
Chinese Journal OF Rice Science ›› 2026, Vol. 40 ›› Issue (1): 61-71.DOI: 10.16819/j.1001-7216.2026.250211
• Research Papers • Previous Articles Next Articles
HUANG Qina, JIANG Hongrui, YANG Jie, YU Kunyu, YANG Changdeng, LIANG Yan*( )
)
Received:2025-02-25
Revised:2025-03-21
Online:2026-01-10
Published:2026-01-21
Contact:
LIANG Yan
通讯作者:
梁燕
基金资助:HUANG Qina, JIANG Hongrui, YANG Jie, YU Kunyu, YANG Changdeng, LIANG Yan. Bioinformatics Analysis, Development and Application of Molecular Markers for Seed Dormancy Gene Sdr4 in Rice[J]. Chinese Journal OF Rice Science, 2026, 40(1): 61-71.
黄奇娜, 姜鸿瑞, 杨婕, 于坤宇, 杨长登, 梁燕. 种子休眠基因Sdr4的生物信息学分析与分子标记开发和应用[J]. 中国水稻科学, 2026, 40(1): 61-71.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2026.250211
| 引物名称 Primer | 引物序列(5’-3’) Sequence(5’-3’) | 片段长度 Length(bp) | 基因组位点 Location |
|---|---|---|---|
| Sdr4-F | CTTCTTAACCCCACCACCCC | 1161 | 引物5’端距离ATG上游102 bp 102 bp upstream of ATG |
| Sdr4-R | TTTGCTCCGGCTTGATGCATC | 引物3’端距离TGA下游27 bp 27 bp downstream of TGA | |
| Sdr4-KF | AAGGTCATCTCGCCGCGCG | 197 | 距离ATG 493~511 bp编码区 Coding region 493-511 bp from ATG |
| Sdr4-KR | GTTGCTGGAGTCCGAGACC | 距离ATG 702~720 bp编码区 Coding region 702-720 bp from ATG | |
| Sdr4-PF | AAGCTGCTGGAGCCG TGG | 227 | 距离ATG 493~509 bp编码区(画横线为引入的错配碱基) Coding region 493-509 bp from ATG (underlined bases are introduced mismatches) |
| Sdr4-PR | CTTGTACGCGTCGTTCACC | 距离ATG 672~690 bp编码区 Coding region 672-690 bp from ATG |
Table 1. Whole-genome amplification primers for Sdr4 and functionally validated molecular markers
| 引物名称 Primer | 引物序列(5’-3’) Sequence(5’-3’) | 片段长度 Length(bp) | 基因组位点 Location |
|---|---|---|---|
| Sdr4-F | CTTCTTAACCCCACCACCCC | 1161 | 引物5’端距离ATG上游102 bp 102 bp upstream of ATG |
| Sdr4-R | TTTGCTCCGGCTTGATGCATC | 引物3’端距离TGA下游27 bp 27 bp downstream of TGA | |
| Sdr4-KF | AAGGTCATCTCGCCGCGCG | 197 | 距离ATG 493~511 bp编码区 Coding region 493-511 bp from ATG |
| Sdr4-KR | GTTGCTGGAGTCCGAGACC | 距离ATG 702~720 bp编码区 Coding region 702-720 bp from ATG | |
| Sdr4-PF | AAGCTGCTGGAGCCG TGG | 227 | 距离ATG 493~509 bp编码区(画横线为引入的错配碱基) Coding region 493-509 bp from ATG (underlined bases are introduced mismatches) |
| Sdr4-PR | CTTGTACGCGTCGTTCACC | 距离ATG 672~690 bp编码区 Coding region 672-690 bp from ATG |
| 类别 Type | 顺式作用元件 cis-acting element | 序列 Sequence | 数量 Number | 功能 Function |
|---|---|---|---|---|
| 激素相关作用元件Hormone-related response elements | ABRE | ACGTG/CACGTG | 11 | 脱落酸响应元件 Abscisic acid response element |
| TCA-element | CCATCTTTTT | 1 | 水杨酸途径响应元件 Salicylic acid pathway response element | |
| CGTCA/TGACG-motif | CGTCA | 10 | 茉莉酸甲酯调控元件 Methyl jasmonate regulatory element | |
| 逆境相关作用元件 Stress-related response elements | ARE | AAACCA | 3 | 厌氧诱导调控元件 Anaerobic induction regulatory element |
| MYB/MYC | CAACCA/CATGTG | 4 | 干旱诱导响应元件 Drought-induced response element | |
| 生长发育相关作用元件Growth and development- related response elements | RY-element | CATGCATG | 4 | 种子特异性调控元件 Seed-specific regulatory element |
| Sp1 | GGGCGG | 7 | 光照响应元件 Light-responsive element |
Table 2. cis-acting elements in the promoter region of Sdr4
| 类别 Type | 顺式作用元件 cis-acting element | 序列 Sequence | 数量 Number | 功能 Function |
|---|---|---|---|---|
| 激素相关作用元件Hormone-related response elements | ABRE | ACGTG/CACGTG | 11 | 脱落酸响应元件 Abscisic acid response element |
| TCA-element | CCATCTTTTT | 1 | 水杨酸途径响应元件 Salicylic acid pathway response element | |
| CGTCA/TGACG-motif | CGTCA | 10 | 茉莉酸甲酯调控元件 Methyl jasmonate regulatory element | |
| 逆境相关作用元件 Stress-related response elements | ARE | AAACCA | 3 | 厌氧诱导调控元件 Anaerobic induction regulatory element |
| MYB/MYC | CAACCA/CATGTG | 4 | 干旱诱导响应元件 Drought-induced response element | |
| 生长发育相关作用元件Growth and development- related response elements | RY-element | CATGCATG | 4 | 种子特异性调控元件 Seed-specific regulatory element |
| Sp1 | GGGCGG | 7 | 光照响应元件 Light-responsive element |

Fig. 2. Sequence alignment and protein structure prediction of Sdr4 A, Alignment of DNA sequences at different sites and the location of the forward primer F (highlighted in green) for molecular marker development; B, Alignment of the Sdr4 amino acid sequences in Kasalath and Nipponbare; C, Prediction of the secondary structure of Sdr4 in Kasalath and Nipponbare; D, AlphaFold analysis of the three-dimensional structure of Sdr4 in Kasalath and Nipponbare.
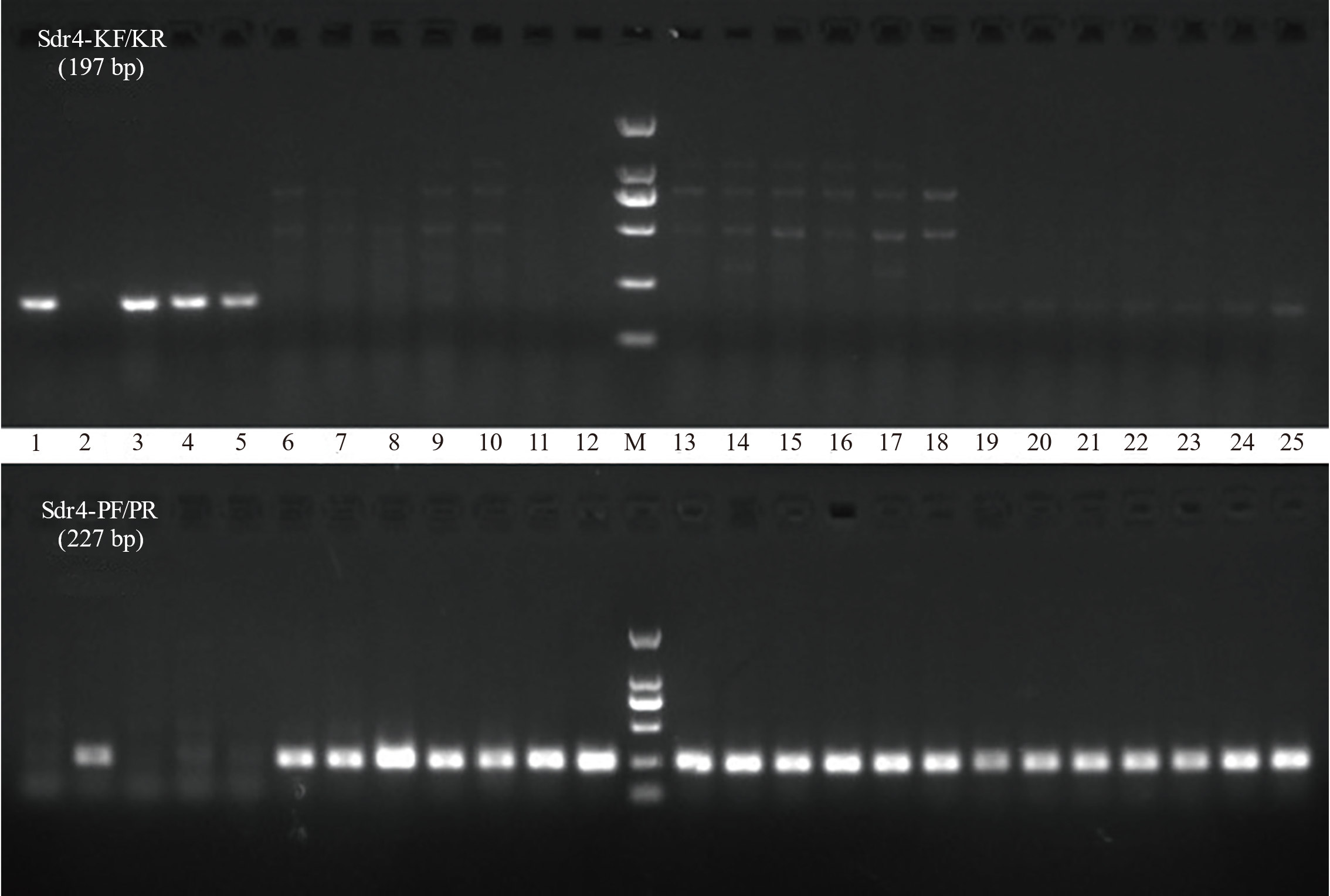
Fig. 4. Analysis of molecular marker detection results in diverse rice varieties 1, Kasalath; 2, Nip; 3, IR24; 4, IR36; 5, IR64; 6, Zhongzu 1; 7, Zhongzu 3; 8, Zhongzu 9; 9, Zhongzao 22; 10, Zhongzao 35; 11, Zhongzao 39; 12, Zhongzu 18; 13, Zhongleng 23; 14, Zhongjiazao 10; 15, Zhe 106; 16, Zhenong 131; 17, Zhenong 135; 18, Yongxian 15; 19, Yongxian 57; 20, Yongxian 378; 21, Wen 723; 22, Taizao 502; 23, Taizao 816; 24, Jinzao 47; 25, Zaoxian 310; M, Easy 2K DNA marker.

Fig. 6. Breeding process of new materials resistant to panicle sprouting (A), molecular marker detection results and their phenotypes (B) Genomic DNA from the rice varieties ZZ18, IR36, K17, and K88 was amplified using the Sdr4-KF/KR (197 bp) and Sdr4-PF/PR (227 bp) primers, respectively. Seeds of ZZ18, K17, and K88 were soaked for 48 at room temperature and subsequently incubated at 37℃ for 48 h to promote germination. Following germination, phenotypic characterization was performed, and images were captured for documentation. M, Marker.
| [1] | Finch-savage W E, Leubner-metzger G. Seed dormancy and the control of germination[J]. New Phytologist, 2006, 171: 501-523. |
| [2] | Sohn S I, Pandian S, Kumar T S, Zoclanclounon Y A B, Muthuramalingam P, Shilpha J, Satish L, Ramesh M. Seed dormancy and pre-harvest sprouting in rice: An updated overview[J]. International Journal of Molecular Sciences, 2021, 22(21): 11804. |
| [3] | Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy[J]. Annual Review of Plant Biology, 2008, 59: 387-415. |
| [4] | Chen W, Wang W, Lyu Y, Wu Y, Huang P, Hu S, Wei X, Jiao G, Sheng Z, Tang S, Shao G, Luo J. OsVP1 activates Sdr4 expression to control rice seed dormancy via the ABA signaling pathway[J]. The Crop Journal, 2021, 9: 68-78. |
| [5] | He D, Yang P. Proteomics of rice seed germination[J]. Frontiers in Plant Science, 2013, 4: 246. |
| [6] | Liu S, Sehgal S K, Li J, Lin M, Trick H N, Yu J, Gill B S, Bai G. Cloning and characterization of a critical regulator for preharvest sprouting in wheat[J]. Genetics, 2013, 195(1): 263-273. |
| [7] | Shu K, Meng Y J, Shuai H W, Liu W G, Du J B, Liu J, Yang W Y. Dormancy and germination: How does the crop seed decide?[J]. Plant Biology, 2015, 17(6): 1104-1112. |
| [8] | Falade K O, Semon O M, Fadairo O S, Oladunjoye A O, Orou K K. Functional and physico-chemical properties of flours and starches of African rice cultivars[J]. Food Hydrocolloids, 2014, 39: 41-50. |
| [9] | Lee H S, Choi M G, Hwang W H, Jeong J H, Lee C G. Occurrence of rice preharvest sprouting varies greatly depending on past weather conditions during grain filling[J]. Field Crops Research, 2021, 264: 108087. |
| [10] | 胡伟民, 马华升, 樊龙江, 阮松林. 杂交水稻制种不育系穗上发芽特性[J]. 作物学报, 2003, 29(3): 441-446. |
| Hu W M, Ma H S, Fan L J, Ruan S L. Characteristics of pre-harvest sprouting in sterile lines in hybrid rice seeds production[J]. Acta Agronomica Sinica, 2003, 29(3): 441-446. (in Chinese with English abstract) | |
| [11] | Liao Y, Bai Q, Xu P, Wu T, Guo D, Peng Y, Zhang H, Deng X, Chen X, Luo M, Ali A, Wang W, Wu X. Mutation in rice abscisic Acid2 results in cell death, enhanced disease-resistance, altered seed dormancy and development[J]. Frontiers in Plant Science, 2018, 9: 405. |
| [12] | Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination[J]. Plant Cell, 2011, 23(9): 3215-3229. |
| [13] | Nakamura S, Pourkheirandish M, Morishige H, Kubo Y, Nakamura M, Ichimura K, Seo S, Kanamori H, Wu J, Ando T, Hensel G, Sameri M, Stein N, Sato K, Matsumoto T, Yano M, Komatsuda T. Mitogen-activated protein Kinase Kinase 3 regulates seed dormancy in barley[J]. Current Biology, 2016, 26(6): 775-781. |
| [14] | Sato K, Yamane M, Yamaji N, Kanamori H, Tagiri A, Schwerdt J G, Fincher G B, Matsumoto T, Takeda K, Komatsuda T. Alanine aminotransferase controls seed dormancy in barley[J]. Nature Communications, 2016, 7: 11625. |
| [15] | Bentsink L, Jowett J, Hanhart C J, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(45): 17042-17047. |
| [16] | Cao H, Han Y, Li J, Ding M, Li Y, Li X, Chen F, Soppe W J, Liu Y. Arabidopsis thaliana SEED DORMAN-CY 4-LIKE regulates dormancy and germination by mediating the gibberellin pathway[J]. Journal of Experimental Botany, 2020, 71(3): 919-933. |
| [17] | Carrillo-barral N, Rodríguez-gacio M D C, Matilla A J. Delay of germination-1 (DOG1): A key to understanding seed dormancy[J]. Plants (Basel), 2020, 9(4): 480. |
| [18] | Liu F, Zhang H, Ding L, Soppe W J J, Xiang Y. RE-VERSAL OF RDO5 1, a homolog of rice seed dormancy4, interacts with bHLH57 and controls ABA biosynthesis and seed dormancy in Arabidopsis[J]. Plant Cell, 2020, 32(6): 1933-1948. |
| [19] | Hattori T, Terada T, Hamasuna S T. Sequence and functional analyses of the rice gene homologous to the maize Vp1[J]. Plant Molecular Biology, 1994, 24: 805-810. |
| [20] | Zhao B, Zhang H, Chen T, Ding L, Zhang L, Ding X, Zhang J, Qian Q, Xiang Y. Sdr4 dominates pre-harvest sprouting and facilitates adaptation to local climatic condition in Asian cultivated rice[J]. Journal of Integrative Plant Biology, 2022, 64(6): 1246-1263. |
| [21] | Fan X, Gao F, Liu Y, Huang W, Yang Y, Luo Z, Zhang J, Qi F, Lü J, Su X, Wang L, Song S, Ren G, Xing Y. The transcription factor CCT30 promotes rice preharvest sprouting by regulating sugar signalling to inhibit the ABA-mediated pathway[J]. Plant Biotechnology Journal, 2025, 23(2): 579-591. |
| [22] | Sugimoto K, Takeuchi Y, Ebana K, Miyao A, Hirochika H, Hara N, Ishiyama K, Kobayashi M, Ban Y, Hattori T, Yano M. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(13): 5792-5797. |
| [23] | Du L, Xu F, Fang J, Gao S, Tang J, Fang S, Wang H, Tong H, Zhang F, Chu J, Wang G, Chu C. Endosperm sugar accumulation caused by mutation of PHS8/ ISA1 leads to pre-harvest sprouting in rice[J]. Plant Journal, 2018, 95(3): 545-556. |
| [24] | Magwa R A, Zhao H, Xing Y. Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.)[J]. BMC Genetics, 2016, 17: 1-13. |
| [25] | Meyer R S, Purugganan M D. Evolution of crop species: Genetics of domestication and diversification[J]. Nature Reviews Genetics, 2013, 14(12): 840-852. |
| [26] | Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis[J]. Nucleic Acids Research, 1991, 19(6): 1349. |
| [27] | Hilhorst H W M, Karssen C M. Seed dormancy and germination: The role of abscisic acid and gibberellins and the importance of hormone mutants[J]. Plant Growth Regulation, 1992, 11: 225-238. |
| [28] | Omoarelojie L O, Kulkarni M G, Finnie J F, van Staden J. Smoke-derived cues in the regulation of seed germination: Are Ca2+-dependent signals involved?[J]. Plant Growth Regulation, 2022, 97(2): 343-355. |
| [29] | 张毛宁. 基于花生RIL群体的鲜种子发芽性状QTL定位及分子标记开发[D]. 郑州: 郑州大学, 2022. |
| Zhang M N. QTL mapping for fresh seed germination traits based on a peanut RIL population and molecular marker development[D]. Zhengzhou: Zhengzhou University, 2022. (in Chinese with English abstract) |
| [1] | LI Xingyi, CHEN Ling, SHAO Jiantao, XIAO Suqin, LI Jinlu, FU Huixian, YIN Fuyou, ZHANG Jianhong, CHENG Zaiquan, LIU Li. Progress in Molecular Genetic Research on Co-regulation of Grain Yield and Starch Quality in Rice [J]. Chinese Journal OF Rice Science, 2026, 40(1): 1-17. |
| [2] | MIAO Zhening, CHEN Jijin, LI Ziming, LIU Yi, LUO Lijun. Advances and Prospects in Breeding Bacterial Blight-resistant Rice in South China [J]. Chinese Journal OF Rice Science, 2026, 40(1): 18-26. |
| [3] | YUE Xuanyu, XIE Wenya, FENG Zhiming, CHEN Zongxiang, HU Keming, ZUO Shimin. Function of OsERF93 in Regulating Resistance to Sheath Blight in Rice [J]. Chinese Journal OF Rice Science, 2026, 40(1): 37-50. |
| [4] | WANG Yixin, LIN Shen, MA Liuyang, CHEN Long, FENG Baohua, NI Shen, WEI Xiangjin, HE Jiwai, CHEN Tianxiao. Regulation of Nitrogen Uptake and Yield in Rice by the Alanine Aminotransferase Gene OsAlaAT4 [J]. Chinese Journal OF Rice Science, 2026, 40(1): 51-60. |
| [5] | CHENG Zhaoping, HE Niqing, BAI Kangcheng, LIN Shaojun, HUANG Fenghuang, LIU Junhua, CHENG Zuxin, HUANG Chengzhi, YANG Dewei. Breeding and Utilization of the Three-Line Male Sterile Line Fumeng A with Pyramided Rice Blast Resistance Genes Pigm-1 and Pid2 [J]. Chinese Journal OF Rice Science, 2026, 40(1): 72-84. |
| [6] | LIU Yaping, DONG Yici, ZHENG Junyan, QIU Xuan, LIU Pengcheng, YE Yafeng, LIU Binmei, CHEN Xifeng, MA Bojun. Phenotypic Identification and Gene Fine Mapping of the Lesion Mimic and Early Senescence Mutant lmes7 in Rice [J]. Chinese Journal OF Rice Science, 2026, 40(1): 85-94. |
| [7] | LIAO Zhengming, GUO Liang, PAN Xiaowu, LI Yongchao, DONG Zheng, LI Xiaoxiang. Identification of Genes for Rice Seed Storability and Transcriptome Analysis Under Different Aging Conditions [J]. Chinese Journal OF Rice Science, 2026, 40(1): 95-105. |
| [8] | ZHANG Cheng, SHAO Guojun, ZHANG Xue, TIAN Shujun, SUN Chi, GUO Yanying, ZHOU Ran, HAN Yong, ZHENG Wenjing, SUN Lianping. QTL Mapping and Pyramiding Effect Analysis of Diurnal Floret Opening Time Traits in japonica Rice [J]. Chinese Journal OF Rice Science, 2026, 40(1): 106-117. |
| [9] | XIAO Wuwei, ZHANG Yuqing, ZHU Chenguang, TIAN Guisheng, CAI Yuehong, WANG Fei, XIONG Dongliang, HUANG Jianliang, PENG Shaobing, CUI Kehui. Effects of Application Time and Rate of Bud-promoting Fertilizer on Main Crop Quality and Annual Yield of Ratoon Rice [J]. Chinese Journal OF Rice Science, 2026, 40(1): 118-130. |
| [10] | XIE Shimin, ZHOU Yuzhu, XUE Xiaodi, ZHU Guangfei, SUN Liang, CHEN Jianneng. Design and Experiment of an Integrated Picking and Planting Mechanism for Rice Pot Seedlings [J]. Chinese Journal OF Rice Science, 2026, 40(1): 131-144. |
| [11] | CHEN Ling, LIN Wenying, LIANG Limei, OUYANG Younan, YE Shenghai, JI Zhijuan. Flowering Habits of Rice and Its Application in Breeding japonica Cytoplasmic Male Sterile Lines [J]. Chinese Journal OF Rice Science, 2025, 39(6): 731-743. |
| [12] | WANG Juan, WU Lijuan, HONG Haibo, YAO Zhiwen, WANG Lei, E Zhiguo. Research Progress on Biological Functions of Ubiquitin-conjugating Enzymes in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 744-750. |
| [13] | TAO Shibo, XU Na, XU Zhengjin, LIU Chang, XU Quan. Cloning of Cold6 Conferring Cold Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 751-759. |
| [14] | CHEN Wei, YE Yuanmei, ZHAO Jianhua, FENG Zhiming, CHEN Zongxiang, HU Keming, ZUO Shimin. Modifying Heading Date of Nanjing 46 via CRISPR/Cas9-mediated Genome Editing [J]. Chinese Journal OF Rice Science, 2025, 39(6): 760-770. |
| [15] | HOU Guihua, ZHOU Liguo, LEI Jianguo, CHEN Hong, NIE Yuanyuan. Preliminary Analysis of Function and Mechanism of OsRDR5 Gene in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 779-788. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||