
Chinese Journal OF Rice Science ›› 2021, Vol. 35 ›› Issue (4): 326-341.DOI: 10.16819/j.1001-7216.2021.210307
• Orginal Article • Previous Articles Next Articles
Juan ZHANG1, Baixiao NIU1, Zhiguo E2, Chen CHEN1,*( )
)
Received:2021-03-09
Revised:2021-04-25
Online:2021-07-10
Published:2021-07-10
Contact:
Chen CHEN
通讯作者:
陈忱
基金资助:Juan ZHANG, Baixiao NIU, Zhiguo E, Chen CHEN. Towards Understanding the Genetic Regulations of Endosperm Development in Rice[J]. Chinese Journal OF Rice Science, 2021, 35(4): 326-341.
张娟, 牛百晓, 鄂志国, 陈忱. 水稻胚乳发育遗传调控的研究进展[J]. 中国水稻科学, 2021, 35(4): 326-341.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2021.210307
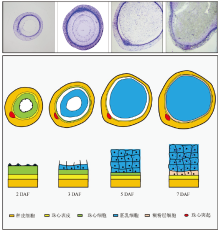
Fig. 1. Schematic illustration of the early endosperm development in rice.The primary endosperm cell is syncytial at 2 days after flowering (DAF). One endosperm cell consists of many free nuclei owing to that the nuclear divisions are uncoupled with cytoplasm divisions during the cell cycles. The syncytial cell initiates to cellularize around 3 DAF to form a layer of endosperm cells. This layer of cells further divide periclinally. The endosperm cells fill up the embryo sac around 5 DAF and start to accumulate storage compounds. Around 7 DAF, the aleurone layer is visible in rice seeds. Aleurone cells are differentiated from the outmost layer of the endosperm cells.
| 基因 Gene | 基因登录号 Gene ID | 编码蛋白 Encoded protein | 是否印记 Imprinted or not | 突变体种子表型 Penotype of mutant | 文献 Reference | |
|---|---|---|---|---|---|---|
| OsFIE2 | LOC_Os08g04270 | PRC2复合体Esc家族 | No | 胚乳细胞化受阻 | [ | |
| OsFIE1 | LOC_Os08g04290 | PRC2复合体Esc家族 | MEG | 休眠性降低 | [ | |
| OsEMF2a | LOC_Os04g08034 | PRC2复合体Su(z)12家族 | MEG | 胚乳细胞化受阻 | [ | |
| OsEMF2b | LOC_Os09g13630 | PRC2复合体Su(z)12家族 | No | 休眠性降低 | [ | |
| OsMADS78 | LOC_Os09g02830 | I型MADS成员 | Unknown | 胚乳细胞化提前 | [ | |
| OsMADS79 | LOC_Os01g74440 | I型MADS成员 | Unknown | 胚乳细胞化提前 | [ | |
| OsMADS87 | LOC_Os03g38610 | I型MADS成员 | MEG | 胚乳细胞化提前 | [ | |
| Orysa;CycB1;1 | LOC_Os01g59120 | 细胞周期蛋白 | No | 胚乳细胞化障碍导致无胚乳表型,胚增大 | [ | |
| Oryza;KRP1 | LOC_Os02g52480 | 细胞周期蛋白依赖性激酶抑制因子 | No | 过量表达时种子灌浆异常 | [ | |
| ENL1 | LOC_Os04g59624 | SNF2解旋酶蛋白 | No | 无胚乳 | [ | |
| OsGCD1 | LOC_Os01g58750 | GCD1同源基因 | No | 细胞化推迟 | [ | |
| RPBF/OsDof3 | LOC_Os02g15350 | 醇溶谷蛋白框结合蛋白, Dof转录因子 | No | 糊粉细胞层数变多 | [ | |
| RISBZ1/bZIP58 | LOC_Os07g08420 | bZIP转录因子 | No | 单突无明显表型 | [ | |
| OsROS1a | LOC_Os01g11900 | DNA去甲基化酶 | No | 糊粉细胞层数显著增多 | [ | |
| GFR1 | LOC_Os10g36400 | 膜定位蛋白 | No | 显著降低了灌浆率 | [ | |
| OsSUT1 | LOC_Os03g07480 | 蔗糖转运蛋白 | No | 籽粒不能正常灌浆 | [ | |
| OsSWEET4 | LOC_Os02g19820 | 蔗糖转运蛋白 | No | 胚乳变小,结实率下降 | [ | |
| OsSWEET11 | LOC_Os08g42350 | 蔗糖转运蛋白 | MEG | 籽粒不充实 | [ | |
| OsSWEET15 | LOC_Os02g30910 | 蔗糖转运蛋白 | No | 籽粒不充实 | [ | |
| GIF1 | LOC_Os04g33740 | 细胞壁转化酶 | No | 灌浆不完全,粒重降低,粉质胚乳表型 | [ | |
| GIF2 | LOC_Os01g44220 | AGP酶大亚基 | No | 种子淀粉合成障碍 | [ | |
| SPK | LOC_Os10g39420 | 蔗糖合酶激酶 | MEG | 抑制淀粉在籽粒中的合成 | [ | |
| OsMADS29 | LOC_Os02g07430 | II型MADS-box转录因子 | No | 细胞不能正常积累储藏物质,种子干瘪、败育 | [ | |
| RSR1 | LOC_Os05g03040 | AP2家族转录因子 | No | 种子变大,直链淀粉含量升高,产量增加 | [ | |
| OsNF-YB1 | LOC_Os02g49410 | NF-YB家族转录因子 | No | 结实率和千粒重均显著降低,种子空瘪萎缩 | [ | |
| OsNF-YB9 | LOC_Os06g17480 | NF-YB家族转录因子 | No | 粒型变长,垩白增多 | [ | |
| OsNF-YC12 | LOC_Os10g11580 | NF-YC家族转录因子 | No | 籽粒变小,千粒重下降 | [ | |
| OsNF-YC10 | LOC_Os01g24460 | NF-YC家族转录因子 | No | 籽粒表现为细窄、变轻 | [ | |
| OsbZIP76 | LOC_Os09g34880 | bZIP转录因子 | MEG | 细胞化进程提前,籽粒变小 | [ | |
| OsNAC20 | LOC_Os01g01470 | NAC转录因子 | No | 和NAC26双突变体表型为粉质胚乳,粒宽、粒厚、千粒重降低 | [ | |
| OsNAC26 | LOC_Os01g29840 | NAC转录因子 | No | 和NAC20双突变体表型为粉质胚乳,粒宽、粒厚、千粒重降低 | [ | |
| ONAC127 | LOC_Os11g31340 | NAC转录因子 | PEG | 抑制种子中储藏物质的合成 | [ | |
| ONAC129 | LOC_Os11g31380 | NAC转录因子 | PEG | 抑制种子中储藏物质的合成 | [ | |
| OsFAD3 | LOC_Os12g01370 | 脂肪酸去饱和酶 | No | 过表达导致亚麻酸(C18:3)含量显著提高 | [ | |
| OsFAD2 | LOC_Os02g48560 | 脂肪酸去饱和酶 | No | 籽粒中油脂的含量显著提高 | [ | |
| OsYUC9 | LOC_Os01g16714 | 黄素单加氧酶 | No | 种子粒重降低、垩白增加 | [ | |
| OsYUC11 | LOC_Os12g08780 | 黄素单加氧酶 | PEG | 种子灌浆缓慢 | [ | |
| OsTAR1 | LOC_Os05g07720 | 色氨酸转氨酶 | No | 种子较小、垩白增加,籽粒灌浆延迟 | [ | |
| TGW6 | LOC_Os06g41850 | IAA-葡萄糖水解酶 | No | 显著提高千粒重 | [ | |
| DG1 | LOC_Os03g12790 | ABA外排转运蛋白 | No | 灌浆延迟,垩白增加,高温下有空瘪籽粒 | [ | |
| OsbHLH144 | LOC_Os04g35010 | bHLH转录因子 | No | 稻谷品质发生改变 | [ | |
| OsCLF/SDG711 | LOC_Os06g16390 | PRC2复合体E(z)家族成员 | No | 抑制淀粉和储藏蛋白积累 | [ | |
| SDG728/OsSET22 | LOC_Os05g41172 | 组蛋白H3K9甲基转移酶 | No | 籽粒变小和粒重降低 | [ | |
| OsLFR | LOC_Os07g41900 | 染色质重塑因子 | No | 胚乳早期游离核数目减少、不能细胞化 | [ | |
| OsSRT1 | LOC_Os04g20270 | 组蛋白去乙酰化酶基因 | No | 籽粒淀粉合成障碍 | [ | |
| OsMET1b | LOC_Os07g08500 | DNA甲基转移酶基因 | PEG | 籽粒皱缩败育 | [ | |
| ROS1c/DNG701 | LOC_Os05g37350 | DNA去甲基化酶 | No | 15%的种子皱缩,不能正常灌浆 | [ | |
| PEG1 | LOC_Os01g08570 | 依赖酮戊二酸和铁的加氧酶 | PEG | 成熟籽粒空瘪 | [ | |
Table 1 .Identified genes for endosperm development in rice.
| 基因 Gene | 基因登录号 Gene ID | 编码蛋白 Encoded protein | 是否印记 Imprinted or not | 突变体种子表型 Penotype of mutant | 文献 Reference | |
|---|---|---|---|---|---|---|
| OsFIE2 | LOC_Os08g04270 | PRC2复合体Esc家族 | No | 胚乳细胞化受阻 | [ | |
| OsFIE1 | LOC_Os08g04290 | PRC2复合体Esc家族 | MEG | 休眠性降低 | [ | |
| OsEMF2a | LOC_Os04g08034 | PRC2复合体Su(z)12家族 | MEG | 胚乳细胞化受阻 | [ | |
| OsEMF2b | LOC_Os09g13630 | PRC2复合体Su(z)12家族 | No | 休眠性降低 | [ | |
| OsMADS78 | LOC_Os09g02830 | I型MADS成员 | Unknown | 胚乳细胞化提前 | [ | |
| OsMADS79 | LOC_Os01g74440 | I型MADS成员 | Unknown | 胚乳细胞化提前 | [ | |
| OsMADS87 | LOC_Os03g38610 | I型MADS成员 | MEG | 胚乳细胞化提前 | [ | |
| Orysa;CycB1;1 | LOC_Os01g59120 | 细胞周期蛋白 | No | 胚乳细胞化障碍导致无胚乳表型,胚增大 | [ | |
| Oryza;KRP1 | LOC_Os02g52480 | 细胞周期蛋白依赖性激酶抑制因子 | No | 过量表达时种子灌浆异常 | [ | |
| ENL1 | LOC_Os04g59624 | SNF2解旋酶蛋白 | No | 无胚乳 | [ | |
| OsGCD1 | LOC_Os01g58750 | GCD1同源基因 | No | 细胞化推迟 | [ | |
| RPBF/OsDof3 | LOC_Os02g15350 | 醇溶谷蛋白框结合蛋白, Dof转录因子 | No | 糊粉细胞层数变多 | [ | |
| RISBZ1/bZIP58 | LOC_Os07g08420 | bZIP转录因子 | No | 单突无明显表型 | [ | |
| OsROS1a | LOC_Os01g11900 | DNA去甲基化酶 | No | 糊粉细胞层数显著增多 | [ | |
| GFR1 | LOC_Os10g36400 | 膜定位蛋白 | No | 显著降低了灌浆率 | [ | |
| OsSUT1 | LOC_Os03g07480 | 蔗糖转运蛋白 | No | 籽粒不能正常灌浆 | [ | |
| OsSWEET4 | LOC_Os02g19820 | 蔗糖转运蛋白 | No | 胚乳变小,结实率下降 | [ | |
| OsSWEET11 | LOC_Os08g42350 | 蔗糖转运蛋白 | MEG | 籽粒不充实 | [ | |
| OsSWEET15 | LOC_Os02g30910 | 蔗糖转运蛋白 | No | 籽粒不充实 | [ | |
| GIF1 | LOC_Os04g33740 | 细胞壁转化酶 | No | 灌浆不完全,粒重降低,粉质胚乳表型 | [ | |
| GIF2 | LOC_Os01g44220 | AGP酶大亚基 | No | 种子淀粉合成障碍 | [ | |
| SPK | LOC_Os10g39420 | 蔗糖合酶激酶 | MEG | 抑制淀粉在籽粒中的合成 | [ | |
| OsMADS29 | LOC_Os02g07430 | II型MADS-box转录因子 | No | 细胞不能正常积累储藏物质,种子干瘪、败育 | [ | |
| RSR1 | LOC_Os05g03040 | AP2家族转录因子 | No | 种子变大,直链淀粉含量升高,产量增加 | [ | |
| OsNF-YB1 | LOC_Os02g49410 | NF-YB家族转录因子 | No | 结实率和千粒重均显著降低,种子空瘪萎缩 | [ | |
| OsNF-YB9 | LOC_Os06g17480 | NF-YB家族转录因子 | No | 粒型变长,垩白增多 | [ | |
| OsNF-YC12 | LOC_Os10g11580 | NF-YC家族转录因子 | No | 籽粒变小,千粒重下降 | [ | |
| OsNF-YC10 | LOC_Os01g24460 | NF-YC家族转录因子 | No | 籽粒表现为细窄、变轻 | [ | |
| OsbZIP76 | LOC_Os09g34880 | bZIP转录因子 | MEG | 细胞化进程提前,籽粒变小 | [ | |
| OsNAC20 | LOC_Os01g01470 | NAC转录因子 | No | 和NAC26双突变体表型为粉质胚乳,粒宽、粒厚、千粒重降低 | [ | |
| OsNAC26 | LOC_Os01g29840 | NAC转录因子 | No | 和NAC20双突变体表型为粉质胚乳,粒宽、粒厚、千粒重降低 | [ | |
| ONAC127 | LOC_Os11g31340 | NAC转录因子 | PEG | 抑制种子中储藏物质的合成 | [ | |
| ONAC129 | LOC_Os11g31380 | NAC转录因子 | PEG | 抑制种子中储藏物质的合成 | [ | |
| OsFAD3 | LOC_Os12g01370 | 脂肪酸去饱和酶 | No | 过表达导致亚麻酸(C18:3)含量显著提高 | [ | |
| OsFAD2 | LOC_Os02g48560 | 脂肪酸去饱和酶 | No | 籽粒中油脂的含量显著提高 | [ | |
| OsYUC9 | LOC_Os01g16714 | 黄素单加氧酶 | No | 种子粒重降低、垩白增加 | [ | |
| OsYUC11 | LOC_Os12g08780 | 黄素单加氧酶 | PEG | 种子灌浆缓慢 | [ | |
| OsTAR1 | LOC_Os05g07720 | 色氨酸转氨酶 | No | 种子较小、垩白增加,籽粒灌浆延迟 | [ | |
| TGW6 | LOC_Os06g41850 | IAA-葡萄糖水解酶 | No | 显著提高千粒重 | [ | |
| DG1 | LOC_Os03g12790 | ABA外排转运蛋白 | No | 灌浆延迟,垩白增加,高温下有空瘪籽粒 | [ | |
| OsbHLH144 | LOC_Os04g35010 | bHLH转录因子 | No | 稻谷品质发生改变 | [ | |
| OsCLF/SDG711 | LOC_Os06g16390 | PRC2复合体E(z)家族成员 | No | 抑制淀粉和储藏蛋白积累 | [ | |
| SDG728/OsSET22 | LOC_Os05g41172 | 组蛋白H3K9甲基转移酶 | No | 籽粒变小和粒重降低 | [ | |
| OsLFR | LOC_Os07g41900 | 染色质重塑因子 | No | 胚乳早期游离核数目减少、不能细胞化 | [ | |
| OsSRT1 | LOC_Os04g20270 | 组蛋白去乙酰化酶基因 | No | 籽粒淀粉合成障碍 | [ | |
| OsMET1b | LOC_Os07g08500 | DNA甲基转移酶基因 | PEG | 籽粒皱缩败育 | [ | |
| ROS1c/DNG701 | LOC_Os05g37350 | DNA去甲基化酶 | No | 15%的种子皱缩,不能正常灌浆 | [ | |
| PEG1 | LOC_Os01g08570 | 依赖酮戊二酸和铁的加氧酶 | PEG | 成熟籽粒空瘪 | [ | |
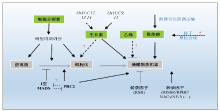
Fig. 2. Schematic illustration of the molecular controls of rice endosperm development.Arrows indicate positive regulations, “T” bars indicate negative regulations; Solid lines indicate the regulations are well supported by different studies; Dash lines indicate the possible regulations that require further evidence.
| [1] | Wu X, Liu J, Li D, Liu C M.Rice caryopsis development: I. Dynamic changes in different cell layers[J]. Journal of Integrative Plant Biology, 2016, 58(9): 772-785. |
| [2] | Zhang M, Xie S, Dong X, Zhao X, Zeng B, Chen J, Li H, Yang W, Zhao H, Wang G, Chen Z, Sun S, Hauck A, Jin W, Lai J.Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize[J]. Genome Research, 2014, 24(1): 167-176. |
| [3] | Zemach A, Kim M Y, Silva P, Rodrigues J A, Dotson B, Brooks M D, Zilberman D.Local DNA hypomethylation activates genes in rice endosperm[J]. Proceedings of the National Academy of Sciences, 2010, 107(43): 18729-18734. |
| [4] | Hsieh T F, Ibarra C I, Silva P, Zemach A, Eshed-Williams L, Fischer R L, Zilberman D.Genome- wide demethylation of Arabidopsis endosperm[J]. Science, 2009, 324: 1451-1454. |
| [5] | Wu X, Liu J, Li D, Liu C M.Rice caryopsis development II: Dynamic changes in the endosperm[J]. Journal of Integrative Plant Biology, 2016, 58(9): 786-798. |
| [6] | Chen C, Begcy K, Liu K, Folsom J J, Wang Z, Zhang C, Walia H.Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity[J]. Plant Physiology, 2016, 171(1): 606-622. |
| [7] | Paul P, Dhatt B K, Sandhu J, Hussain W, Irvin L, Morota G, Staswick P, Walia H.Divergent phenotypic response of rice accessions to transient heat stress during early seed development[J]. Plant Direct, 2020, 4(1): 1-13. |
| [8] | Zhang H, Luo M, Johnson S D, Zhu X, Liu L, Huang F, Liu Y, Xu P, Wu X.Parental genome imbalance causes post-zygotic seed lethality and deregulates imprinting in rice[J]. Rice, 2016, 9(1): 43. |
| [9] | Sekine D, Ohnishi T, Furuumi H, Ono A, Yamada T, Kurata N, Kinoshita T.Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm[J]. The Plant Journal, 2013, 76(5): 792-799. |
| [10] | Tonosaki K, Sekine D, Ohnishi T, Ono A, Furuumi H, Kurata N, Kinoshita T.Overcoming the species hybridization barrier by ploidy manipulation in the genus Oryza[J]. Plant Journal, 2018, 93(3): 534-544. |
| [11] | Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, Kinoshita T.Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division[J]. The Plant Journal, 2011, 65(5): 798-806. |
| [12] | Mozgova I, Hennig L.The polycomb group protein regulatory network[J]. Annual Review of Plant Biology, 2015, 66(1): 269-296. |
| [13] | Holec S, Berger F.Polycomb group complexes mediate developmental transitions in plants[J]. Plant Physiology, 2012, 158(1): 35-43. |
| [14] | Ohad N, Margossian L, Hsu Y C, Williams C, Repetti P, Fischer R L.A mutation that allows endosperm development without fertilization[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(11): 5319-5324. |
| [15] | Eacock W J A P, Chaudhury A M, Ming L, Miller C, Craig S, Dennis E S, Peacock W J. Fertilization- independent seed development in Arabidopsis thaliana[J]. Proceedings of the National Academy of Sciences of the United States of America, 1997, 94: 4223-4228. |
| [16] | Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada J J, Goldberg R B, Fischer R L.Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis[J]. Proceedings of the National Academy of Sciences, 1999, 96(7): 4186-4191. |
| [17] | Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W.Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development[J]. The EMBO Journal, 2003, 22(18): 4804-4814. |
| [18] | Luo M, Platten D, Chaudhury A, Peacock W J, Dennis E S.Expression, imprinting, and evolution of rice homologs of the polycomb group genes[J]. Molecular Plant, 2009, 2(4): 711-723. |
| [19] | Li S, Zhou B, Peng X, Kuang Q, Huang X, Yao J, Du B, Sun M X.OsFIE2 plays an essential role in the regulation of rice vegetative and reproductive development[J]. New Phytologist, 2014, 201(1): 66-79. |
| [20] | Zhang L, Cheng Z, Qin R, Qiu Y, Wang J L, Cui X, Gu L, Zhang X, Guo X, Wang D, Jiang L, Wu C Y, Wang H, Cao X, Wan J.Identification and characterization of an Epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice[J]. The Plant Cell, 2012, 24(11): 4407-4421. |
| [21] | Cheng X, Pan M, E Z, Zhou Y, Niu B, Chen C, Cheng X, Niu B, Chen C, Pan M, Zhiguo E, Zhou Y, Niu B, Chen C,. Functional divergence of two duplicated fertilization independent endosperm genes in rice with respect to seed development[J]. The Plant Journal, 2020, 104(1): 124-137. |
| [22] | Furihata H Y, Suenaga K, Kawanabe T, Yoshida T, Kawabe A.Gene duplication, silencing and expression alteration govern the molecular evolution of PRC2 genes in plants[J]. Genes & Genetic Systems, 2016, 91(2): 85-95. |
| [23] | Tonosaki K, Kinoshita T.Possible roles for polycomb repressive complex 2 in cereal endosperm[J]. Frontiers in Plant Science, 2015, 6: 1-5. |
| [24] | Cheng X, Pan M, E Z G, Zhou Y, Niu B, Chen C. The maternally expressed polycomb group gene OsEMF2a is essential for endosperm cellularization and imprinting in rice[J]. Plant Communications, 2020, 2: 100092. |
| [25] | Tonosaki K, Ono A, Kunisada M, Nishino M, Nagata H, Sakamoto S, Kijima S T, Furuumi H, Nonomura K I, Sato Y, Ohme-Takagi M, Endo M, Comai L, Hatakeyama K, Kawakatsu T, Kinoshita T.Mutation of the imprinted gene OsEMF2a induces autonomous endosperm development and delayed cellularization in rice[J]. The Plant Cell, 2021, 33(1): 85-103. |
| [26] | Deng L, Zhang S, Wang G, Fan S, Li M, Chen W, Tu B, Tan J, Wang Y, Ma B, Li S, Qin P.Down-regulation of OsEMF2b caused semi-sterility due to anther and pollen development defects in rice[J]. Frontiers in Plant Science, 2017, 8: 1998. |
| [27] | Xie S, Chen M, Pei R, Ouyang Y, Yao J.OsEMF2b acts as a regulator of flowering transition and floral organ identity by mediating H3K27me3 deposition at OsLFL1 and OsMADS4 in rice[J]. Plant Molecular Biology Reporter, 2015, 33(1): 121-132. |
| [28] | Conrad L J, Khanday I, Johnson C, Guiderdoni E, An G, Vijayraghavan U, Sundaresan V.The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice[J]. Plant Journal, 2014, 80(5): 883-894. |
| [29] | Chen M, Xie S, Ouyang Y, Yao J.Rice PcG gene OsEMF2b controls seed dormancy and seedling growth by regulating the expression of OsVP1[J]. Plant Science, 2017, 260: 80-89. |
| [30] | Zhong J, Peng Z, Peng Q, Cai Q, Peng W, Chen M, Yao J.Regulation of plant height in rice by the Polycomb group genes OsEMF2b, OsFIE2 and OsCLF[J]. Plant Science, 2018, 267: 157-167. |
| [31] | Smaczniak C, Immink R G H, Angenent G C, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies[J]. Development, 2012, 139(17): 3081-3098. |
| [32] | Masiero S, Colombo L, Grini P E, Schnittger A, Kater M M.The emerging importance of type I MADS box transcription factors for plant reproduction[J]. The Plant Cell, 2011, 23(3): 865-872. |
| [33] | Kang I H, Steffen J G, Portereiko M F, Lloyd A, Drews G N.The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis[J]. The Plant Cell, 2008, 20(3): 635-647. |
| [34] | Portereiko M F, Lloyd A, Steffen J G, Punwani J A, Otsuga D, Drews G N.AGL80 is required for central cell and endosperm development in Arabidopsis[J]. The Plant Cell, 2006, 18(8): 1862-1872. |
| [35] | Kohler C.The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1[J]. Genes & Development, 2003, 17(12): 1540-1553. |
| [36] | Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L.Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility[J]. Current Biology, 2009, 19(13): 1128-1132. |
| [37] | Nam J, Kim J, Lee S, An G, Ma H, Nei M.Type I MADS-box genes have experienced faster birth-and -death evolution than type II MADS-box genes in angiosperms[J]. Proceedings of the National Academy of Sciences, 2004, 101(7): 1910-1915. |
| [38] | Gramzow L, Theißen G.Phylogenomics of MADS-box genes in plants: Two opposing life styles in one gene family[J]. Biology, 2013, 2(3): 1150-1164. |
| [39] | Paul P, Dhatt B K, Miller M, Folsom J J, Wang Z, Krassovskaya I, Liu K, Sandhu J, Yu H, Zhang C, Obata T, Staswick P, Walia H.MADS78 and MADS79 are essential regulators of early seed development in rice[J]. Plant Physiology, 2020, 182(2): 933-948. |
| [40] | Olsen O A.ENDOSPERM DEVELOPMENT: Cellularization and cell fate specification[J]. Annual Review of Plant Physiology and Plant Molecular Biology, 2001, 52(1): 233-267. |
| [41] | Dante R, Larkins B, Sabelli P.Cell cycle control and seed development[J]. Frontiers in Plant Science, 2014, 5: 493. |
| [42] | Guo J, Wang F, Song J, Sun W, Zhang X S.The expression of Orysa;CycB1;1 is essential for endosperm formation and causes embryo enlargement in rice[J]. Planta, 2010, 231(2): 293-303. |
| [43] | Mizutani M, Naganuma T, Tsutsumi K, Saitoh Y.The syncytium-specific expression of the Orysa;KRP3 CDK inhibitor: Implication of its involvement in the cell cycle control in the rice (Oryza sativa L.) syncytial endosperm[J]. Journal of Experimental Botany, 2010, 61(3): 791-798. |
| [44] | Barrôco R M, Peres A, Droual A M, De Veylder L, Nguyen L S L, De Wolf J, Mironov V, Peerbolte R, Beemster G T S, Inzé D, Broekaert W F, Frankard V. The cyclin-dependent kinase inhibitor Orysa;KRP1 plays an important role in seed development of rice[J]. Plant Physiology, 2006, 142(3): 1053-1064. |
| [45] | Hara T, Katoh H, Ogawa D, Kagaya Y, Sato Y, Kitano H, Nagato Y, Ishikawa R, Ono A, Kinoshita T, Takeda S, Hattori T.Rice SNF2 family helicase ENL1 is essential for syncytial endosperm development[J]. The Plant Journal, 2015, 81(1): 1-12. |
| [46] | Huang X, Peng X, Sun M X. OsGCD1 is essential for rice fertility and required for embryo dorsal-ventral pattern formation and endosperm development[J]. New Phytologist, 2017, 215(3): 1039-1058. |
| [47] | Becraft P W, Yi G.Regulation of aleurone development in cereal grains[J]. Journal of Experimental Botany, 2011, 62(5): 1669-1675. |
| [48] | Yan D, Duermeyer L, Leoveanu C, Nambara E.The functions of the endosperm during seed germination[J]. Plant & Cell Physiology, 2014, 55(9): 1521-1533. |
| [49] | Wu H, Gontarek B C, Yi G, Beall B D, Neelakandan A K, Adhikari B, Chen R, McCarty D R, Severin A J, Becraft P W. The thick aleurone1 gene encodes a NOT1 subunit of the CCR4-NOT complex and regulates cell patterning in endosperm[J]. Plant Physiology, 2020, 184: 00703.2020. DOI: 10.1104/pp.20.00703 |
| [50] | Lid S E, Gruis D, Jung R, Lorentzen J A, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen O A.The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(8): 5460-5465. |
| [51] | Yi G, Neelakandan A K, Gontarek B C, Vollbrecht E, Becraft P W.The naked endosperm genes encode duplicate INDETERMINATE domain transcription factors required for maize endosperm cell patterning and differentiation[J]. Plant Physiology, 2015, 167(2): 443-456. |
| [52] | Gontarek B C, Neelakandan A K, Wu H, Becraft P W.NKD transcription factors are central regulators of maize endosperm development[J]. Plant Cell, 2016, 28(12): 2916-2936. |
| [53] | Pu C X, Ma Y, Wang J, Zhang Y C, Jiao X W, Hu Y H, Wang L L, Zhu Z G, Sun D, Sun Y.Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation[J]. The Plant Journal, 2012, 70(6): 940-953. |
| [54] | Hibara K ichiro, Obara M, Hayashida E, Abe M, Ishimaru T, Satoh H, Itoh J, Nagato Y. The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice[J]. Developmental Biology, 2009, 334(2): 345-354. |
| [55] | Kawakatsu T, Yamamoto M P, Touno S M, Yasuda H, Takaiwa F.Compensation and interaction between RISBZ1 and RPBF during grain filling in rice[J]. The Plant Journal, 2009, 59(6): 908-920. |
| [56] | Qi X, Li S, Zhu Y, Zhao Q, Zhu D, Yu J.ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm[J]. Plant Molecular Biology, 2017, 93(1-2): 7-20. |
| [57] | Liu J, Wu X, Yao X, Yu R, Larkin P J, Liu C M.Mutations in the DNA demethylase OsROS1 result in a thickened aleurone and improved nutritional value in rice grains[J]. Proceedings of the National Academy of Sciences, 2018, 115(44): 11327-11332. |
| [58] | Liu E, Zeng S, Zhu S, Liu Y, Wu G, Zhao K, Liu X, Liu Q, Dong Z, Dang X, Xie H, Li D, Hu X, Hong D.Favorable alleles of GRAIN-FILLING RATE1 increase the grain-filling rate and yield of rice[J]. Plant Physiology, 2019, 181(3): 1207-1222. |
| [59] | Scofield G N, Hirose T, Gaudron J A, Upadhyaya N M, Ohsugi R, Furbank R T.Antisense suppression of the rice sucrose transporter gene, OsSUT1, leads to impaired grain filling and germination but does not affect photosynthesis[J]. Functional Plant Biology, 2002, 29(7): 815-826. |
| [60] | Sosso D, Luo D, Li Q B, Sasse J, Yang J, Gendrot G, Suzuki M, Koch K E, McCarty D R, Chourey P S, Rogowsky P M, Ross-Ibarra J, Yang B, Frommer W B. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport[J]. Nature Genetics, 2015, 47(12): 1489-1493. |
| [61] | Ma L, Zhang D, Miao Q, Yang J, Xuan Y, Hu Y.Essential Role of Sugar Transporter OsSWEET11 during the early stage of rice grain filling[J]. Plant and Cell Physiology, 2017, 58(5): 863-873. |
| [62] | Yang J, Luo D, Yang B, Frommer W B, Eom J S.SWEET11 and 15 as key players in seed filling in rice[J]. The New Phytologist, 2018, 218(2): 604-615. |
| [63] | Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, Ma H, Zhang G, He Z.Control of rice grain-filling and yield by a gene with a potential signature of domestication[J]. Nature Genetics, 2008, 40(11): 1370-1374. |
| [64] | Asano T, Kunieda N, Omura Y, Ibe H, Kawasaki T, Takano M, Sato M, Furuhashi H, Mujin T, Takaiwa F, Wu Cy C, Tada Y, Satozawa T, Sakamoto M, Shimada H.Rice SPK, a calmodulin-like domain protein kinase, is required for storage product accumulation during seed development: phosphorylation of sucrose synthase is a possible factor[J]. The Plant Cell, 2002, 14(3): 619-628. |
| [65] | Jeon J S, Ryoo N, Hahn T R, Walia H, Nakamura Y.Starch biosynthesis in cereal endosperm[J]. Plant Physiology and Biochemistry, Elsevier Masson SAS, 2010, 48(6): 383-392. |
| [66] | Wei X, Jiao G, Lin H, Sheng Z, Shao G, Xie L, Tang S, Xu Q, Hu P.GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development[J]. Journal of Integrative Plant Biology, 2017, 59(2): 134-153. |
| [67] | Wang J C C, Xu H, Zhu Y, Liu Q Q Q, Cai X L L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm[J]. Journal of Experimental Botany, 2013, 64(11): 3453-3466. |
| [68] | Yin L L, Xue H W.The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development[J]. The Plant Cell, 2012, 24(3): 1049-1065. |
| [69] | Fu F F, Xue H W.Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator[J]. Plant Physiology, 2010, 154(2): 927-938. |
| [70] | Laloum T, De Mita S, Gamas P, Baudin M, Niebel A.CCAAT-box binding transcription factors in plants: Y so many?[J]. Trends in Plant Science, 2013, 18(3): 157-166. |
| [71] | Zhiguo E, Li T, Zhang H, Liu Z, Deng H, Sharma S, Wei X, Wang L, Niu B, Chen C.A group of nuclear factor y transcription factors are sub-functionalized during endosperm development in monocots[J]. Journal of Experimental Botany, 2018, 69(10): 2495-2510. |
| [72] | Xu J J, Zhang X F, Xue H W.Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor[J]. Journal of Experimental Botany, 2016, 67(22): 6399-6411. |
| [73] | Bello B K, Hou Y, Zhao J, Jiao G, Wu Y, Li Z, Wang Y, Tong X, Wang W, Yuan W, Wei X, Zhang J.NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.)[J]. Plant Biotechnology Journal, 2019, 17(7): 1222-1235. |
| [74] | Niu B, Deng H, Li T, Sharma S, Yun Q, Li Q, E Z G, Chen C. OsbZIP76 interacts with OsNF-YBs and regulates endosperm cellularization in rice(Oryza sativa)[J]. Journal of Integrative Plant Biology, 2020, 62(12): 1983-1996. |
| [75] | Bai A N, Lu X D, Li D Q, Liu J X, Liu C M.NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm[J]. Cell Research, 2016, 26(3): 384-388. |
| [76] | Pelletier J M, Kwong R W, Park S, Le B H, Baden R, Cagliari A, Hashimoto M, Munoz M D, Fischer R L, Goldberg R B, Harada J J.LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development[J]. Proceedings of the National Academy of Sciences, 2017, 114(32): E6710-E6719. |
| [77] | Jo L, Pelletier J M, Hsu S, Baden R, Goldberg R B, Harada J J.Combinatorial interactions of the LEC1 transcription factor specify diverse developmental programs during soybean seed development[J]. Proceedings of the National Academy of Sciences, 2020, 117(2): 1223-1232. |
| [78] | Zhang Z, Dong J, Ji C, Wu Y, Messing J.NAC-type transcription factors regulate accumulation of starch and protein in maize seeds[J]. Proceedings of the National Academy of Sciences, 2019, 116(23): 11223-11228. |
| [79] | Wang J, Chen Z, Zhang Q, Meng S, Wei C.The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis[J]. Plant Physiology, 2020, 184(4): 1775-1791. |
| [80] | Ren Y, Huang Z, Jiang H, Wang Z, Wu F, Xiong Y, Yao J.A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling[J]. Journal of Experimental Botany, 2021, 72(8): 2947-2964. |
| [81] | Mathew I E, Priyadarshini R, Mahto A, Jaiswal P, Parida S K, Agarwal P.SUPER STARCHY1/ONAC025 participates in rice grain filling[J]. Plant Direct, 2020, 4(9): 1-25. |
| [82] | Zhou H, Xia D, He Y Q.Rice grain quality-traditional traits for high quality rice and health-plus substances[J]. Molecular Breeding, 2020, 40(1): 1. DOI:10.1007/ s11032-019-1080-6. |
| [83] | Liu L, Waters D L E, Rose T J, Bao J, King G J. Phospholipids in rice: Significance in grain quality and health benefits: A review[J]. Food Chemistry, 2013, 139(1-4): 1133-1145 |
| [84] | Hu Z L, Li P, Zhou M Q, Zhang Z H, Wang L X, Zhu L H, Zhu Y G.Mapping of quantitative trait loci (QTLs) for rice protein and fat content using doubled haploid lines[J]. Euphytica, 2004, 135(1-2): 47-54. |
| [85] | Liu W, Zeng J, Jiang G, He Y.QTLs identification of crude fat content in brown rice and its genetic basis analysis using DH and two backcross populations[J]. Euphytica, 2009, 169(2): 197-205. |
| [86] | Qin Y, Kim S M, Zhao X, Lee H S, Jia B, Kim K M, Eun M Y, Sohn J K.QTL detection and MAS selection efficiency for lipid content in brown rice (Oryza sativa L.)[J]. Genes and Genomics, 2010, 32(6): 506-512. |
| [87] | Liu H L, Yin Z J, Xiao L, Xu Y N, Qu L Q.Identification and evaluation of ω-3 fatty acid desaturase genes for hyperfortifying α-linolenic acid in transgenic rice seed[J]. Journal of Experimental Botany, 2012, 63(8): 3279-3287. |
| [88] | Zaplin E S, Liu Q, Li Z, Butardo V M, Blanchard C L, Rahman S.Production of high oleic rice grains by suppressing the expression of the OsFAD2-1 gene[J]. Functional Plant Biology, 2013, 40(10): 996-1004. |
| [89] | Zhou H, Xia D, Li P, Ao Y, Xu X, Wan S, Li Y, Wu B, Shi H, Wang K, Gao G, Zhang Q, Wang G, Xiao J, Li X, Yu S, Lian X, He Y.Genetic architecture and key genes controlling the diversity of oil composition in rice grains[J]. Molecular Plant, 2021, 14(3): 456-469. |
| [90] | Zhang X F, Tong J H, Bai A N, Liu C M, Xiao L T, Xue H W.Phytohormone dynamics in developing endosperm influence rice grain shape and quality[J]. Journal of Integrative Plant Biology, 2020, 62(10): 1625-1637. |
| [91] | Zhao Y.Auxin biosynthesis[J]. The Arabidopsis Book, 2014, 12: e0173. |
| [92] | Figueiredo D D, Batista R A, Roszak P J, Köhler C.Auxin production couples endosperm development to fertilization[J]. Nature Plants, 2015, 1: 15184. |
| [93] | Batista R A, Figueiredo D D, Santos-González J, Köhler C.Auxin regulates endosperm cellularization in Arabidopsis[J]. Genes & Development, 2019, 33(7-8): 466-476. |
| [94] | Abu-Zaitoon Y M, Bennett K, Normanly J, Nonhebel H M. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA[J]. Physiologia Plantarum, 2012, 146(4): 487-499. |
| [95] | Xu X, Zhang D, Niu B, Chen C, Yun Q, Zhou Y.OsYUC11-mediated auxin biosynthesis is essential for endosperm development of rice[J]. Plant Physiology, 2021, 185(3): 934-950. |
| [96] | Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B I, Onishi A, Miyagawa H, Katoh E.Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield[J]. Nature Genetics, 2013, 45(6): 707-711. |
| [97] | Lur H S, Setter T L.Role of auxin in maize endosperm development (timing of nuclear DNA endoreduplication, zein expression, and cytokinin)[J]. Plant Physiology, 1993, 103(1): 273-280. |
| [98] | Bernardi J, Lanubile A, Li Q B, Kumar D, Kladnik A, Cook S D, Ross J J, Marocco A, Chourey P S.Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize[J]. Plant Physiology, 2012, 160(3): 1318-1328. |
| [99] | Forestan C, Meda S, Varotto S.ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development[J]. Plant Physiology, 2010, 152(3): 1373-1390. |
| [100] | Wang Z, Xu Y, Chen T, Zhang H, Yang J, Zhang J.Abscisic acid and the key enzymes and genes in sucrose-to-starch conversion in rice spikelets in response to soil drying during grain filling[J]. Planta, 2015, 241(5): 1091-1107. |
| [101] | Zhang D, Zhang M, Zhou Y, Wang Y, Shen J, Chen H, Zhang L, Lü B, Liang G, Liang J.The rice G protein γ subunit DEP1/qPE9-1 positively regulates grain-filling process by increasing auxin and cytokinin content in rice grains[J]. Rice, 2019, 12(1): 91. |
| [102] | Qin P, Zhang G, Hu B, Wu J, Chen W, Ren Z, Liu Y, Xie J, Yuan H, Tu B, Ma B, Wang Y, Ye L, Li L, Xiang C, Li S. Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism[J]. Science Advances, 2021, 7(3): eabc8873. |
| [103] | Sreenivasulu N, Radchuk V, Alawady A, Borisjuk L, Weier D, Staroske N, Fuchs J, Miersch O, Strickert M, Usadel B, Wobus U, Grimm B, Weber H, Weschke W.De-regulation of abscisic acid contents causes abnormal endosperm development in the barley mutant seg8[J]. Plant Journal, 2010, 64(4): 589-603. |
| [104] | Xing M Q, Zhang Y J, Zhou S R, Hu W Y, Wu X X, Ye Y J, Wu X T, Xiao Y P, Li X, Xue H W.Global analysis reveals the crucial roles of DNA methylation during rice seed development[J]. Plant Physiology, 2015, 168(4): 1417-1432. |
| [105] | Yang C, Ma B, He S, Xiong Q, Duan K, Yin C, Chen H, Lu X, Chen S, Zhang J.MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3- LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice[J]. Plant Physiology, 2015, 169(1): 148-165. |
| [106] | Yin C, Zhao H, Ma B, Chen S, Zhang J.Diverse roles of ethylene in regulating agronomic traits in rice[J]. Frontiers in Plant Science, 2017, 8: 1676. DOI:10.3389/fpls.2017.01676. |
| [107] | Huang X, Lu Z, Wang X, Ouyang Y, Chen W, Xie K, Wang D, Luo M, Luo J, Yao J.Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3[J]. The Plant Journal, 2016, 87(3): 305-317. |
| [108] | Liu X, Wang P.SDG711 is involved in rice seed development through regulation of starch metabolism gene expression in coordination with other histone modi cations[J]. Rice, 2021, 14(1): 25. |
| [109] | Qin F J, Sun Q W, Huang L M, Chen X S, Zhou D X.Rice SUVH histone methyltransferase genes display specific functions in chromatin modification and retrotransposon repression[J]. Molecular Plant, 2010, 3(4): 773-782. |
| [110] | Qi D, Wen Q, Meng Z, Yuan S, Guo H, Zhao H, Cui S.OsLFR is essential for early endosperm and embryo development by interacting with SWI/SNF complex members in Oryza sativa[J]. Plant Journal, 2020, 104(4): 901-916. |
| [111] | Zhang H, Lu Y, Zhao Y, Zhou D X.OsSRT1 is involved in rice seed development through regulation of starch metabolism gene expression[J]. Plant Science, 2016, 248: 28-36. |
| [112] | La H, Ding B, Mishra G P, Zhou B, Yang H, Bellizzi M D R, Chen S, Meyers B C, Peng Z, Zhu J K, Wang G L. A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice[J]. Proceedings of the National Academy of Sciences USA, 2011, 108(37): 15498-15503. |
| [113] | Köhler C, Wolff P, Spillane C.Epigenetic mechanisms underlying genomic imprinting in plants[J]. Annual Review of Plant Biology, 2012, 63(1): 331-352. |
| [114] | Kermicle J L.Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission[J]. Genetics, 1970, 66(1): 69-85. |
| [115] | Gehring M, Missirian V, Henikoff S.Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds[J]. PLoS ONE, 2011, 6(8): e23687. DOI: 10.1371/journal.pone.0023687 |
| [116] | Yang G, Liu Z, Gao L, Yu K, Feng M, Yao Y, Peng H, Hu Z, Sun Q, Ni Z, Xin M.Genomic imprinting was evolutionarily conserved during wheat polyploidization[J]. Plant Cell, 2018, 30(1): 37-47. |
| [117] | Luo M, Taylor J M, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A.A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm[J]. PLoS Genet, 2011, 7(6): e1002125. |
| [118] | Chen C, Li T, Zhu S, Liu Z, Shi Z, Zheng X, Chen R, Huang J, Shen Y, Luo S, Wang L, Liu Q Q, E Z G. Characterization of imprinted genes in rice reveals conservation of regulation and imprinting with other plant species[J]. Plant Physiology, 2018, 177(4): 1754-1771. |
| [119] | Florez-Rueda A M, Paris M, Schmidt A, Widmer A, Grossniklaus U, Städler T. Genomic imprinting in the endosperm is systematically perturbed in abortive hybrid tomato seeds[J]. Molecular Biology and Evolution, 2016, 33(11): 2935-2946. |
| [120] | Xu W, Dai M, Li F, Liu A.Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean[J]. Nucleic Acids Research, 2014, 42(11): 6987-6998. |
| [121] | Hsieh T-F F, Shin J, Uzawa R, Silva P, Cohen S, Bauer M J, Hashimoto M, Kirkbride R C, Harada J J, Zilberman D, Fischer R L. Regulation of imprinted gene expression in Arabidopsis endosperm[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(5): 1755-1762. |
| [122] | Waters A J, Bilinski P, Eichten S R, Vaughn M W, Ross-Ibarra J, Gehring M, Springer N M.Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species[J]. Proceedings of the National Academy of Sciences, 2013, 110(48): 19639-19644. |
| [123] | Zhang M, Li N, He W, Zhang H, Yang W, Liu B.Genome-wide screen of genes imprinted in sorghum endosperm and the roles of allelic differential cytosine methylation[J]. The Plant Journal, 2015: 424-436. |
| [124] | Waters A J, Makarevitch I, Eichten S R, Swanson-Wagner R A, Yeh C T, Xu W, Schnable P S, Vaughn M W, Gehring M, Springer N M. Parent-of -origin effects on gene expression and DNA methylation in the maize endosperm[J]. The Plant Cell, 2011, 23(12): 4221-4233. |
| [125] | Hatorangan M R, Laenen B, Steige K, Slotte T, Köhler C.Rapid evolution of genomic imprinting in two species of the brassicaceae[J]. The Plant Cell, 2016, 28(8): 1815-1827. |
| [126] | Pignatta D, Erdmann R M, Scheer E, Picard C L, Bell G W, Gehring M.Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting[J]. eLife, 2014: e03198. |
| [127] | Batista R A, Köhler C.Genomic imprinting in plants-revisiting existing models[J]. Genes & Development, 2020, 34(1-2): 24-36. |
| [128] | Huh J H, Bauer M J, Hsieh T F, Fischer R L.Cellular programming of plant gene imprinting[J]. Cell, 2008, 132(5): 735-744. |
| [129] | Burkart-Waco D, Ngo K, Lieberman M, Comai L.Perturbation of parentally biased gene expression during interspecific hybridization[J]. PloS ONE, 2015, 10(2): e0117293. |
| [130] | Kradolfer D, Wolff P, Jiang H, Siretskiy A, Kohler C.An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana[J]. Development Cell, 2013, 26(5): 525-535. |
| [131] | Jullien P E, Berger F.Parental genome dosage imbalance deregulates imprinting in Arabidopsis[J]. PLoS Genetics, 2010, 6(3): e1000885. |
| [132] | Kinoshita T, Yadegari R, Harada J J, Goldberg R B, Fischer R L.Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm[J]. The Plant Cell, 1999, 11(10): 1945-1952. |
| [133] | Liu P, Qi M, Wang Y, Chang M, Liu C, Sun M, Yang W, Ren H.Arabidopsis RAN1 mediates seed development through its parental ratio by affecting the onset of endosperm cellularization[J]. Molecular Plant, 2014, 7(8): 1316-1328. |
| [134] | Wolff P, Jiang H, Wang G, Santos-González J, Köhler C.Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana[J]. eLife, 2015, 4: 1-14. |
| [135] | Wang G, Jiang H, Del Toro de León G, Martinez G, Köhler C. Sequestration of a transposon-derived sirna by a target mimic imprinted gene induces postzygotic reproductive isolation in Arabidopsis[J]. Developmental Cell, 2018, 46(6): 696-705. |
| [136] | Yuan J, Chen S, Jiao W, Wang L, Wang L, Ye W, Lu J, Hong D, You S, Cheng Z, Yang D L, Chen Z J.Both maternally and paternally imprinted genes regulate seed development in rice[J]. New Phytologist, 2017, 216(2): 373-387. |
| [137] | Folsom J J, Begcy K, Hao X, Wang D, Walia H.Rice fertilization-independent endosperm1 regulates seed size under heat stress by controlling early endosperm development[J]. Plant Physiology, 2014, 165(1): 238-248. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||