
Chinese Journal OF Rice Science ›› 2017, Vol. 31 ›› Issue (3): 223-231.DOI: 10.16819/j.1001-7216.2017.7029 223
• Orginal Article • Next Articles
Lan SHEN1,2,#, Jian LI3,#, Yaping FU2, Junjie WANG2, Yufeng HUA2, Xiaozhen JIAO2, Changjie YAN1,*( ), Kejian WANG2,*(
), Kejian WANG2,*( )
)
Received:2017-03-09
Revised:2017-03-20
Online:2017-05-10
Published:2017-05-10
Contact:
Changjie YAN, Kejian WANG
沈兰1,2,#, 李健3,#, 付亚萍2, 王俊杰2, 华宇峰2, 焦晓真2, 严长杰1,*( ), 王克剑2,*(
), 王克剑2,*( )
)
通讯作者:
严长杰,王克剑
基金资助:CLC Number:
Lan SHEN, Jian LI, Yaping FU, Junjie WANG, Yufeng HUA, Xiaozhen JIAO, Changjie YAN, Kejian WANG. Orientation Improvement of Grain Length and Grain Number in Rice by Using CRISPR/Cas9 System[J]. Chinese Journal OF Rice Science, 2017, 31(3): 223-231.
沈兰, 李健, 付亚萍, 王俊杰, 华宇峰, 焦晓真, 严长杰, 王克剑. 利用CRISPR/Cas9系统定向改良水稻粒长和穗粒数性状[J]. 中国水稻科学, 2017, 31(3): 223-231.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2017.7029 223
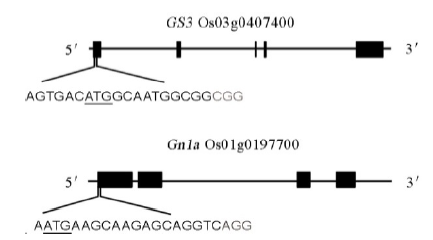
Fig. 1. Schematic diagram of the targeted sites in GS3 and Gn1a. The underlined letters are the initiation codons. The gray letters are the protospacer adjacent motif (PAM) sequences.

Fig. 2. Flow diagram of CRISPR/Cas9 system for construction. The intermediate vector SK-gRNA contains the U3 promotor and sgRNA scaffold. Binary vector pC1300-Cas9 contains the 2×35S promotor and a Cas9 protein. Two gRNA scaffolds with GS3, Gn1a gene targets are respectively digested with KpnⅠ/ SalⅠ, and XhoⅠ/ BglⅡ, and cloned into pC1300-Cas9 between the KpnⅠand BamHⅠsites in a one-step ligation.
| 引物名称 Primer name | 引物序列 Sequence(5′-3′) | |
|---|---|---|
| GS3-g++ | GGCAGTGACATGGCAATGGCGG | |
| GS3-g–– | AAACCCGCCATTGCCATGTCAC | |
| Gn1a-g++ | GGCAATGAAGCAAGAGCAGGTC | |
| Gn1a-g–– | AAACGACCTGCTCTTGCTTCAT | |
| GS3-JC-F | CTATACATAGCTGCTGCAC | |
| GS3-JC-R | GACAGATAGCAAGCCGTAC | |
| Gn1a-JC-F | TTCCATCGTCAGCACACAAA | |
| Gn1a-JC-R | ACGGAGAGGTTGCCAAAGTC | |
| Hyg-F1 | GCTGTTATGCGGCCATTGTC | |
| Hyg-R1 | GACGTCTGTCGAGAAGTTTC | |
| Cas9-F2 | ACCAGACACGAGACGACTAA | |
| pC1300-R2 | ATCGGTGCGGGCCTCTTC | |
| Actin-F | TGCTATGTACGTCGCCATCCA | |
| Actin-R | AATGAGTAACCACGCTCCGTC | |
| Gn1a-F | CCATGGTATGCATGCAACACCATG | |
| Gn1a-R | CGTTGTCACGTACTCCCTCCGTA | |
| GS3-F | CTATACATAGCTGCTGCACCGTCT | |
| GS3-R | CAATCACGTACTCATCATGGCAGCA | |
| T3 | ATTAACCCTCACTAAAGGGA | |
Table 1 Primers used in this research.
| 引物名称 Primer name | 引物序列 Sequence(5′-3′) | |
|---|---|---|
| GS3-g++ | GGCAGTGACATGGCAATGGCGG | |
| GS3-g–– | AAACCCGCCATTGCCATGTCAC | |
| Gn1a-g++ | GGCAATGAAGCAAGAGCAGGTC | |
| Gn1a-g–– | AAACGACCTGCTCTTGCTTCAT | |
| GS3-JC-F | CTATACATAGCTGCTGCAC | |
| GS3-JC-R | GACAGATAGCAAGCCGTAC | |
| Gn1a-JC-F | TTCCATCGTCAGCACACAAA | |
| Gn1a-JC-R | ACGGAGAGGTTGCCAAAGTC | |
| Hyg-F1 | GCTGTTATGCGGCCATTGTC | |
| Hyg-R1 | GACGTCTGTCGAGAAGTTTC | |
| Cas9-F2 | ACCAGACACGAGACGACTAA | |
| pC1300-R2 | ATCGGTGCGGGCCTCTTC | |
| Actin-F | TGCTATGTACGTCGCCATCCA | |
| Actin-R | AATGAGTAACCACGCTCCGTC | |
| Gn1a-F | CCATGGTATGCATGCAACACCATG | |
| Gn1a-R | CGTTGTCACGTACTCCCTCCGTA | |
| GS3-F | CTATACATAGCTGCTGCACCGTCT | |
| GS3-R | CAATCACGTACTCATCATGGCAGCA | |
| T3 | ATTAACCCTCACTAAAGGGA | |
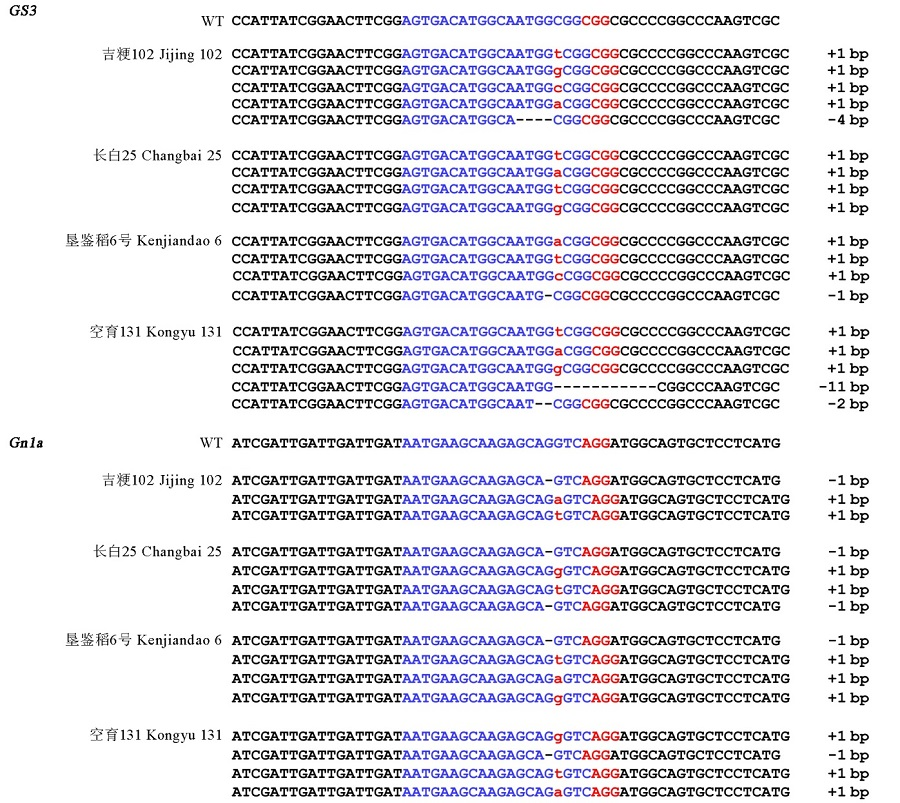
Fig. 3. Mutation types at the GS3 and Gn1a loci of the four varieties in T0 generation. The targeted sequence is highlighted in blue and the PAM sequence in red. Mutations with 1 bp insertion are represented by red lowercase letters. The deleted sequences are shown by black hyphens.
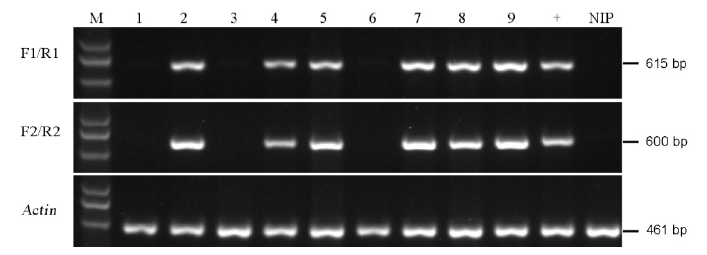
Fig. 4. PCR identification of the marker-free transgenic plants. M, Marker D5000; Lanes 1 to 9, T1 transgenic lines; +, Positive control of transgenic line, NIP, Negative control(Nipponbare).

Fig. 5. Grass morphology of the plants of the four varieties in T1 generation. J102, Jijing 102; Ch 25, Changbai 25; Ken 6, Kenjiandao 6; Ko 131, Kongyu 131.

Fig. 6. Grain size of the four varieties and their mutants in T1 generation. A, D, G, J, Grain shapes of Jijing 102, Changbai 25, Kenjiandao 6 and Kongyu 131 and their mutants, respectively. B, E, H, K and C, F, I, L show the grain length and grain width, respectively. *, **, Significant difference at 0.05 and 0.01 levels, respectively. The same as in Fig 7 and 8.
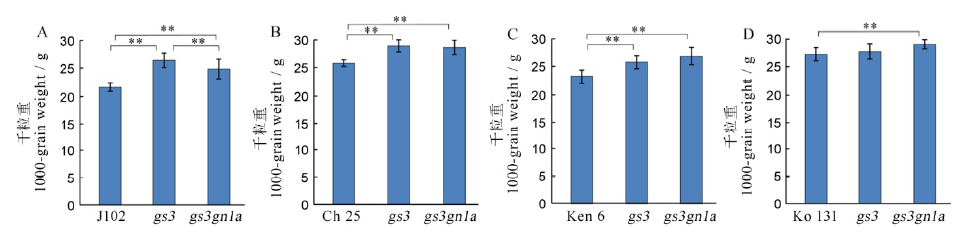
Fig. 7. 1000-grain weight among the four varieties and their mutants in T1 generation. A, B, C, D shows the 1000-grain weight of Jijing 102, Changbai 25, Kenjiandao 6 and Kongyu 131 and their mutants, respectively.
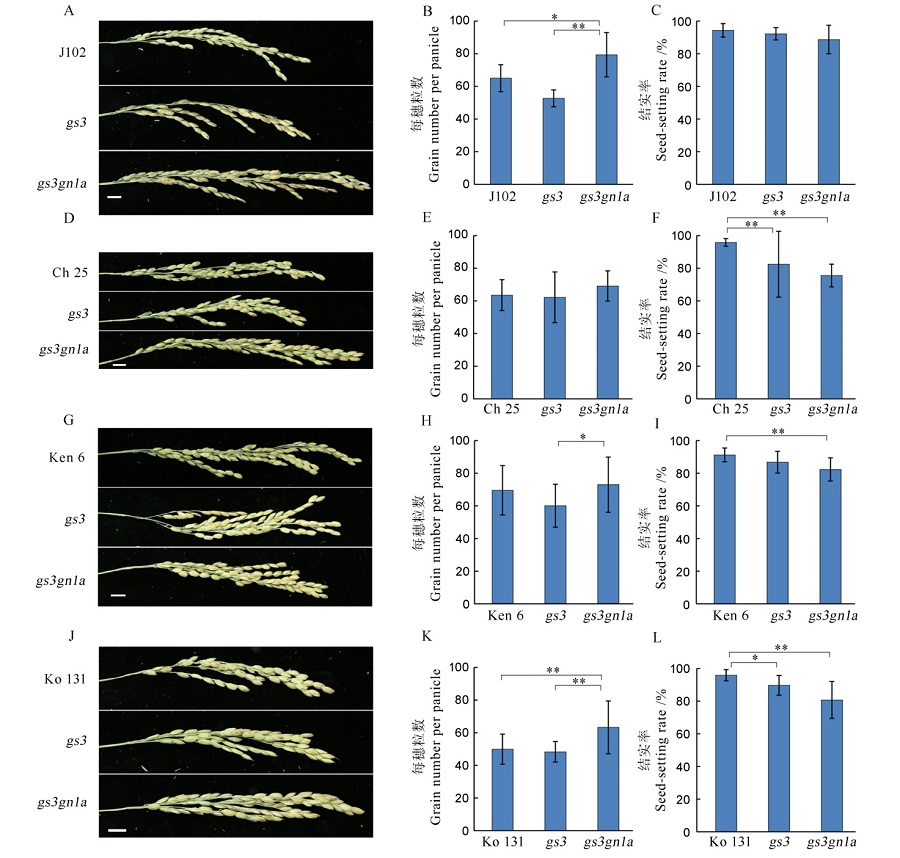
Fig. 8. Grain number per panicle and the seed-setting rate among the four varieties in T1 generation and their mutants. A, D, G, J indicate the panicle phenotype of Jijing 102, Changbai 25, Kenjiandao 6 and Kongyu 131 and their mutants, respectively. B, E, H, K and C, F, I, L show the grain number per panicle and the seed-setting rate, respectively.
| [1] | Wiedenheft B, Sternberg S H, Doudna J A.RNA-guided genetic silencing systems in bacteria and archaea.Nature, 2012, 482: 331-338. |
| [2] | Cong L, Ran F A, Cox D, Lin S, Barretto R, Habib N, Hsu P D, Wu X, Jiang W, Marraffini L A, Zhang F.Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339: 819-823. |
| [3] | Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi J J, Qiu J L, Gao C.Targeted genome modification of crop plants using a CRISPR/Cas system. Nat Biotechnol, 2013, 31: 686-688. |
| [4] | DiCarlo J E, Norville J E, Mali P, Rios X, Aach J, Church G M. Genome engineering in Saccharomyces cerevisiae using CRISPR/Cas systems.Nucleic Acids Res, 2013, 41: 4336-4343. |
| [5] | Jakociunas T, Bonde I, Herrgard M, Harrison SJ, Kristensen M, Pedersen L E, Jensen M K, Keasling J D.Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae.Metab Eng, 2015, 28: 213-222. |
| [6] | Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S, Yi X, Guo L, Esteban M A, Zeng Y, Yang H, Lai L.Generation of CRISPR/Cas 9-mediated gene-targeted pigs via somatic cell nuclear transfer.Cell Mol Life Sci, 2015, 72: 1175-1184. |
| [7] | Zou Q, Wang X, Liu Y, Ouyang Z, Long H, Wei S, Xin J, Zhao B, Lai S, Shen J, Ni Q, Yang H, Zhong H, Li L, Hu M, Zhang Q, Zhou Z, He J, Yan Q, Fan N, Zhao Y, Liu Z, Guo L, Huang J, Zhang G, Ying J, Lai L, Gao X.Generation of gene-target dogs using CRISPR/Cas 9 system.J Mol Cell Biol, 2015, 7: 580-583. |
| [8] | Wang H, Yang H, Shivalila C S, Dawlaty M M, Cheng A W, Zhang F, Jaenisch R.One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas- mediated genome engineering.Cell, 2013, 153: 910-918. |
| [9] | Hoshijima K, Jurynec M J, Grunwald D J.Precise editing of the zebrafish genome made simple and efficient.Dev Cell, 2016, 36: 654-667. |
| [10] | Gratz S J, Harrison M M, Wildonger J, O'Connor-Giles K M. Precise genome editing of drosophila with CRISPR/Cas RNA-guided Cas9.Methods Mol Biol, 2015, 1311: 335-348. |
| [11] | Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu J K.Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant, 2013, 6: 2008-2011. |
| [12] | Puchta H.Applying crispr/cas for genome engineering in plants: The best is yet to come.Curr Opin Plant Biol, 2016, 36: 1-8. |
| [13] | Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks D P.Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice.Nucleic Acids Res, 2013, 41: e188. |
| [14] | Feng Z, Zhang B, Ding W, Liu X, Yang D L, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu J K.Efficient genome editing in plants using a CRISPR/Cas system.Cell Res, 2013, 23: 1229-1232. |
| [15] | Wang C, Shen L, Fu Y, Yan C, Wang K.A simple crispr/cas9 system for multiplex genome editing in rice. J Genet Genom, 2015, 42: 703-706. |
| [16] | Zhang Z J, Mao Y F, Ha S, Liu W S, Botella J R, Zhu J K.A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis.Plant Cell Rep, 2016, 35: 1519-1533. |
| [17] | Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G.A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants.Mol Plant, 2015, 8: 1274-1284. |
| [18] | Ma X, Chen L, Zhu Q, Chen Y, Liu Y G.Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products.Mol Plant, 2015, 8: 1285-1287. |
| [19] | Xu R F, Li H, Qin R Y, Li J, Qiu C H, Yang Y C, Ma H, Li L, Wei P C, Yang J B.Generation of inheritable and "transgene clean" targeted genome-modified rice in later generations using the CRISPR/Cas9 system.Sci Rep, 2015, 5: 11491 |
| [20] | Li J, Sun Y, Du J, Zhao Y, Xia L.Generation of targeted point mutations in rice by a modified CRISPR/CAS9 system. Mol Plant, 2017, 10(3): 526-529. |
| [21] | Lu Y, Zhu J K.Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system.Mol Plant, 2017, 10(3): 523-525. |
| [22] | Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q.Gs3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet, 2006, 112: 1164-1171. |
| [23] | Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q.Linking differential domain functions of theGS3 protein to natural variation of grain size in rice.Proc Natl Acad Sci U S A, 2010, 107: 19579-19584. |
| [24] | Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles E R, Qian Q, Kitano H, Matsuoka M.Cytokinin oxidase regulates rice grain production.Science, 2005, 309: 741-745. |
| [25] | Shen L, Wang C, Fu Y, Wang J, Liu Q, Zhang X, Yan C, Qian Q, Wang K.QTL editing confers opposing yield performance in different rice varieties.J Integr Plant Biol, 2016, DOI: 10.1111/jipb.12501 |
| [26] | Hiei Y, Ohta S, Komari T, Kumashiro T.Efficient transformation of rice(Oryza sativa L.) mediated by agrobacterium and sequence analysis of the boundaries of the t-DNA.Plant J, 1994, 6: 271-282. |
| [27] | Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G, Li H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci, 2016, 7: |
| [28] | Feng C, Yuan J, Wang R, Liu Y, Birchler J A, Han F.Efficient targeted genome modification in maize using CRISPR/Cas 9 system. J Genet Genom, 2016, 43: 37-43. |
| [29] | Svitashev S, Young J K, Schwartz C, Gao H, Falco S C, Cigan A M.Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA.Plant Physiol, 2015, 169: 931-945 |
| [30] | Shan Q, Wang Y, Li J, Gao C.Genome editing in rice and wheat using the CRISPR/Cas system.Nat Protoc, 2014, 9: 2395-2410. |
| [31] | Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J L.Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew.Nat Biotechnol, 2014, 32: 947-951. |
| [32] | Lawrenson T, Shorinola O, Stacey N, Li C, Ostergaard L, Patron N, Uauy C, Harwood W.Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guidedCas9 nuclease. Genome Biol, 2015, 16: 258. |
| [33] | Cai Y, Chen L, Liu X, Sun S, Wu C, Jiang B, Han T, Hou W.CRISPR/Cas 9-mediated genome editing in soybean hairy roots.PLoS One, 2015, 10: e0136064. |
| [34] | Jacobs T B, LaFayette P R, Schmitz R J, Parrott W A. Targeted genome modifications in soybean with CRISPR/Cas 9.BMC Biotechnol, 2015, 15: 16. |
| [35] | Xing Y, Zhang Q.Genetic and molecular bases of rice yield.Annu Rev Plant Biol, 2010, 61: 421-442. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||