12-10
修回日期:2024-01-10
出版日期:2024-11-10
发布日期:2024-11-15
通讯作者:
*email: qmlv@hhrrc.ac.cn
基金资助:
WANG Qing1,2, WANG Yanru2,3, ZHANG Xiuli1,2, LÜ Qiming1,2,*( )
)
Received:2023-12-10
Revised:2024-01-10
Online:2024-11-10
Published:2024-11-15
Contact:
*email: qmlv@hhrrc.ac.cn
摘要:
【目的】在水稻卵细胞中异位表达BBM1能够诱导孤雌生殖,挖掘BBM1优异等位变异和功能类似基因对水稻杂种优势固定具有重要意义。【方法】利用383份水稻核心种质的三代测序数据,对BBM1基因进行结构变异分析、序列多态性分析、单倍型分析,并根据序列相似度以及表达模式相似度筛选BBM1功能类似基因。【结果】BBM1基因在3份种质中存在两个拷贝,在2份野生稻中存在微型颠倒重复转座元件(MITEs)插入变异;具有非常明显的籼粳分化,94.80%的粳稻和3.80%的籼稻中BBM1因单碱基变异造成翻译提前终止,缺少26个氨基酸残基;BBM1编码区序列十分保守,绝大多数材料中具有相同的单倍型;筛选到BBM1序列和表达模式相似基因LOC_Os06g44750,其可能具有BBM1诱导孤雌生殖的功能。【结论】明确了水稻中BBM1基因变异情况,挖掘出重要单倍型和功能类似候选基因,为进一步提高水稻孤雌生殖频率奠定基础。
汪晴, 王艳茹, 张秀丽, 吕启明. 水稻孤雌生殖诱导基因BBM1序列变异分析[J]. 中国水稻科学, 2024, 38(6): 627-637.
WANG Qing, WANG Yanru, ZHANG Xiuli, LÜ Qiming. Sequence Variation Analysis of the Parthenogeny-inducing Gene BBM1 in Rice[J]. Chinese Journal OF Rice Science, 2024, 38(6): 627-637.
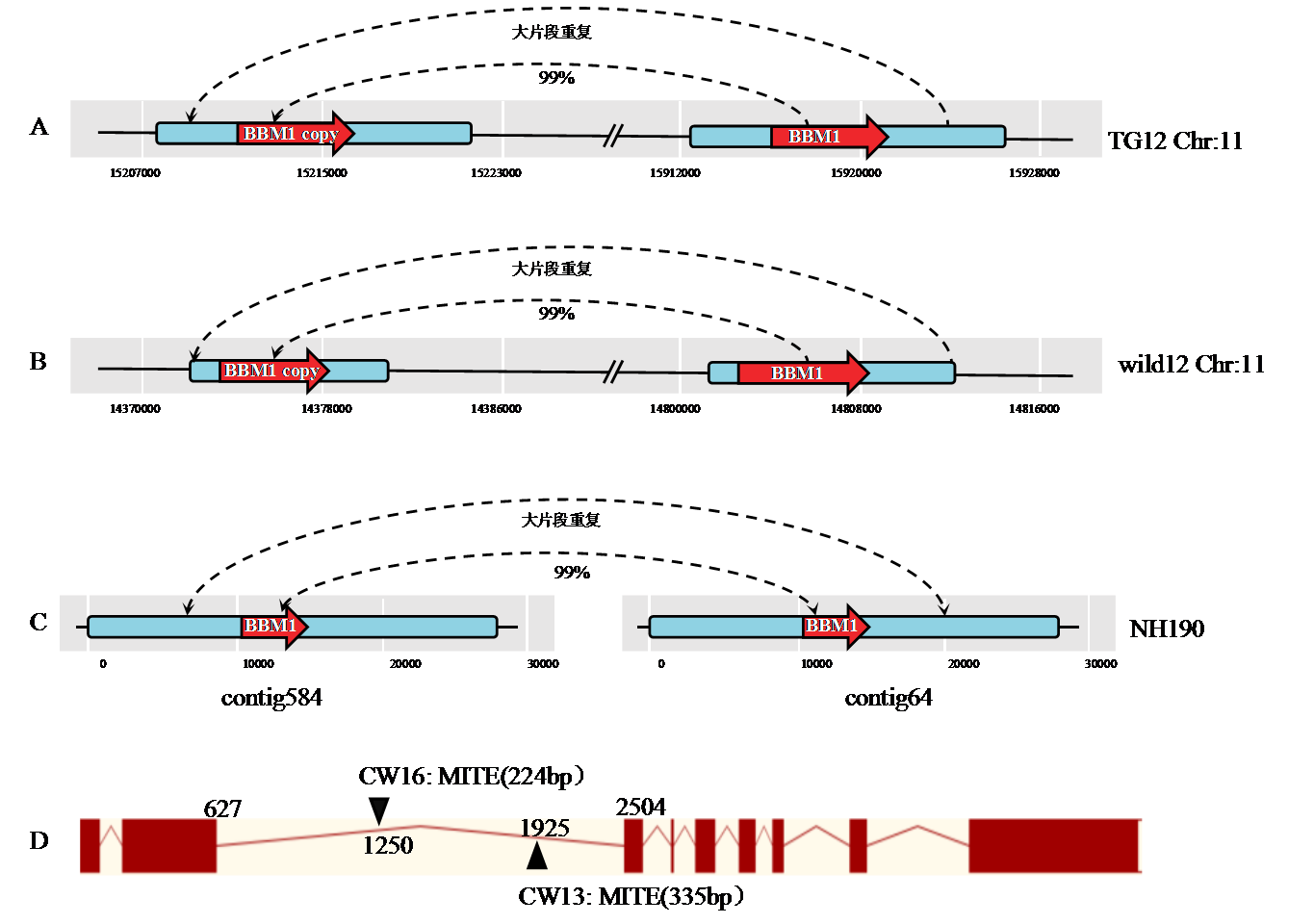
图1 BBM1结构变异 A、B、C分别为TG12、wild12和NH190的BBM1结构变异示意图。其中蓝色代表BBM1侧翼大片段重复序列,红色代表BBM1基因序列。D: CW13和CW16中BBM1基因结构图。其中酒红色代表外显子,黄色代表内含子,▲代表MITE插入位点。
Fig. 1. Structural variation of BBM1 A, B, C show the BBM1 structural variants of TG12, wild12 and NH190, blue represents the BBM1 flanking large fragment repeat sequence and red represents the BBM1 gene sequence. D: Map of BBM1 gene structure of CW13 and CW16, burgundy represents exons, yellow represents introns, and ▲ represents MITE insertion sites.

图2 野生稻、籼稻和粳稻中BBM1序列核苷酸多态性 A、B、C分别表示野生稻、籼稻和粳稻中BBM1序列核苷酸多性。酒红色表示BBM1外显子,橙色表示BBM1内含子。
Fig. 2. Nucleotide diversity of BBM1 sequences in different rice accessions A, B and C show the nucleotide polymorphisms of BBM1 sequence in wild rice, indica and japonica rice, respectively. The BBM1 exon is shown in burgundy and the BBM1 intron in orange.
| 水稻类型 Rice type | 对比序列数Number of sequences used | 多态性位点Number of polymorphic (segregating) sites | 突变总数 Total number of mutations | 单倍体数Number of haplotypes | 单倍型多样性Haplotype (gene) diversity | 单倍型多态性 方差 Variance of haplotype diversity | 单倍型多态性 标准差 Standard deviation of haplotype diversity | 核苷酸多态性Nucleotide diversity |
|---|---|---|---|---|---|---|---|---|
| 核心种质 Core collections | 383 | 268 | 285 | 69 | 0.868 | 0.0001 | 0.010 | 0.00506 |
| 野生稻 Wild rice | 30 | 196 | 200 | 29 | 0.998 | 0.00009 | 0.009 | 0.00623 |
| 籼稻 indica | 204 | 91 | 94 | 22 | 0.786 | 0.00045 | 0.021 | 0.00333 |
| 粳稻 japonica | 103 | 77 | 79 | 11 | 0.318 | 0.00359 | 0.060 | 0.00131 |
表1 不同水稻材料中BBM1序列多态性分析
Table 1. Analysis of BBM1 sequence polymorphism in different rice accessions
| 水稻类型 Rice type | 对比序列数Number of sequences used | 多态性位点Number of polymorphic (segregating) sites | 突变总数 Total number of mutations | 单倍体数Number of haplotypes | 单倍型多样性Haplotype (gene) diversity | 单倍型多态性 方差 Variance of haplotype diversity | 单倍型多态性 标准差 Standard deviation of haplotype diversity | 核苷酸多态性Nucleotide diversity |
|---|---|---|---|---|---|---|---|---|
| 核心种质 Core collections | 383 | 268 | 285 | 69 | 0.868 | 0.0001 | 0.010 | 0.00506 |
| 野生稻 Wild rice | 30 | 196 | 200 | 29 | 0.998 | 0.00009 | 0.009 | 0.00623 |
| 籼稻 indica | 204 | 91 | 94 | 22 | 0.786 | 0.00045 | 0.021 | 0.00333 |
| 粳稻 japonica | 103 | 77 | 79 | 11 | 0.318 | 0.00359 | 0.060 | 0.00131 |

图5 BBM1结构域示意图 BBM1含有两个AP2结构域,红色代表AP2结构域,蓝色代表氨基酸序列,数字代表氨基酸位置。
Fig. 5. Schematic representation of the BBM1 domain BBM1 contains two AP2 domains, the AP2 domain in red, the amino acid sequence in blue, and the amino acid position in numbers.
| 单倍型 Haplotype | 数量 Number | 16 | 67 | 69 | 107 | 124 | 142 | 146 | 147 | 147.1 | 258 | 351 | 445 | 493 | 521 | 537 | 561 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap-1 | 109 | S | T | A | M | V | G | A | A | - | R | T | T | P | D | L | stop |
| Hap-36 | 92 | S | T | A | M | A | D | A | A | - | R | I | T | P | D | V | +26AA |
| Hap-37 | 51 | S | M | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-22 | 29 | S | T | A | M | A | G | A | A | - | R | T | T | Q | D | V | +26AA |
| Hap-17 | 17 | S | T | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-40 | 12 | S | T | A | M | A | D | A | A | - | R | I | I | P | D | V | +26AA |
| Hap-4 | 10 | S | T | A | M | A | G | - | - | - | R | T | T | P | N | V | +26AA |
| Hap-6 | 5 | S | T | A | M | A | G | - | - | - | R | T | T | P | D | V | +26AA |
| Hap-43 | 4 | Y | T | D | M | V | G | A | A | A | R | T | T | P | D | L | stop |
| Hap-23 | 3 | S | T | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-8 | 2 | S | T | A | M | A | G | - | - | - | \ | \ | \ | \ | \ | \ | \ |
| Hap-44 | 2 | S | T | A | T | A | G | A | A | - | R | T | T | P | D | V | +26AA |
表2 不同单倍型中非同义突变位点
Table 2. Nonsynonymous mutation sites in different haplotypes
| 单倍型 Haplotype | 数量 Number | 16 | 67 | 69 | 107 | 124 | 142 | 146 | 147 | 147.1 | 258 | 351 | 445 | 493 | 521 | 537 | 561 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap-1 | 109 | S | T | A | M | V | G | A | A | - | R | T | T | P | D | L | stop |
| Hap-36 | 92 | S | T | A | M | A | D | A | A | - | R | I | T | P | D | V | +26AA |
| Hap-37 | 51 | S | M | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-22 | 29 | S | T | A | M | A | G | A | A | - | R | T | T | Q | D | V | +26AA |
| Hap-17 | 17 | S | T | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-40 | 12 | S | T | A | M | A | D | A | A | - | R | I | I | P | D | V | +26AA |
| Hap-4 | 10 | S | T | A | M | A | G | - | - | - | R | T | T | P | N | V | +26AA |
| Hap-6 | 5 | S | T | A | M | A | G | - | - | - | R | T | T | P | D | V | +26AA |
| Hap-43 | 4 | Y | T | D | M | V | G | A | A | A | R | T | T | P | D | L | stop |
| Hap-23 | 3 | S | T | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-8 | 2 | S | T | A | M | A | G | - | - | - | \ | \ | \ | \ | \ | \ | \ |
| Hap-44 | 2 | S | T | A | T | A | G | A | A | - | R | T | T | P | D | V | +26AA |
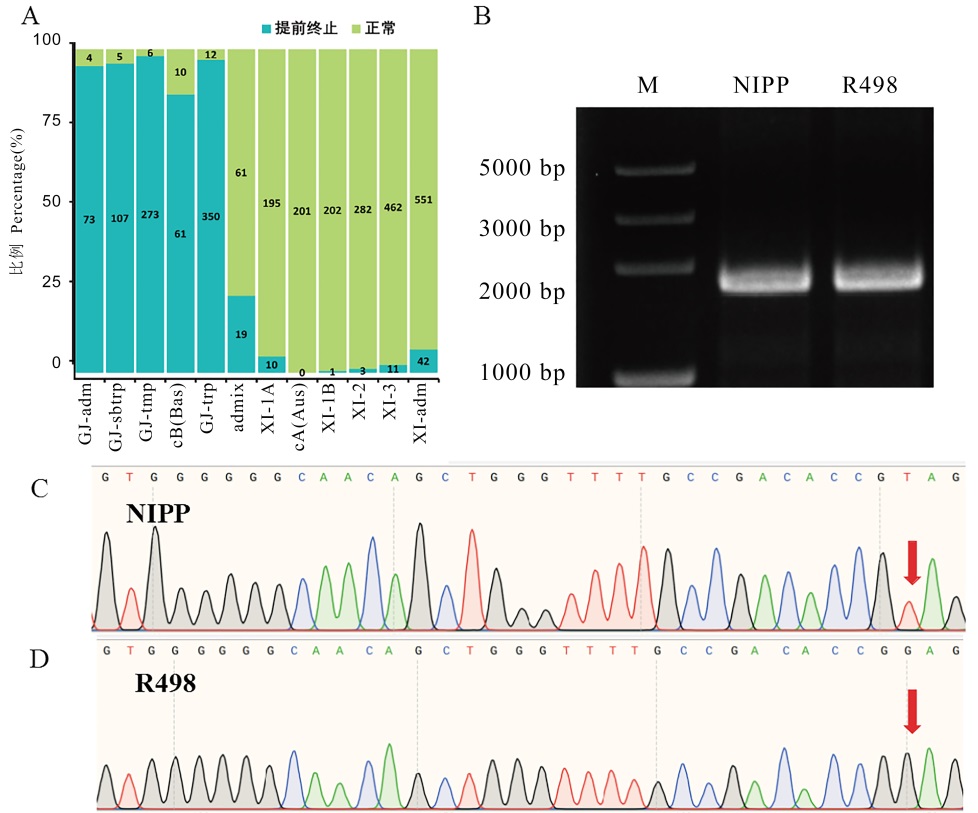
图6 不同类型BBM1序列验证 A: 2941份水稻材料中不同BBM1类型分布,青色表示发生提前终止BBM1,绿色表示正常的BBM1;B: NIPP和R498中BBM1编码区序列全长扩增琼脂糖电泳结果;C: NIPP中BBM1编码区测序峰图;D: R498中BBM1序列编码区测序峰图。
Fig. 6. Validation of different types of BBM1 sequences A, Distribution map of different BBM1 types in 2941 rice accessions, cyan indicates the occurrence of premature termination of BBM1, and green indicates normal BBM1; B, Results of agarose electrophoresis of full-length amplified BBM1 coding region sequences in NIPP and R498; C, Sequencing peak map of BBM1 coding region in NIPP; D, Sequencing peak map of BBM1 coding region in R498.
| [1] | Ye X X, Ma Y B, Sun B. Influence of soil type and genotype on Cd bioavailability and uptake by rice and implications for food safety[J]. Journal of Environmental Sciences, 2012, 24(9): 1647-1654. |
| [2] | 任光俊, 颜龙安, 谢华安. 三系杂交水稻育种研究的回顾与展望[J]. 科学通报, 2016, 61(35): 3748-3760. |
| Ren G J, Yan L A, Xie H A. Retrospective and perspective on indica three-line hybrid rice breeding research in China[J]. Chinese Science Bulletin, 2016, 61(35): 3748-3760. (in Chinese with English abstract) | |
| [3] | 袁隆平. 杂交水稻的育种战略设想[J]. 杂交水稻, 1987(1): 1-3. |
| Yuan L P. Strategy of hybrid rice breeding[J]. Hybrid Rice, 1987(1): 1-3. (in Chinese with English abstract) | |
| [4] | Bicknell R A, Koltunow A M. Understanding apomixis: Recent advances and remaining conundrums[J]. The Plant Cell, 2004, 16: 228-245. |
| [5] | Koltunow A M. Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules[J]. The Plant Cell, 1993, 5(10): 1425-1437. |
| [6] | 蔡得田, 陈冬玲. 论无融合生殖固定水稻杂种优势的策略[J]. 杂交水稻, 1989(6): 1-3. |
| Cai D T, Chen D L. Trying discussion in the tactics of fixing hybrid vigor of rice by apomixes[J]. Hybrid Rice, 1989(6): 1-3. (in Chinese with English abstract) | |
| [7] | 蔡得田, 马平福, 关和新, 姚家琳, 王灶安, 祝虹. 高频率无融合生殖水稻的研究[J]. 华中农业大学学报, 1991, 10(3): 223-227. |
| Cai D T, Ma P F, Gan H X, Yao J L, Wang Z A, Zhu H. The study of high frequency of apomictic rice material (HDAR). Journal of Huazhong Agricultural University, 1991, 10(3): 223-227. (in Chinese with English abstract) | |
| [8] | Jefferson R A. Apomixis: A social revolution for agriculture[J]. Biotechnology and Development Monitor, 1994, 19: 14-16. |
| [9] | Spillane C, Curtis M D, Grossniklaus U. Apomixis technology development-virgin births in farmers' fields?[J]. Nature Biotechnology, 2004, 22(6): 687-691. |
| [10] | Ondřej P, Michal S, Martin D. Reciprocal hybridization between diploid Ficaria calthifolia and tetraploid Ficaria verna subsp verna: Evidence from experimental crossing, genome size and molecular markers[J]. Botanical Journal of the Linnean Society, 2019, 189(3): 293-310. |
| [11] | Vijverberg K, Ozias-Akins P, Schranz M E. Identifying and engineering genes for parthenogenesis in plants[J]. Frontiers in Plant Science, 2019, 10: 128. |
| [12] | Brukhin V. Molecular and genetic regulation of apomixis[J]. Russian Journal of Genetics, 2017, 53(9): 943-964. |
| [13] | Mieulet D, Jolivet S, Rivard M, Cromer L, Vernet A, Mayonove P, Pereira L, Droc G, Courtois B, Guiderdoni E, Mercier R. Turning rice meiosis into mitosis[J]. Cell Research, 2016, 26(11): 1242-1254. |
| [14] | Rashid M, He G Y, Yang G X, Hussain J, Xu Y. AP2/ERF transcription factor in rice: Genome-wide canvas and syntenic relationships between monocots and eudicots[J]. Evolutionary Bioinformatics Online, 2012, 8: 321-355. |
| [15] | Xie W, Ding C, Hu H, Dong G, Zhang G, Qian Q, Ren D. Molecular events of rice AP2/ERF transcription factors[J]. International Journal of Molecular Sciences, 2022, 23(19): 12013. |
| [16] | Chen B, Maas L, Figueiredo D, Zhong Y, Reis R, Li M, Horstman A, Riksen T, Weemen M, Liu H, Siemons C, Chen S, Angenent G C, Boutilier K. BABY BOOM regulates early embryo and endosperm development[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(25): e2201761119. |
| [17] | Anderson S N, Johnson C S, Chesnut J, Jones D S, Khanday I, Woodhouse M, Li C, Conrad L J, Russell S D, Sundaresan V. The zygotic transition is initiated in unicellular plant zygotes with asymmetric activation of parental genomes[J]. Developmental Cell, 2017, 43: 349-358. |
| [18] | Anderson S N, Johnson C S, Jones D S, Conrad L J, Gou X, Russell S D, Sundaresan V. Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: Evidence for distinct sex-dependent chromatin and epigenetic states before fertilization[J]. Plant Journal, 2013, 76(5): 729-741. |
| [19] | Conner J A, Mookkan M, Huo H, Chae K, Ozias-Akins P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112: 11205-11210. |
| [20] | Conner J A, Podio M, Ozias-Akins P. Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes[J]. Plant Reproduction, 2017, 30: 41-52. |
| [21] | Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds[J]. Nature, 2019, 565(7737): 91-95. |
| [22] | Vernet A, Meynard D, Lian Q, Mieulet D, Gibert O, Bissah M, Rivallan R, Autran D, Leblanc O, Meunier A C, Frouin J, Taillebois J, Shankle K, Khanday I, Mercier R, Sundaresan V, Guiderdoni E. High-frequency synthetic apomixis in hybrid rice[J]. Nature Communications, 2022, 13(1): 7963. |
| [23] | Wei X, Liu C, Chen X, Lu H, Wang J, Yang S, Wang K. Synthetic apomixis with normal hybrid rice seed production[J]. Molecular Plant, 2023, 16(3): 489-492. |
| [24] | Wang C, Liu Q, Shen Y, Hua Y, Wang J, Lin J, Wu M, Sun T, Cheng Z, Mercier R, Wang K. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes[J]. Nature Biotechnology, 2019, 37(3): 283-286. |
| [25] | Wang C C, Yu H, Huang J, Wang W S, Faruquee M, Zhang F, Zhao X Q, Fu B Y, Chen K, Zhang H L, Tai S S, Wei C, McNally K L, Alexandrov N, Gao X Y, Li J, Li Z K, Xu J L, Zheng T Q. Towards a deeper haplotype mining of complex traits in rice with RFGB v2.0[J]. Plant Biotechnology Journal, 2020, 18(1): 14-16. |
| [26] | Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes R R, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis K C, Xu J, Sun C, Fu B, Zhang H, Gao Y, Zhao X, Shen F, Cui X, Yu H, Li Z, Chen M, Detras J, Zhou Y, Zhang X, Zhao Y, Kudrna D, Wang C, Li R, Jia B, Lu J, He X, Dong Z, Xu J, Li Y, Wang M, Shi J, Li J, Zhang D, Lee S, Hu W, Poliakov A, Dubchak I, Ulat V J, Borja F N, Mendoza J R, Ali J, Li J, Gao Q, Niu Y, Yue Z, Naredo M E B, Talag J, Wang X, Li J, Fang X, Yin Y, Glaszmann J C, Zhang J, Li J, Hamilton R S, Wing R A, Ruan J, Zhang G, Wei C, Alexandrov N, McNally K L, Li Z, Leung H. Genomic variation in 3,010 diverse accessions of Asian cultivated rice[J]. Nature, 2018, 557(7703): 43-49. |
| [27] | Lü Q, Li W, Sun Z, Ouyang N, Jing X, He Q, Wu J, Zheng J, Zheng J, Tang S, Zhu R, Tian Y, Duan M, Tan Y, Yu D, Sheng X, Sun X, Jia G, Gao H, Zeng Q, Li Y, Tang L, Xu Q, Zhao B, Huang Z, Lu H, Li N, Zhao J, Zhu L, Li D, Yuan L, Yuan D. Resequencing of 1,143 indica rice accessions reveals important genetic variations and different heterosis patterns[J]. Nature Communication, 2020, 11(1): 4778. |
| [28] | Yu H, Kou L, Li J. 10k-level integrated rice database shows power for exploiting rare variants[J]. Journal of Integrative Plant Biology, 2023, 65(12): 2539-2540. |
| [29] | Shang L, Li X, He H, Yuan Q, Song Y, Wei Z, Lin H, Hu M, Zhao F, Zhang C, Li Y, Gao H, Wang T, Liu X, Zhang H, Zhang Y, Cao S, Yu X, Zhang B, Zhang Y, Tan Y, Qin M, Ai C, Yang Y, Zhang B, Hu Z, Wang H, Lv Y, Wang Y, Ma J, Wang Q, Lu H, Wu Z, Liu S, Sun Z, Zhang H, Guo L, Li Z, Zhou Y, Li J, Zhu Z, Xiong G, Ruan J, Qian Q. A super pan-genomic landscape of rice[J]. Cell Research, 2022, 32(10): 878-896. |
| [30] | Qin P, Lu H, Du H, Wang H, Chen W, Chen Z, He Q, Ou S, Zhang H, Li X, Li X, Li Y, Liao Y, Gao Q, Tu B, Yuan H, Ma B, Wang Y, Qian Y, Fan S, Li W, Wang J, He M, Yin J, Li T, Jiang N, Chen X, Liang C, Li S. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations[J]. Cell, 2021, 184(13): 3542-3558. |
| [31] | Zhang F, Xue H, Dong X, Li M, Zheng X, Li Z, Xu J, Wang W, Wei C. Long-read sequencing of 111 rice genomes reveals significantly larger pan-genomes[J]. Genome Research, 2022, 32(5): 853-863. |
| [32] | 项聪英, 蔡年俊, 李静, 羊健, 陈剑平, 张恒木. 一个水稻小热休克蛋白基因的克隆和鉴定[J]. 中国水稻科学, 2016, 30(6): 587-592. |
| Xiang C Y, Cai N J, Li J, Yang J, Chen J P, Zhang H M. Cloning and characterization of a small heat shock protein (SHSP) gene in rice plant[J]. Chinese Journal of Rice Science, 2016, 30(6): 587-592. (in Chinese with English abstract) | |
| [33] | Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11[J]. Molecular Biology and Evolution, 2021, 38(7): 3022-3027. |
| [34] | Rozas J, Ferrer-Mata A, Sánchez-DelBarrio J C, Guirao-Rico S, Librado P, Ramos-Onsins S E, Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets[J]. Molecular Biology Evolution, 2017, 34(12): 3299-3302. |
| [35] | Kawahara Y, de la Bastide M, Hamilton J P, Kanamori H, McCombie W R, Ouyang S, Schwartz D C, Tanaka T, Wu J, Zhou S, Childs K L, Davidson R M, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee S S, Kim J, Numa H, Itoh T, Buell C R, Matsumoto T. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data[J]. Rice, 2013, 6(1): 4. |
| [36] | Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8): 1194-1202. |
| [37] | 赵凤利. 水稻全基因组拷贝数变异研究揭示复制基因的非对称进化[D]. 北京: 中国农业科学院, 2021. |
| Zhao F L. A genome-wide survey of copy number variations reveals an asymmetric evolution of duplicated genes in rice[D]. Beijing: Agricultural Genomics Institute Graduate School, 2021. (in Chinese with English abstract) | |
| [38] | Guillet-Claude C, Birolleau-Touchard C, Manicacci D, Rogowsky P M, Rigau J, Murigneux A, Martinant J P, Barrière Y. Nucleotide diversity of the ZmPox3 maize peroxidase gene: Relationships between a MITE insertion in exon 2 and variation in forage maize digestibility[J]. BMC Genetics, 2004, 16(5): 19. |
| [1] | 随晶晶, 赵桂龙, 金欣, 卜庆云, 唐佳琦. 水稻孕穗期耐冷调控的分子及生理机制研究进展 [J]. 中国水稻科学, 2025, 39(1): 1-10. |
| [2] | 任宁宁, 孙永建, 申聪聪, 朱双兵, 李慧菊, 张志远, 陈凯. 水稻中胚轴研究进展 [J]. 中国水稻科学, 2025, 39(1): 11-23. |
| [3] | 张丰勇, 应晓平, 张健, 杨隆维, 应杰政. 半矮秆基因sd1调控水稻重要农艺性状的研究进展 [J]. 中国水稻科学, 2025, 39(1): 24-32. |
| [4] | 陈智慧, 陶亚军, 范方军, 许扬, 王芳权, 李文奇, 古丽娜尔·巴合提别克, 蒋彦婕, 朱建平, 李霞, 杨杰. 水稻抽穗期调控基因Hd6功能标记的开发及应用 [J]. 中国水稻科学, 2025, 39(1): 47-54. |
| [5] | 胡风越, 王健, 王春, 王克剑, 刘朝雷. 水稻DMP1、DMP2、DMP3基因突变体的创制及其单倍体诱导能力鉴定 [J]. 中国水稻科学, 2025, 39(1): 55-66. |
| [6] | 陈书融, 朱练峰, 秦碧蓉, 王婕, 朱旭华, 田文昊, 朱春权, 曹小闯, 孔亚丽, 张均华, 金千瑜. 增氧灌溉下配施硝化抑制剂对水稻生长、产量和氮肥利用的影响 [J]. 中国水稻科学, 2025, 39(1): 92-100. |
| [7] | 吴猛, 倪川, 康钰莹, 毛雨欣, 叶苗, 张祖建. 水稻分蘖早发特性的品种间差异及其氮素响应 [J]. 中国水稻科学, 2025, 39(1): 101-114. |
| [8] | 王晓茜, 蔡创, 宋练, 周伟, 杨雄, 顾歆悦, 朱春梧. 开放式大气CO2浓度升高和温度升高对扬稻6号稻米品质的影响 [J]. 中国水稻科学, 2025, 39(1): 115-127. |
| [9] | 江敏, 王广伦, 李明璐, 苗波, 李明煊, 石春林. 基于模型的水稻高温热害风险评估与动态预警 [J]. 中国水稻科学, 2025, 39(1): 128-142. |
| [10] | 冯向前, 王爱冬, 洪卫源, 李子秋, 覃金华, 詹丽钏, 陈里鹏, 张运波, 王丹英, 陈松. 基于低空无人机遥感的水稻产量估测方法研究进展[J]. 中国水稻科学, 2024, 38(6): 604-616. |
| [11] | 叶苗, 毛雨欣, 张德海, 康钰莹, 袁榕, 张祖建. 高光效水稻品种的叶片和冠层生理生态特征及其氮素调控机制研究进展[J]. 中国水稻科学, 2024, 38(6): 617-626. |
| [12] | 钟智慧, 秦璐, 黎志力, 杨珍, 贺晓鹏, 蔡怡聪. 水稻IDD基因家族的全基因组鉴定及综合分析[J]. 中国水稻科学, 2024, 38(6): 638-652. |
| [13] | 杜彦修, 孙文玉, 袁泽科, 张倩倩, 李富豪, 李俊周, 孙红正. 利用QTL-Seq结合分子标记定位粳稻垩白粒率控制位点qChalk8[J]. 中国水稻科学, 2024, 38(6): 665-671. |
| [14] | 毋翔, 张义凯, 张鹏, 马昕伶, 陈玉林, 陈惠哲, 张玉屏, 向镜, 王亚梁, 王志刚, 李良涛. 2,4-表油菜素内酯对生物炭基质育秧水稻秧苗根系生长及生理特性的影响[J]. 中国水稻科学, 2024, 38(6): 685-694. |
| [15] | 汪邑晨, 朱本顺, 周磊, 朱骏, 杨仲南. 光/温敏核不育系的不育机理及两系杂交稻的发展与展望[J]. 中国水稻科学, 2024, 38(5): 463-474. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||