
Chinese Journal OF Rice Science ›› 2018, Vol. 32 ›› Issue (3): 219-225.DOI: 10.16819/j.1001-7216.2018.7106
• Research Papers • Previous Articles Next Articles
Xuejiao HU1,2, Jia YANG1,3, Can CHENG1, Jihua ZHOU1, Fuan NIU1, Xinqi WANG1, Meiliang ZHANG2, Liming CAO1,*( ), Huangwei CHU1,*(
), Huangwei CHU1,*( )
)
Received:2017-09-01
Revised:2017-12-27
Online:2018-05-10
Published:2018-05-10
Contact:
Liming CAO, Huangwei CHU
胡雪娇1,2, 杨佳1,3, 程灿1, 周继华1, 牛付安1, 王新其1, 张美良2, 曹黎明1,*( ), 储黄伟1,*(
), 储黄伟1,*( )
)
通讯作者:
曹黎明,储黄伟
基金资助:CLC Number:
Xuejiao HU, Jia YANG, Can CHENG, Jihua ZHOU, Fuan NIU, Xinqi WANG, Meiliang ZHANG, Liming CAO, Huangwei CHU. Targeted Editing of Rice SD1 Gene UsingCRISPR/Cas9 System[J]. Chinese Journal OF Rice Science, 2018, 32(3): 219-225.
胡雪娇, 杨佳, 程灿, 周继华, 牛付安, 王新其, 张美良, 曹黎明, 储黄伟. 利用CRISPR/Cas9系统定向编辑水稻SD1基因[J]. 中国水稻科学, 2018, 32(3): 219-225.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2018.7106
| 引物名称 Primer name | 引物序列(5′-3′) Primer sequence (5′-3′) |
| Oligo-F | TGGCGGAGGATGGAGCCCAAGATCC |
| Oligo-R | AAACGGATCTTGGGCTCCATCCTC |
| M13-F SEQ1-F SEQ1-R EXON1-F EXON1-R EXON3-F EXON3-R HYG-F HYG-R CAS9-F CAS9-R | TGTAAAACGACGGCCAGT GGGTCATTGATTCGACCATC GTGCTCGGACACCTGGAAGAAC GTCATTGATTCGACCATCATGTCTGTC GAATTACTTGTTCTGTTGCTTCGAAGCA CTCTCCGTTGATGAATGATGATG CTTCTGTTCGTTCCGTTTCGTTC CGTGCTTTCAGCTTCGATGTAGGA GCATATGAAATCACGCCATGTAGTGT CTTCATCGAGCGGATGACCAACT AGATCGTGGTATGTGCCCAGGGA |
Table 1 Primers used in this research.
| 引物名称 Primer name | 引物序列(5′-3′) Primer sequence (5′-3′) |
| Oligo-F | TGGCGGAGGATGGAGCCCAAGATCC |
| Oligo-R | AAACGGATCTTGGGCTCCATCCTC |
| M13-F SEQ1-F SEQ1-R EXON1-F EXON1-R EXON3-F EXON3-R HYG-F HYG-R CAS9-F CAS9-R | TGTAAAACGACGGCCAGT GGGTCATTGATTCGACCATC GTGCTCGGACACCTGGAAGAAC GTCATTGATTCGACCATCATGTCTGTC GAATTACTTGTTCTGTTGCTTCGAAGCA CTCTCCGTTGATGAATGATGATG CTTCTGTTCGTTCCGTTTCGTTC CGTGCTTTCAGCTTCGATGTAGGA GCATATGAAATCACGCCATGTAGTGT CTTCATCGAGCGGATGACCAACT AGATCGTGGTATGTGCCCAGGGA |
| 靶位点编号 Targetnumber | 靶位点序列 Sequence of target site | 候选识别位点的毗邻基序Protospacer adjacent motif | 位置 Site / bp | DNA链 DNA strand |
| 1 | GAGGATGGAGCCCAAGATCC | CGG | 108 | + |
| 2 | AGATCCCGGAGCCATTCGTG | TGG | 122 | + |
| 3 | GAGCCATTCGTGTGGCCGAA | CGG | 130 | + |
| 4 | GGATCTTGGGCTCCATCCTC | AGG | 107 | – |
| 5 | ACGAATGGCTCCGGGATCTT | GGG | 120 | – |
| 6 | CACGAATGGCTCCGGGATCT | TGG | 121 | – |
| 7 | TTCGGCCACACGAATGGCTC | CGG | 129 | – |
Table 2 Putative CRISPR/Cas9 target sites in rice SD1 gene.
| 靶位点编号 Targetnumber | 靶位点序列 Sequence of target site | 候选识别位点的毗邻基序Protospacer adjacent motif | 位置 Site / bp | DNA链 DNA strand |
| 1 | GAGGATGGAGCCCAAGATCC | CGG | 108 | + |
| 2 | AGATCCCGGAGCCATTCGTG | TGG | 122 | + |
| 3 | GAGCCATTCGTGTGGCCGAA | CGG | 130 | + |
| 4 | GGATCTTGGGCTCCATCCTC | AGG | 107 | – |
| 5 | ACGAATGGCTCCGGGATCTT | GGG | 120 | – |
| 6 | CACGAATGGCTCCGGGATCT | TGG | 121 | – |
| 7 | TTCGGCCACACGAATGGCTC | CGG | 129 | – |

Fig. 3. Mutation type in SD1 gene of T0transgenic lines. The grey letters indicated the target sequence. The underline indicated protospacer adjacent motif(PAM) sequence. The box represented the insert base. The transverse line indicated deletion sequence. Arrow indicated the Cas9 cleavage site.
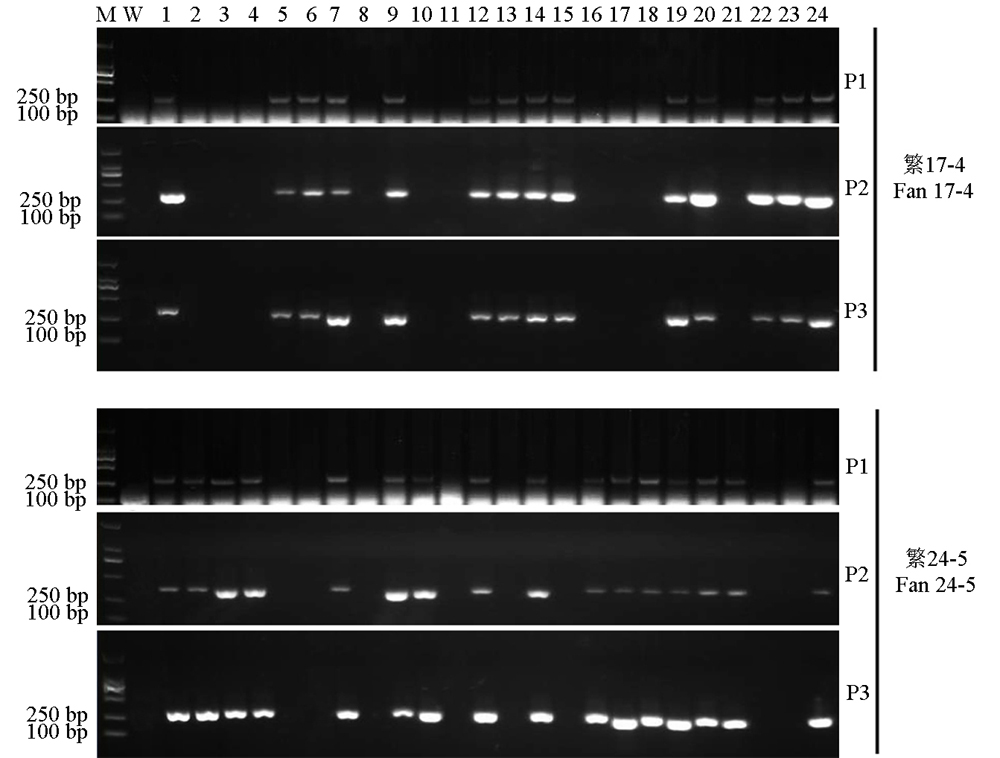
Fig. 4. PCR screening of transgenic-free plants in T1 generation. M, D2000marker, W, Wild type;Lanes 1-24, T1 transgenic plants; P1, Primer M13F and Oligo-R;P2, Primer HYG-F and HYG-R; P3, Primer Cas9-F and Cas9-R.
| [1] | Doebley J F, Gaut B S, Smith B D.The molecular genetics of crop domestication.Cell, 2006, 127(7): 1309-1321. |
| [2] | Liu Q, Xu X, Ren X, Fu H, Wu D, Shu Q.Generation and characterization of low phytic acid germplasm in rice ( Oryza sativa L.).Theor Appl Genet, 2007, 114(5): 803-814. |
| [3] | Cho S W, Kim S, Kim J M, Kim J S.Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease.Nat Biotechnol, 2013, 31(3): 230-232. |
| [4] | Cong L, Ran F A, Cox D, Lin S, Barretto R, Habib N, Hsu P D, Wu X, Jiang W, Marraffini L A, Zhang F.Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339(6121): 819-23. |
| [5] | Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J.RNA-programmed genome editing in human cells.Elife, 2013, 2: e00471. |
| [6] | Mali P, Aach J, Stranges P B, Esvelt K M, Moosburner M, Kosuri S, Yang L, Church G M.CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering.Nat Biotechnol, 2013, 31(9): 833-838. |
| [7] | Mani M, Kandavelou K, Dy F J, Durai S, Chandrasegaran S.Design, engineering, and characterization of zinc finger nucleases.Biochem Biophys Res Commun, 2005, 335(2): 447-457. |
| [8] | Bogdanove A J, Voytas D F.TAL effectors: Customizable proteins for DNA targeting.Science, 2011, 333(6051): 1843-1846. |
| [9] | Samanta M K, Dey A, Gayen S.CRISPR/Cas9: An advanced tool for editing plant genomes.Transgenic Res, 2016, 25(5): 561-573. |
| [10] | Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu J K.Application of the CRISPR-Cas system for efficient genome engineering in plants.Mol Plant, 2013, 6(6): 2008-2011. |
| [11] | Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi J J, Qiu J L, Gao C.Targeted genome modification of crop plants using a CRISPR-Cas system.Nat Biotechnol, 2013, 31(8): 686-688. |
| [12] | Jiang W Z, Zhou H B, Bi H H, Fromm M, Yang B, Weeks D P.Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice.Nucleic Acids Res, 2013, 41(20): e188. |
| [13] | Liang Z, Zhang K, Chen K, Gao C.Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system.J Genet Genom, 2014, 41(2): 63-68. |
| [14] | Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J L.Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew.Nat Biotechnol, 2014, 32(9): 947-951. |
| [15] | Jacobs T B, LaFayette P R, Schmitz R J, Parrott W A. Targeted genome modifications in soybean with CRISPR/Cas9.BMC Biotechnol, 2015, 15: 16. |
| [16] | Li Z, Liu Z B, Xing A, Moon B P, Koellhoffer J P, Huang L, Ward R T, Clifton E, Falco S C, Cigan A M.Cas9-Guide RNA directed genome editing in soybean.Plant Physiol, 2015, 169(2): 960-970. |
| [17] | Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y.Targeted mutagenesis in soybean using the CRISPR-Cas9 system.Sci Rep, 2015, 5: 10342. |
| [18] | Brooks C, Nekrasov V, Lippman Z B, Van Eck J.Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system.Plant Physiol, 2014, 166(3): 1292-1297. |
| [19] | Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu J K.The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation.Plant Biotechnol J, 2014, 12(6): 797-807. |
| [20] | Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y.Positional cloning of rice semidwarfing gene,sd-1: Rice “Green Revolution Gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res, 2002, 9(1): 11-17. |
| [21] | Spielmeyer W, Ellis M H, Chandler P M.Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA, 2002, 99(13): 9043-9048. |
| [22] | Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush G S, Kitano H, Matsuoka M.Green revolution: A mutant gibberellin-synthesis gene in rice.Nature, 2002, 416(6882): 701-702. |
| [23] | 谷福林, 翟虎渠, 万建民, 张红生. 水稻矮秆性状研究及矮源育种利用. 江苏农业学报, 2003, 19(1): 48-54. |
| Gu F L, Zhai H Q, Wan L M, Zhang H S.Study on Inheritance of dwarf character and its utilization in rice(Oryza sativa L.) breeding. Jiangsu JAgric Sci, 2003, 19(1):48-54. (in Chinese with English abstract) | |
| [24] | Asano K, Yamasaki M, Takuno S, Miura K, Katagiri S, Ito T, Doi K, Wu J, Ebana K, Matsumoto T, Innan H, Kitano H, Ashikari M, Matsuoka M. Artificial selection for a green revolution gene during japonica rice domestication.Proc Natl Acad Sci USA#, 2011, 108(27): 11034-11039. |
| [25] | Yan M, Zhou S R, Xue H W.CRISPR Primer Designer: Design primers for knockout and chromosome imaging CRISPR-Cas system.J Integr Plant Biol, 2015, 57(7): 613-617. |
| [26] | Qiao F, Zhao K J.The Influence of RNAi targeting of OsGA20ox2gene on plant height in rice. Plant Mol Biol Rep, 2011, 29(4): 952. |
| [27] | Harrison M M, Jenkins B V, O’Connor-Giles K M, Wildonger J. A CRISPR view of development.Genes Dev, 2014, 28(17): 1859-1872. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||