
中国水稻科学 ›› 2024, Vol. 38 ›› Issue (2): 150-159.DOI: 10.16819/j.1001-7216.2024.230305
收稿日期:2023-03-17
修回日期:2023-06-16
出版日期:2024-03-10
发布日期:2024-03-14
通讯作者:
* email: hejiwai@hunau.edu.cn
基金资助:
LIU Zhongqi1,2, ZHANG Haiqing1, HE Jiwai1,*( ), GUI Jinxin1
), GUI Jinxin1
Received:2023-03-17
Revised:2023-06-16
Online:2024-03-10
Published:2024-03-14
Contact:
* email: hejiwai@hunau.edu.cn
摘要:
【目的】籽粒脱水速率直接影响种子的安全收获和快速干燥。选育成熟期籽粒含水量低、脱水速率快的品种,可保障种子质量,降低生产成本。【方法】采用来自82个不同国家和地区的165份水稻核心种质作为试验材料,将成熟期种子脱水速率表型与基因型相结合进行GWAS分析,挖掘调控种子脱水的关键基因,为培育和创制脱水快速品种奠定基础。【结果】1)对165份水稻核心种质群体脱水速率性状进行描述性统计分析,结果显示2年脱水速率性状均呈连续性偏正态分布,2年快速脱水的脱水速率性状在年际间具有显著相关性。不同亚群之间快速脱水与慢速脱水速率正相关。2)GWAS关联分析共获得与脱水速率显著关联的SNP 170个,QTL 36个。通过LD分析定位到6个与脱水速率密切相关的QTL,分别为qGDR2.3、qGDR4.1、qGDR4.2、qGDR6.1、qGDR6.4、qGDR10.1。【结论】这些QTL区间内主要候选基因OsPIP1;1、OsTIFY9、OsbZIP48、OsATG8b、OsDREB1C、OsSCP46与水分转运活性、信号转导、转录调控、抗氧化防御密切相关,推测它们与成熟期水稻种子脱水速率相关,可作为候选基因。
刘忠奇, 张海清, 贺记外, 桂金鑫. 成熟期水稻种子脱水速率全基因组关联分析[J]. 中国水稻科学, 2024, 38(2): 150-159.
LIU Zhongqi, ZHANG Haiqing, HE Jiwai, GUI Jinxin. Genome-wide Association Analysis of Rice Seed Dehydration Rate at Maturity Stage[J]. Chinese Journal OF Rice Science, 2024, 38(2): 150-159.
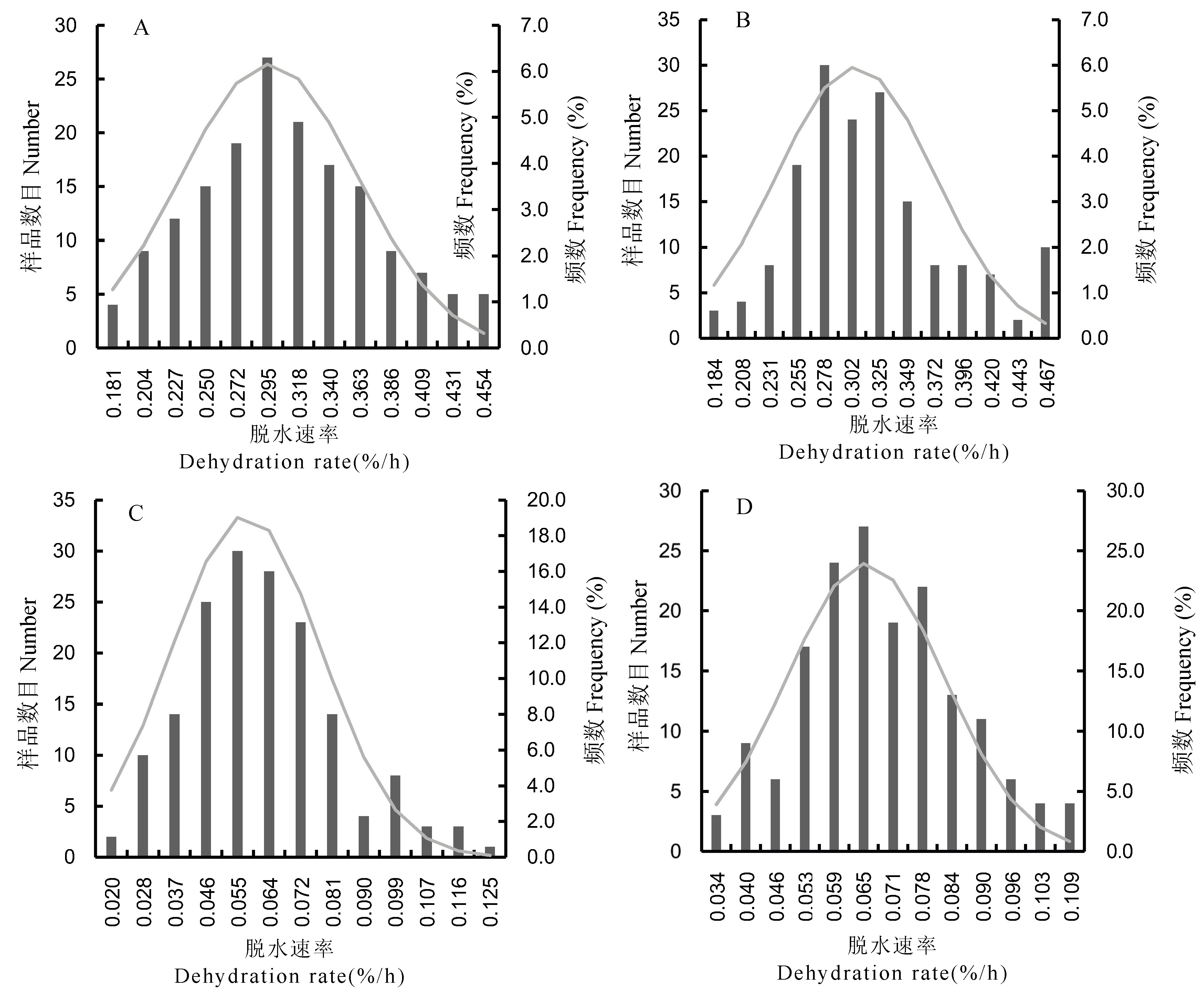
图1 供试水稻材料脱水速率群体分布 A:快速脱水处理(2019年);B:快速脱水处理(2020年);C:慢速脱水处理(2019年);D:慢速脱水处理(2020年)。其中,快速脱水条件为恒温45脱水℃,慢速脱水条件为恒温40脱水℃。
Fig. 1. Distribution of rice seed dehydration rates of tested rice material A, Rapid dehydration treatment(2019); B, Rapid dehydration treatment (2020); C, Slow dehydration treatment(2019); D, Slow dehydration treatment(2020). The fast dehydration condition is constant temperature 45 ± 2℃, the slow dehydration condition is constant temperature 40 ± 2℃.
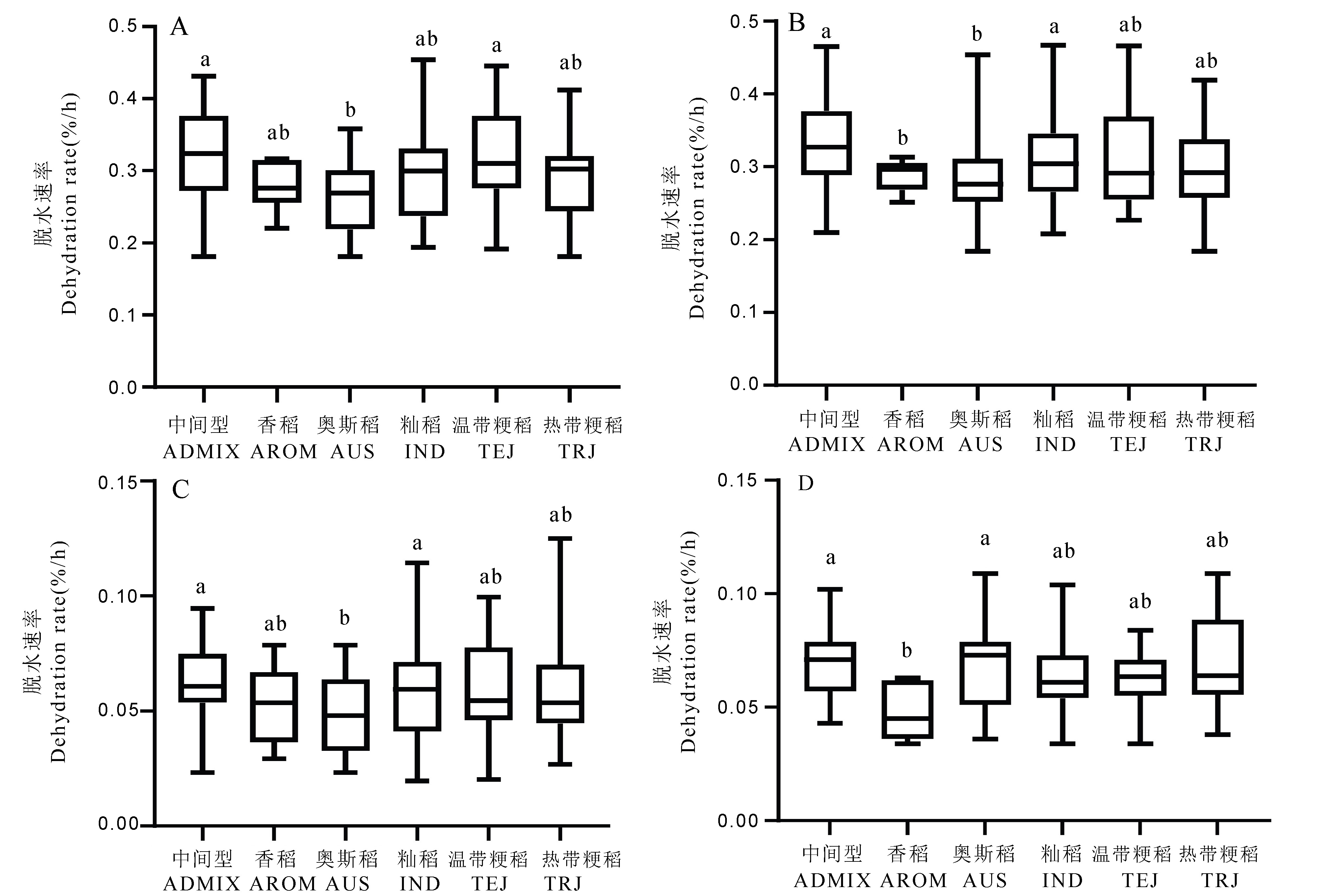
图2 不同亚群水稻种质群体在2年不同脱水方式下脱水速率差异的箱形图 A:快速脱水处理(2019年);B:快速脱水处理(2020年);C:慢速脱水处理(2019年);D:慢速脱水处理(2020年)。箱体上标有不同小写字母者表示不同亚群间差异达0.05显著水平(Duncan多重比较)。
Fig. 2. Box plots of dehydration rate of different sub-groups of rice under different dehydration conditions in two years A, Rapid dehydration treatment(2019); B, Rapid dehydration treatment(2020); C, Slow dehydration treatment(2019); D, Slow dehydration treatment(2020). Different lowercase letters indicate significant difference of 0.05 level among different subgroups (Duncan multiple comparison). ADMIX, Intermediate type; AROM, Aromatic rice; AUS, Aus rice; IND, indica rice; TEJ, Temperate japonica rice; TRJ, Tropical japonica rice.

图3 水稻成熟种子快速脱水与慢速脱水条件下脱水速率QTL位点的曼哈顿图与QQ图 A:2019年快速脱水条件下脱水速率QTL位点的曼哈顿图,图中红色实线以上区域代表在P<0.001水平上极显著,下同;B:2019年快速脱水条件下脱水速率QTL位点的QQ图;C和D:2020年快速脱水;E和F:2019年慢速脱水;G和H:2020年慢速脱水。
Fig. 3. Manhattan map and QQ map of QTLs for seed dehydration rate at maturity under different dehydration treatments A, Manhattan plot of QTLs for seed dehydration rate at maturity under the fast dehydration conditions in 2019. The areas above the red solid line represent significant difference at P < 0.001 level, and the same below; B, QQ map of QTLs for seed dehydration rate at maturity under the fast dehydration conditions in 2019; C and D, Under the fast dehydration conditions in 2020; E and F, Under the low dehydration conditions in 2019; G and H, Under the low dehydration conditions in 2020.
| 染色体Chromosome | 位点 Locus | 候选基因 Candidate gene | 功能注释 Functional annotation | 参考基因 Reference gene | |
|---|---|---|---|---|---|
| 1 | qGDR1.2 | LOC_Os01g49710 | 谷胱甘肽S-转移酶 Glutathione S-transferase | ||
| 2 | qGDR2.2 | LOC_Os02g36974 | 14-3-3蛋白 14-3-3 protein | ||
| 2 | qGDR2.3 | LOC_Os02g44630 | 水通道蛋白 Aquaporin protein | OsPIP1;1[ | |
| 4 | qGDR4.1 | LOC_Os04g32480 | 锌指蛋白 Zinc-finger protein | OsTIFY9[ | |
| 4 | qGDR4.2 | LOC_Os04g53240 | 自噬相关蛋白 Autophagy-related protein | OsATG8b[ | |
| 6 | qGDR6.1 | LOC_Os06g03670 | 脱水反应元件结合蛋白 Dehydration-responsive element-binding protein | OsDREB1C[ | |
| 6 | qGDR6.2 | LOC_Os06g12350 | 氨基酸转运蛋白 Amino acid transporter | ||
| 6 | qGDR6.4 | LOC_Os06g39960 | bZIP转录因子结构域含蛋白 bZIP transcription factor domain containing protein | OsbZIP48[ | |
| 7 | qGDR7.3 | LOC_Os07g37890 | 蛋白磷酸酶2C Protein phosphatase 2C | ||
| 7 | qGDR7.4 | LOC_Os07g38580 | 锌指家族蛋白 Zinc finger family protein | ||
| 7 | qGDR7.6 | LOC_Os07g47250 | 脂肪酶前体 Lipase precursor | ||
| 8 | qGDR8.2 | LOC_Os08g06240 | MYB家族转录因子 MYB family transcription factor | ||
| 8 | qGDR8.3 | LOC_Os08g15149 | 硫氧还蛋白结构域蛋白9 Thioredoxin domain-containing protein 9 | ||
| 9 | qGDR9.1 | LOC_Os09g20990 | 海藻糖-6-磷酸合成酶 Trehalose-6-phosphate synthase | ||
| 9 | qGDR9.2 | LOC_Os09g26170 | MYB家族转录因子 MYB family transcription factor | ||
| 10 | qGDR10.1 | LOC_Os10g01134 | OsSCP46-推定丝氨酸羧肽酶同源物 OsSCP46-putative Serine Carboxypeptidase homologue | OsSCP46[ | |
| 10 | qGDR10.3 | LOC_Os10g25010 | OsCML8-钙调素相关钙传感器蛋白 OsCML8-calmodulin-related calcium sensor protein | ||
| 11 | qGDR11.1 | LOC_Os11g37000 | 热激蛋白DnaJ Heat shock protein DnaJ | ||
表1 预测水稻成熟期种子脱水速率相关候选基因及其功能注释
Table 1. Candidate genes and their functional annotations for predicting the rate of seed dehydration in maturity
| 染色体Chromosome | 位点 Locus | 候选基因 Candidate gene | 功能注释 Functional annotation | 参考基因 Reference gene | |
|---|---|---|---|---|---|
| 1 | qGDR1.2 | LOC_Os01g49710 | 谷胱甘肽S-转移酶 Glutathione S-transferase | ||
| 2 | qGDR2.2 | LOC_Os02g36974 | 14-3-3蛋白 14-3-3 protein | ||
| 2 | qGDR2.3 | LOC_Os02g44630 | 水通道蛋白 Aquaporin protein | OsPIP1;1[ | |
| 4 | qGDR4.1 | LOC_Os04g32480 | 锌指蛋白 Zinc-finger protein | OsTIFY9[ | |
| 4 | qGDR4.2 | LOC_Os04g53240 | 自噬相关蛋白 Autophagy-related protein | OsATG8b[ | |
| 6 | qGDR6.1 | LOC_Os06g03670 | 脱水反应元件结合蛋白 Dehydration-responsive element-binding protein | OsDREB1C[ | |
| 6 | qGDR6.2 | LOC_Os06g12350 | 氨基酸转运蛋白 Amino acid transporter | ||
| 6 | qGDR6.4 | LOC_Os06g39960 | bZIP转录因子结构域含蛋白 bZIP transcription factor domain containing protein | OsbZIP48[ | |
| 7 | qGDR7.3 | LOC_Os07g37890 | 蛋白磷酸酶2C Protein phosphatase 2C | ||
| 7 | qGDR7.4 | LOC_Os07g38580 | 锌指家族蛋白 Zinc finger family protein | ||
| 7 | qGDR7.6 | LOC_Os07g47250 | 脂肪酶前体 Lipase precursor | ||
| 8 | qGDR8.2 | LOC_Os08g06240 | MYB家族转录因子 MYB family transcription factor | ||
| 8 | qGDR8.3 | LOC_Os08g15149 | 硫氧还蛋白结构域蛋白9 Thioredoxin domain-containing protein 9 | ||
| 9 | qGDR9.1 | LOC_Os09g20990 | 海藻糖-6-磷酸合成酶 Trehalose-6-phosphate synthase | ||
| 9 | qGDR9.2 | LOC_Os09g26170 | MYB家族转录因子 MYB family transcription factor | ||
| 10 | qGDR10.1 | LOC_Os10g01134 | OsSCP46-推定丝氨酸羧肽酶同源物 OsSCP46-putative Serine Carboxypeptidase homologue | OsSCP46[ | |
| 10 | qGDR10.3 | LOC_Os10g25010 | OsCML8-钙调素相关钙传感器蛋白 OsCML8-calmodulin-related calcium sensor protein | ||
| 11 | qGDR11.1 | LOC_Os11g37000 | 热激蛋白DnaJ Heat shock protein DnaJ | ||

图4 候选QTL区间内关联位点的LD分析 A: qGDR2.3(OsPIP1;1的LD分析); B: qGDR6.1(OsDREB1C的LD分析); C: qGDR10.1(OsSCP46的LD分析); D: qGDR6.4(OsbZIP48的LD分析); E: qGDR4.1(OsTIFY9的LD分析); F: qGDR4.2(OsATG8b的LD分析)。
Fig. 4. LD analysis of association loci within candidate QTL intervals
| [1] | 刘忠奇, 贺记外, 张海清, 刘爱民. 植物种子脱水耐性的研究现状分析与展望[J]. 中国农学通报, 2020, 36(2): 36-41. |
| Liu Z Q, He J W, Zhang H Q, Liu A M. Dehydration tolerance of plant seeds: Current research situation and prospects[J]. Chinese Agricultural Science Bulletin, 2020, 36(2): 36-41. (in Chinese with English abstract) | |
| [2] | Zhou G F, Hao D R, Xue L, Chen G Q, Lu H H, Zhang Z L, Shi M L, Mao Y X. Genome-wide association study of kernel moisture content at harvest stage in maize[J]. Breeding Science, 2018, 68(5): 622-628. |
| [3] | Jia T J, Wang L F, Li J J, Ma J, Cao Y Y, Lübbersted T, Li H Y. Integrating a genome-wide association study with transcriptomic analysis to detect genes controlling grain drying rate in maize (Zea mays L)[J]. Theoretical and Applied Genetics, 2020, 133(2): 623-634. |
| [4] | 刘显君, 王振华, 王霞, 李庭锋, 张林. 玉米籽粒生理成熟后自然脱水速率QTL的初步定位[J]. 作物学报, 2010, 36(1): 47-52. |
| Liu X J, Wang Z H, Wang X, Li T F, Zhang L. Primary mapping of QTL for dehydration rate of maize kernel after physiological maturing[J]. Acta Agronomica Sinica, 2010, 36(1): 47-52. (in Chinese with English abstract) | |
| [5] | Sala R G, Andrade F H, Camadro E L, Cerono J C. Quantitative trait loci for grain moisture at harvest and field grain drying rate in maize (Zea mays L.)[J]. Theoretical and Applied Genetics, 2006, 112: 462-471. |
| [6] | Li Y L, Dong Y B, Yang M L, Wang Q L, Shi Q, Zhou Q, Deng F, Ma Z Y, Qiao D H, Xu H. QTL detection for grain water relations and genetic correlations with grain matter accumulation at four stages after pollination in maize[J]. Journal of Plant Biochemistry and Physiology, 2014, 2(1): 1-9. |
| [7] | Capelle V, Remoué C, Moreau L, Reyss A, MahéahssMassonneau A, Falque M, Charcosset A, Thévenot C, Rogowsky P, Coursol S, Prioul J L. QTLs and candidate genes for desiccation and abscisic acid content in maize kernels[J]. BMC Plant Biology, 2010, 10(1): 1-22. |
| [8] | 孙乐秀. 玉米自交系机械化收获相关性状及其SNP标记关联分析[D]. 泰安: 山东农业大学, 2015. |
| Sun L X. Traits related to mechanical harvesting and their association analysis with SNP markers in maize inbred lines[D]. Tai’an: Shandong Agricultural University, 2015. (in Chinese with English abstract) | |
| [9] | Li W Q, Yu Y H, Wang L X, Luo Y, Peng Y, Xu Y C, Liu X G, Wu S S, Jian L M, Xu J T, Xiao Y J, Yan J B. The genetic architecture of the dynamic changes in grain moisture in maize[J]. Plant Biotechnology Journal, 2021, 19: 1195-1205. |
| [10] | 朱冬梅, 胡文静, 别同德, 陆成彬, 赵仁慧, 高德荣. 利用四交RIL群体定位小麦籽粒脱水速率 QTL[J]. 麦类作物学报, 2020, 40(1): 49-54. |
| Zhu D M, Hu W J, Bie T D, Lu C B, Zhao R H, Gao D R. QTL mapping for kernel dehydration rate after physiological maturity using in four way RIL populations of wheat[J]. Journal of Triticeae Crop, 2020, 40(1): 49-54. (in Chinese with English abstract) | |
| [11] | 于澎湃. 高粱耐密性研究及籽粒脱水速率的QTL定位[D]. 天津: 天津农学院, 2019. |
| Yu P P. Studies on sorghum density tolerance and QTL mapping of grain dehydration rate[D]. Tianjin:Tianjin Agricultural University, 2019. (in Chinese with English abstract) | |
| [12] | 袁静, 任育军, 赵洁. 水稻胚胎发育过程相关基因的鉴定[J]. 分子细胞生物学报, 2008, 41(3): 238-244. |
| Yuan J, Ren Y J, Zhao J. Identification of genes associated with early and late embryo development in rice[J]. Journal of Molecular Cell Biology, 2008, 41(3): 238-244. (in Chinese with English abstract) | |
| [13] | 张娟, 牛百晓, 鄂志国, 陈忱. 水稻胚乳发育遗传调控的研究进展[J]. 中国水稻科学, 2021, 35(4): 326-341. |
| Zhang J, Niu B X, E Z G, Chen C. Towards understanding the genetic regulations of endosperm development in rice[J]. Chinese Journal of Rice Science, 2021, 35(4): 326-341. (in Chinese with English abstract) | |
| [14] | Samarah N H, Mullen R E, Alqudah A M. An index to quantify seed moisture loss rate in relationship with seed desiccation tolerance in common vetch[J]. Seed Science and Technology, 2009, 37: 413-422. |
| [15] | Zhao K Y, Tung C W, Eizenga G C, Wright M H, Ali M L, Price A H, Norton G J, Islam M R, Reynolds A, Mezey J, McClung A M, Bustamante C D, McCouch S R. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa[J]. Nature Communications, 2011, 2: 467-477. |
| [16] | 林晋军, 姚威, 彭沙莎, 刘文韬, 陈文凤, 刘雄伦, 刘金灵. 水稻抽穗期性状QTL的全基因组关联定位分析[J]. 湖南农业大学学报, 2021, 47(3): 283-290. |
| Lin J J, Yao W, Peng S S, Liu W T, Chen W F, Liu X L, Liu J L. Genome-wide association dissection of QTLs for heading date traits in rice[J]. Journal of Hunan Agricultural University, 2021, 47(3): 283-290. (in Chinese with English abstract) | |
| [17] | Liu C W, Tatsuya F, Tadashi M, Gena P, Frascaria D, Kaneko T, Katsuhara M, Zhong S H, Sun X L, Zhu Y M, Iwasaki I, Kitagawa Y L. Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination[J]. Plant Physiology and Biochemistry, 2013, 63: 151-158. |
| [18] | Dubouzet J G, Sakuma Y, Ito Y, Kasuga M, Dubouzet E G, Miura S, Seki M, Shinozak K, Shinozaki Y. OsDREB genes in rice, encode transcription activators that function in drought, high-salt and cold responsive gene expression[J]. The Plant Journal, 2003, 33(4): 751-763. |
| [19] | Li Z Y, Tang L Q, Qiu J H, Zhang W, Wang Y F, Tong X H, Wei X J, Hou Y X, Zhang J. Serine carboxypeptidase 46 regulates grain filling and seed germination in rice[J]. PLoS One, 2016, 11(7): 1-18. |
| [20] | Ji Q, Zhang L S, Wang Y F, Wang J. Genome-wide analysis of basic leucine zipper transcription factor families in Arabidopsis thaliana, Oryza sativa and Populus trichocarpa[J]. Journal of Shanghai University, 2009, 13(2): 174-182. |
| [21] | Ye H, Du H, Tang N, Li X, Xiong L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice[J]. Plant Molecular Biology, 2009, 71(3): 291-305. |
| [22] | Izumi M, Hidema J, Wada S, Kondo E, Makino A, Ishida H. Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation[J]. Plant Physiology, 2015, 167(4): 1307-1320. |
| [23] | 渠建洲. 玉米籽粒含水率和脱水速率的基因挖掘与分析[D]. 杨凌: 西北农林科技大学, 2021. |
| Qu J Z. Gene mining and analysis of maize kernel moisture content and dehydration rate[D]. Yangling: Northwest A&F University, 2021. (in Chinese with English abstract) | |
| [24] | Si J K, Arifa A. Tissue and cell-specific localization of rice aquaporins and their water transport activities[J]. Plant and Cell Physiology, 2008, 49(1): 30-39. |
| [25] | 张凤启, 王邑双, 丁勇, 张君, 赵霞, 赵发欣, 唐保军. 玉米籽粒脱水速率研究进展[J]. 农学学报, 2018, 8(11): 4-11. |
| Zhang F Q, Wang Y S, Ding Y, Zhang J, Zhao X, Zhao F X, Tang B J. Corn kernel dehydration rate: Research progress[J]. Journal of Agriculture, 2018, 8(11): 4-11. (in Chinese with English abstract) | |
| [26] | Peng B, Kong H L, Li Y B, Wang L Q, Zhong M, Sun L, Gao G J, Zhang Q L, Luo L J, Wang G W, Xie W B, Chen J X, Yao W, Peng Y, Lei L, Lian X M, Xiao J H, Xu C G, Li X H, He Y Q. OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice[J]. Nature Communications, 2014, 5: 5847. (in Chinese with English abstract) |
| [27] | 彭波, 孙艳芳, 庞瑞华, 周棋赢, 袁红雨, 宋世枝. 植物氨基酸转运蛋白的研究进展[J]. 热带作物学报, 2016, 37(6): 1238-1243. |
| Peng B, Sun Y F, Pang R H, Zhou Q Y, Yuan H Y, Song S Z. Research progress of amino acid transporters in plants[J]. Chinese Journal of Tropical Crops, 2016, 37(6): 1238-1243. (in Chinese with English abstract) | |
| [28] | 叶玲飞, 罗光宇, 向建华, 刘爱玲, 陈信波. 丝氨酸羧肽酶及其类蛋白的研究进展[J]. 植物生理学报, 2013, 49(2): 122-126. |
| Ye L F, Luo G Y, Xiang J H, Liu A L, Chen X B. Research progress of serine carboxypeptidases and serine carboxypeptidase like proteins[J]. Plant Physiology Journal, 2013, 49(2): 122-126. (in Chinese with English abstract) | |
| [29] | 叶玲飞. 水稻OsSCP基因的耐逆相关功能研究[D]. 长沙: 湖南农业大学, 2013. |
| Ye L F. Studies on the stress resistance functions of OsSCP gene[D]. Changsha: Hunan Agricultural University, 2013. (in Chinese with English abstract) |
| [1] | 吕阳, 刘聪聪, 杨龙波, 曹兴岚, 王月影, 童毅, Mohamed Hazman, 钱前, 商连光, 郭龙彪. 全基因组关联分析(GWAS)鉴定水稻氮素利用效率候选基因 [J]. 中国水稻科学, 2024, 38(5): 516-524. |
| [2] | 郑广杰, 叶昌, 朱均林, 陶怡, 肖德顺, 徐亚楠, 褚光, 徐春梅, 王丹英. 淹水胁迫下水稻种子和胚芽葡萄糖供应差异与胚芽存活的关系[J]. 中国水稻科学, 2024, 38(2): 172-184. |
| [3] | 刘进, 崔迪, 余丽琴, 张立娜, 周慧颖, 马小定, 胡佳晓, 韩冰, 韩龙植, 黎毛毛. 水稻苗期耐热种质资源筛选及QTL定位[J]. 中国水稻科学, 2022, 36(3): 259-268. |
| [4] | 吴婷, 李霞, 黄得润, 黄凤林, 肖宇龙, 胡标林. 应用东乡野生稻回交重组自交系分析水稻耐低氮产量相关性状QTL[J]. 中国水稻科学, 2020, 34(6): 499-511. |
| [5] | 魏秀彩, 刘金栋, 刘利成, 黎用朝, 潘孝武, 董铮, 刘文强, 熊海波, 闵军, 李小湘. 基于MAGIC群体的水稻抽穗期和产量相关性状全基因组 关联分析[J]. 中国水稻科学, 2020, 34(4): 325-331. |
| [6] | 张桂莲, 廖斌, 唐文帮, 陈立云, 肖应辉. 稻米垩白性状对高温耐性的QTL分析[J]. 中国水稻科学, 2017, 31(3): 257-264. |
| [7] | 张杰, 郑蕾娜, 蔡跃, 尤小满, 孔飞, 汪国湘, 燕海刚, 金洁, 王亮, 张文伟, 江玲. 稻米淀粉RVA谱特征值与直链淀粉、蛋白含量的相关性及QTL定位分析[J]. 中国水稻科学, 2017, 31(1): 31-39. |
| [8] | 徐晓明1,2,3,#,张迎信1,3,#,王会民1,4,任翠5,王汝慈1,3,沈希宏1,3,占小登1,3,吴玮勋1,3,程式华1,3,曹立勇1,3,*. 一个水稻根长QTL qRL4的分离鉴定[J]. 中国水稻科学, 2016, 30(4): 363-370. |
| [9] | 王军, 朱金燕, 周勇, 杨杰, 范方军, 李文奇, 王芳权, 仲维功, 梁国华. 不同温光条件下水稻抽穗期QTL的定位与分析[J]. 中国水稻科学, 2016, 30(3): 247-255. |
| [10] | 任洁, 赵秀琴, 丁在松, 项超, 张晶, 王超, 张俊巍, 张强, 庞昀龙, 高用明, 石英尧. 利用选择导入系进行水稻耐低磷鉴定与QTL定位分析[J]. 中国水稻科学, 2015, 29(1): 1-13. |
| [11] | 占小登,于萍,林泽川,陈代波,沈希宏,张迎信,付君林,程式华* ,曹立勇*. 利用大粒籼/小粒粳重组自交系定位水稻生育期及产量相关性状QTL[J]. 中国水稻科学, 2014, 28(6): 570-580. |
| [12] | 姜树坤,张凤鸣* ,白良明,孙世臣,王彤彤,丁国华,姜辉,张喜娟. 水稻移栽后新生根系相关性状的QTL分析[J]. 中国水稻科学, 2014, 28(6): 598-604. |
| [13] | 冯跃,翟荣荣,林泽川,曹立勇,魏兴华,程式华*. 不同供氮水平下水稻产量性状的QTL分析[J]. 中国水稻科学, 2013, 27(6): 577-584. |
| [14] | 杨占烈1,2,戴高兴3,翟荣荣1,林泽川1,王会民4,曹立勇1,*,程式华1,*. 多环境条件下超级杂交稻协优9308 重组自交系群体粒形性状的QTL分析[J]. 中国水稻科学, 2013, 27(5): 482-490. |
| [15] | 周勇1,2 ,朱孝波1 ,袁华1 ,郑英2 ,钦鹏1 ,魏应海1,2 ,王玉平1 ,黄世君2 ,李仕贵1,*. 水稻单片段代换系芽期和苗期耐冷性分析及耐冷性QTL鉴定[J]. 中国水稻科学, 2013, 27(4): 381-388. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||