
Chinese Journal OF Rice Science ›› 2026, Vol. 40 ›› Issue (1): 95-105.DOI: 10.16819/j.1001-7216.2026.241110
• Research Papers • Previous Articles Next Articles
LIAO Zhengming1,#, GUO Liang2,3,#, PAN Xiaowu2,3, LI Yongchao2, DONG Zheng2,3, LI Xiaoxiang2,3,*( )
)
Received:2024-11-18
Revised:2025-01-06
Online:2026-01-10
Published:2026-01-21
Contact:
LI Xiaoxiang
About author:First author contact:These authors contributed equally to this work
廖政明1,#, 郭梁2,3,#, 潘孝武2,3, 黎用朝2, 董铮2,3, 李小湘2,3,*( )
)
通讯作者:
李小湘
作者简介:第一联系人:共同第一作者
基金资助:LIAO Zhengming, GUO Liang, PAN Xiaowu, LI Yongchao, DONG Zheng, LI Xiaoxiang. Identification of Genes for Rice Seed Storability and Transcriptome Analysis Under Different Aging Conditions[J]. Chinese Journal OF Rice Science, 2026, 40(1): 95-105.
廖政明, 郭梁, 潘孝武, 黎用朝, 董铮, 李小湘. 水稻种子耐储性基因的挖掘及不同老化方式的转录组分析[J]. 中国水稻科学, 2026, 40(1): 95-105.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2026.241110
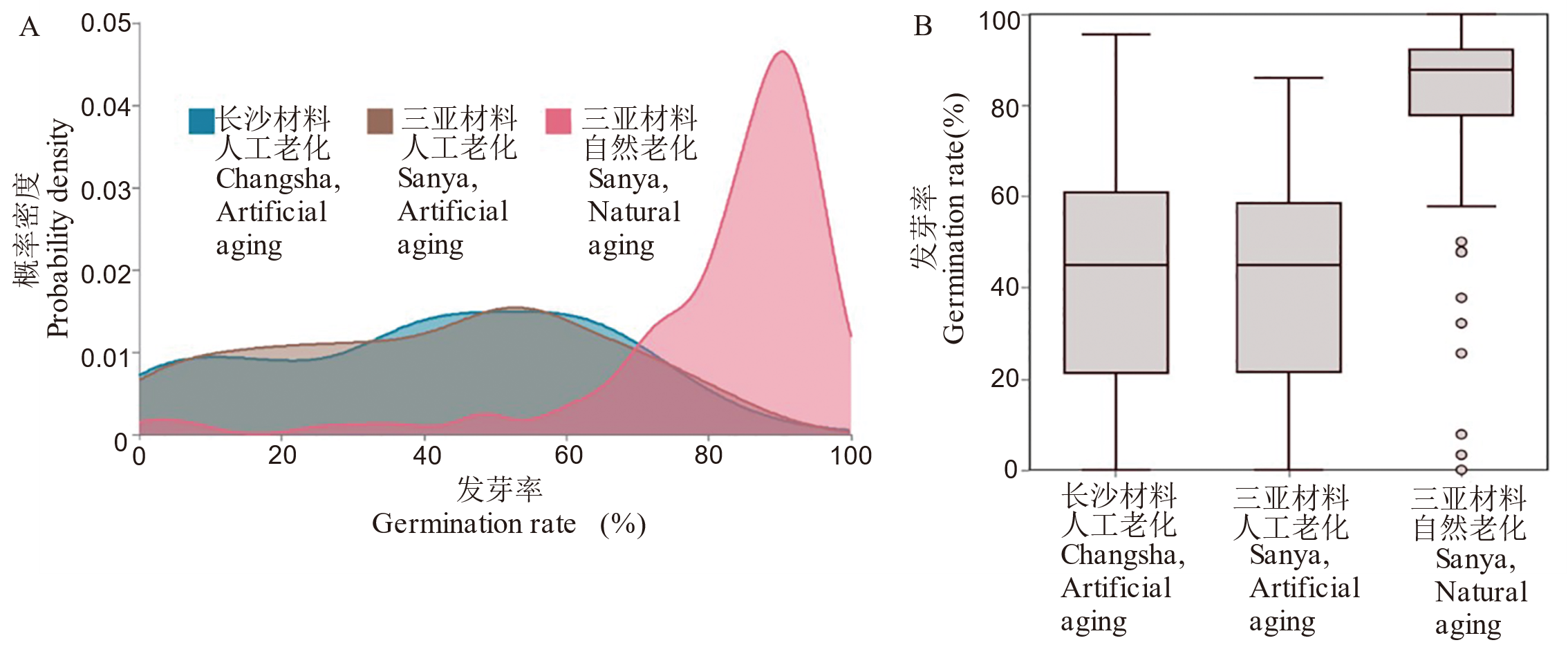
Fig. 2. Distribution and summary statistics of seed germination rate from different origins under different aging treatments A, Kernel density estimation (KDE) curves of germination rates; B, Boxplots of germination rate.
| 染色体 Chr | QTL | 处理 Treatment | 标记区间 Marker region | 物理区间 Interval (Mb) | LOD | R2 (%) | 加性效应 Additive effect | 前人研究 Previous study |
|---|---|---|---|---|---|---|---|---|
| 1 | qASS1.1 | 人工老化(三亚) | Bin147-Bin152 | 37.73-38.68 | 7.28 | 19.0 | 0.13 | qSS1.1[ |
| 1 | qASS1.1 | 人工老化(长沙) | Bin147-Bin152 | 37.73-38.68 | 11.60 | 15.5 | 0.16 | qSS1.1[ |
| 1 | qASS1.2 | 人工老化(长沙) | Bin154-Bin160 | 38.73-40.29 | 3.90 | 4.5 | −0.09 | |
| 1 | qNSS1.1 | 自然老化(三亚) | Bin147-Bin152 | 37.73-38.68 | 19.04 | 21.0 | 0.13 | qSS1.1[ |
| 1 | qNSS1.2 | 自然老化(三亚) | Bin154-Bin160 | 38.73-40.29 | 7.19 | 5.8 | −0.08 | |
| 3 | qASS3 | 人工老化(长沙) | Bin333-Bin340 | 50.15-67.74 | 3.00 | 3.3 | 0.12 | |
| 6 | qNSS6 | 自然老化(三亚) | Bin580-Bin612 | 1.56-13.19 | 9.76 | 9.2 | 0.40 | qSS-6[ |
| 7 | qNSS7 | 自然老化(三亚) | Bin662-Bin669 | 6.16-17.35 | 8.40 | 8.1 | 0.15 | qSS7.1[ |
| 9 | qASS9 | 人工老化(三亚) | Bin813-Bin819 | 8.04-10.05 | 6.96 | 17.7 | 0.16 | qRC9-2[ |
Table 1. Identified QTLs for seed storability under natural or artificial aging treatments in an IL population
| 染色体 Chr | QTL | 处理 Treatment | 标记区间 Marker region | 物理区间 Interval (Mb) | LOD | R2 (%) | 加性效应 Additive effect | 前人研究 Previous study |
|---|---|---|---|---|---|---|---|---|
| 1 | qASS1.1 | 人工老化(三亚) | Bin147-Bin152 | 37.73-38.68 | 7.28 | 19.0 | 0.13 | qSS1.1[ |
| 1 | qASS1.1 | 人工老化(长沙) | Bin147-Bin152 | 37.73-38.68 | 11.60 | 15.5 | 0.16 | qSS1.1[ |
| 1 | qASS1.2 | 人工老化(长沙) | Bin154-Bin160 | 38.73-40.29 | 3.90 | 4.5 | −0.09 | |
| 1 | qNSS1.1 | 自然老化(三亚) | Bin147-Bin152 | 37.73-38.68 | 19.04 | 21.0 | 0.13 | qSS1.1[ |
| 1 | qNSS1.2 | 自然老化(三亚) | Bin154-Bin160 | 38.73-40.29 | 7.19 | 5.8 | −0.08 | |
| 3 | qASS3 | 人工老化(长沙) | Bin333-Bin340 | 50.15-67.74 | 3.00 | 3.3 | 0.12 | |
| 6 | qNSS6 | 自然老化(三亚) | Bin580-Bin612 | 1.56-13.19 | 9.76 | 9.2 | 0.40 | qSS-6[ |
| 7 | qNSS7 | 自然老化(三亚) | Bin662-Bin669 | 6.16-17.35 | 8.40 | 8.1 | 0.15 | qSS7.1[ |
| 9 | qASS9 | 人工老化(三亚) | Bin813-Bin819 | 8.04-10.05 | 6.96 | 17.7 | 0.16 | qRC9-2[ |
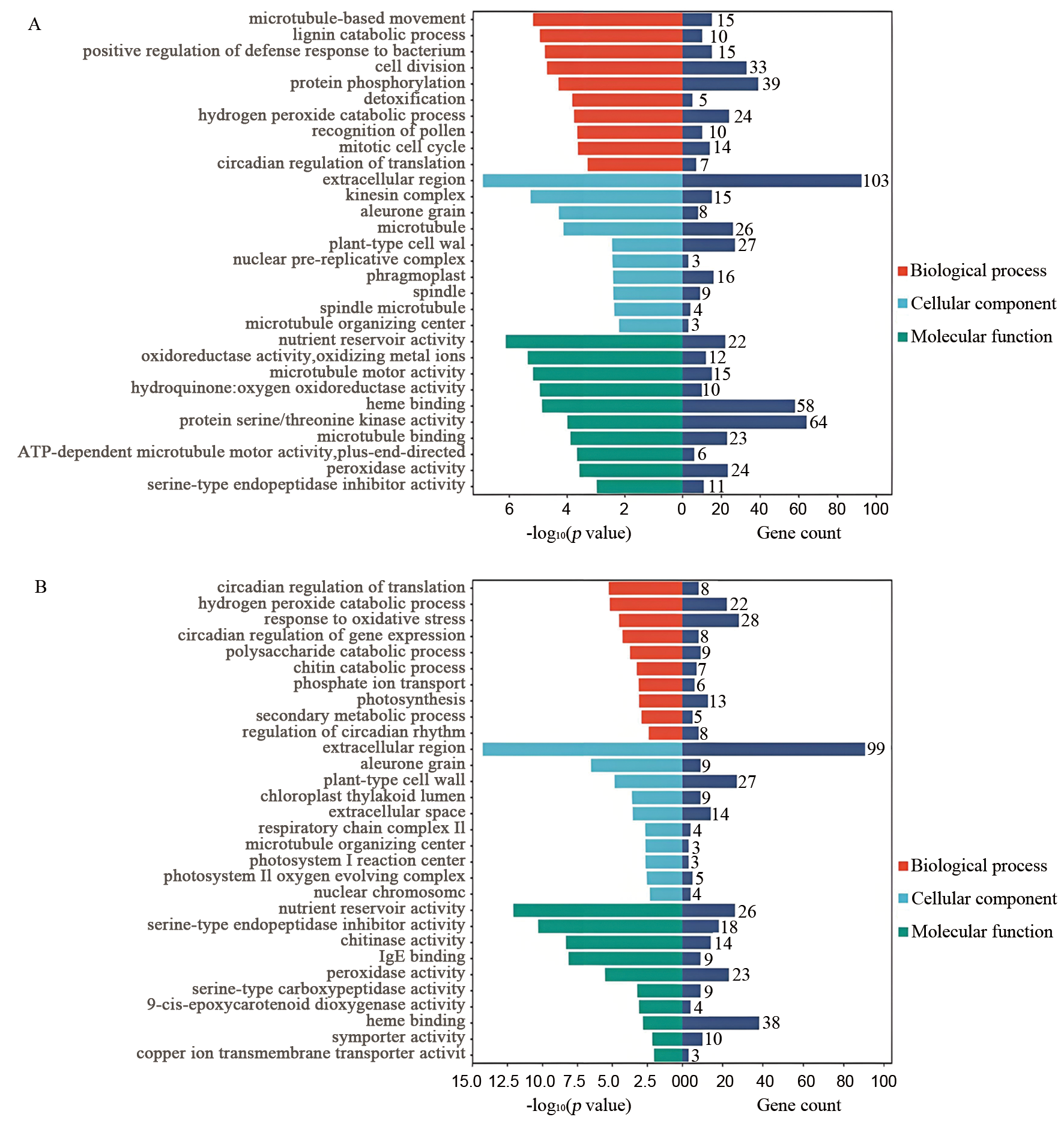
Fig. 3. GO enrichment analysis of differentially expressed genes A, DEGs of Ra32 vs. Ra146 after artificial aging; B, DEGs of Ra32 vs. Ra146 after natural aging.
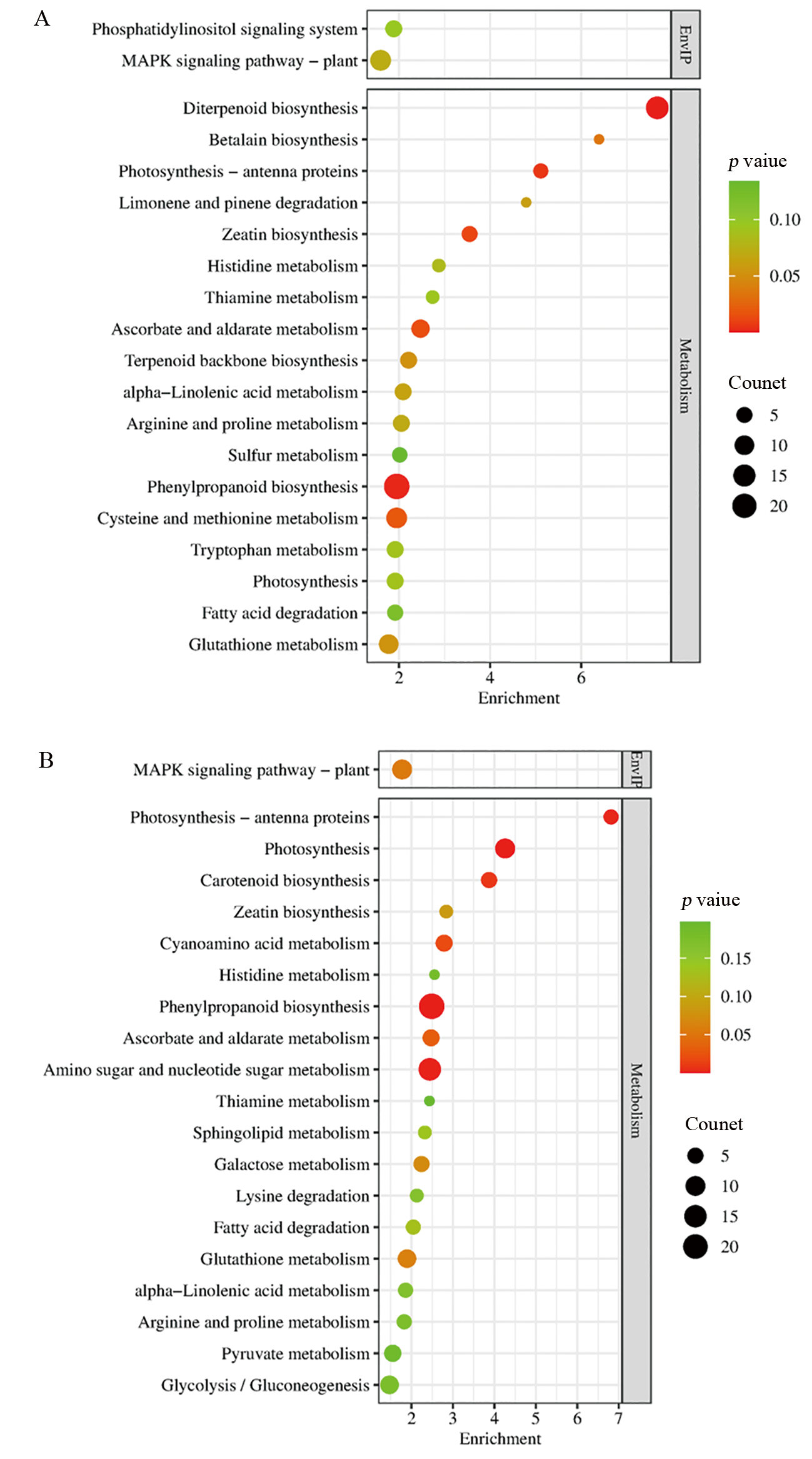
Fig. 4. KEGG enrichment analysis of differentially expressed genes A, DEGs of Ra32 vs. Ra146 after artificial aging; B, DEGs of Ra32 vs. Ra146 after natural aging; EnvIP, Environmental Information Processing.
| 候选基因 Candidate gene | 功能注释 Functional annotation | Ra32 vs Ra146 1) | GO富集通路 GO enriched pathways | KEGG富集通路 KEGG enriched pathways |
|---|---|---|---|---|
| Os01g0842400 | 漆酶前体蛋白 Laccase precursor protein | 下调 Down-regulated | 木质素分解过程、 氧化还原酶活性 Lignin catabolic process, oxidoreductase activity | |
| Os01g0842500 | 漆酶前体蛋白 Laccase precursor protein | 下调 Down-regulated | 木质素分解过程、 氧化还原酶活性 Lignin catabolic process, oxidoreductase activity | |
| Os01g0847800 | 醛酮还原酶家族蛋白 Aldo-keto reductase family protein | 上调 Up-regulated | 糖酵解/糖异生、抗坏血酸和醛糖酸代谢、丙酮酸代谢Glycolysis/gluconeogenesis, ascorbate and aldarate metabolism, pyruvate metabolism | |
| Os01g0855900 | CDC6-DNA复制起始蛋白 CDC6-DNA replication initiation protein | 下调 Down-regulated | 细胞分裂、 有丝分裂细胞周期 Cell division, mitotic cell cycle | |
| Os01g0860400 | 糖基水解酶 Glycoside hydrolase | 上调 Up-regulated | 壳聚糖分解过程、 多糖分解过程 Chitosan catabolic process, polysaccharide catabolic process | 氨基糖和核苷酸糖代谢 Amino sugar and nucleotide sugar metabolism |
Table 2. Candidate genes for qASS1.1/qNSS1.1 controlling seed storability
| 候选基因 Candidate gene | 功能注释 Functional annotation | Ra32 vs Ra146 1) | GO富集通路 GO enriched pathways | KEGG富集通路 KEGG enriched pathways |
|---|---|---|---|---|
| Os01g0842400 | 漆酶前体蛋白 Laccase precursor protein | 下调 Down-regulated | 木质素分解过程、 氧化还原酶活性 Lignin catabolic process, oxidoreductase activity | |
| Os01g0842500 | 漆酶前体蛋白 Laccase precursor protein | 下调 Down-regulated | 木质素分解过程、 氧化还原酶活性 Lignin catabolic process, oxidoreductase activity | |
| Os01g0847800 | 醛酮还原酶家族蛋白 Aldo-keto reductase family protein | 上调 Up-regulated | 糖酵解/糖异生、抗坏血酸和醛糖酸代谢、丙酮酸代谢Glycolysis/gluconeogenesis, ascorbate and aldarate metabolism, pyruvate metabolism | |
| Os01g0855900 | CDC6-DNA复制起始蛋白 CDC6-DNA replication initiation protein | 下调 Down-regulated | 细胞分裂、 有丝分裂细胞周期 Cell division, mitotic cell cycle | |
| Os01g0860400 | 糖基水解酶 Glycoside hydrolase | 上调 Up-regulated | 壳聚糖分解过程、 多糖分解过程 Chitosan catabolic process, polysaccharide catabolic process | 氨基糖和核苷酸糖代谢 Amino sugar and nucleotide sugar metabolism |
| 基因ID Gene ID | 正向引物 Forward primer | 反向引物 Reverse primer |
|---|---|---|
| Os01g0842400 | ATCTTCGGCGAGTGGTGGA | TGTCTTGGGCAGAGCAGTTG |
| Os01g0842500 | CTTCTCCGTTCCTCCATCCT | ACGTTCGTCGTTTTCACATCGA |
| Os01g0847800 | TGGCGTTCATCTCTCTGCGTA | CTGACCCTGCTGAAGACCC |
| Os01g0855900 | CCATACCCGCGAAGCATTAAG | AGAGCCAAGCCCGCCAAAG |
| Os01g0860400 | GACCTCGCCAGGGATCTGA | CGAACAGCCCCGTGCTGAT |
Table 3. Primer sequences used for RT-qPCR validation of candidate genes
| 基因ID Gene ID | 正向引物 Forward primer | 反向引物 Reverse primer |
|---|---|---|
| Os01g0842400 | ATCTTCGGCGAGTGGTGGA | TGTCTTGGGCAGAGCAGTTG |
| Os01g0842500 | CTTCTCCGTTCCTCCATCCT | ACGTTCGTCGTTTTCACATCGA |
| Os01g0847800 | TGGCGTTCATCTCTCTGCGTA | CTGACCCTGCTGAAGACCC |
| Os01g0855900 | CCATACCCGCGAAGCATTAAG | AGAGCCAAGCCCGCCAAAG |
| Os01g0860400 | GACCTCGCCAGGGATCTGA | CGAACAGCCCCGTGCTGAT |
| [1] | 朱俊峰. 我国粮食产后损失的现状、影响因素及改进对策[J]. 江西社会科学, 2023, 43(9): 29-40. |
| Zhu J F. The current situation, influencing factors, and improvement strategies of post-harvest losses in grain production in China[J]. Jianxi Social Sciences, 2023, 43(9): 29-40. (in Chinese with Chinese abstract) | |
| [2] | Zhou T S, Yu D, Wu L B, Xu Y S, Duan M J, Yuan D Y. Seed storability in rice: Physiological foundations, molecular mechanisms, and applications in breeding[J]. Rice Science, 2024, 31(4): 401-416. |
| [3] | 王雪彬, 张健, 韦燕燕, 罗继景, 梁云涛, 蔡中全. 基于BSA-seq的水稻籽粒耐陈化QTL定位分析[J]. 分子植物育种, 2023, 21(16): 5337-5347. |
| Wang X B, Zhang J, Wei Y Y, Luo J J, Liang Y T, Cai Z Q. QTLs mapping analysis of rice grain aging tolerance based on BSA-seq[J]. Molecular Plant Breeding, 2023, 21(16): 5337-5347. (in Chinese with English abstract) | |
| [4] | 曹玉洁, 费月新, 赵文佳, 侯璐燕, 王越, 吴敏, 许珊, 吴洪恺. 以种子电导率为指标定位水稻耐储藏QTL[J]. 中国稻米, 2020, 26(1): 46-49. |
| Cao Y J, Fei Y X, Zhao W J, Hou L Y, Wang Y, Wu M, Xu S, Wu H K. Mapping of QTL for rice storability based on seed electric conductivity[J]. China Rice, 2020, 26(1): 46-49. (in Chinese with English abstract) | |
| [5] | Jin B C, Qi H N, Jia L Q, Tang Q Z, Gao L, Li Z N, Zhao G W. Determination of viability and vigor of naturally-aged rice seeds using hyperspectral imaging with machine learning[J]. Infrared Physics & Technology, 2022, 122: 104097. |
| [6] | Song P, Wang Z Y, Song P, Yue X, Bai Y H, Feng L L. Evaluating the effect of aging process on the physicochemical characteristics of rice seeds by low field nuclear magnetic resonance and its imaging technique[J]. Journal of Cereal Science, 2021, 99: 103190. |
| [7] | Ye J, Wang C J, Chen L, Zhai R R, Wu M M, Lu Y T, Yu F M, Zhang X M, Zhu G F, Ye S H. Golden hull: A potential biomarker for assessing seed aging tolerance in rice[J]. Agronomy. 2024, 14(10): 2357. |
| [8] | Wang W Q, He A B, Peng S B, Huang J L, Cui K H, Nie L X. The effect of storage condition and duration on the deterioration of primed rice seeds[J]. Frontiers in Plant Science, 2018, 9: 172. |
| [9] | 吴方喜, 罗曦, 魏毅东, 郑燕梅, 林强, 谢国生, 谢华安, 张建福. 世界水稻核心种质的耐储藏特性鉴定[J]. 福建稻麦科技, 2021, 39(1): 1-5. |
| Wu F X, Luo X, Wei Y D, Zheng Y M, Lin Q, Xie G S, Xie H A, Zhang J F. Identification of seed storability in rice core collections from 47 countries worldwide[J]. Fujian Science and Technology of Rice and Wheat, 2021, 39(1): 1-5. (in Chinese with English abstract) | |
| [10] | 黄祎雯, 张维谊, 王霞, 王敏, 沈斯文, 高猛峰, 梅博, 汪弘康, 童金蓉, 曹黎明, 丰东升, 孙滨. 33份不同基因型水稻耐储性比较[J/OL]. 分子植物育种, 1-13[2025-03-13]. http://kns.cnki.net/kcms/detail/46.1068.S.20240202.1646.010.html |
| Huang Y W, Zhang W Y, Wang X, Wang M, Shen S W, Gao M F, Mei B, Wang H K, Tong J R, Cao L M, Feng D S, Sun B. Comparison of storage tolerance among 33 different genotypes of rice[J/OL]. Molecular Plant Breeding, 2024, http://kns.cnki.net/kcms/detail/46.1068.S.20240202.1646.010. (in Chinese with English abstract) | |
| [11] | 黄祎雯, 孙滨, 程灿, 牛付安, 周继华, 张安鹏, 涂荣剑, 李瑶, 姚瑶, 代雨婷, 谢开珍, 陈小荣, 曹黎明, 储黄伟. 对水稻种子耐储性QTL的研究[J]. 作物学报, 2022, 48(9): 2255-2264. |
| Huang Y W, Sun B, Chen C, Niu F A, Zhou J H, Zhang A P, Tu R J, Li Y, Yao Y, Dai Y T, Xie K Z, Chen X R, Cao L M, Chu H W. QTL mapping of seed storage tolerance in rice (Oryza sativa L.)[J]. Acta Agronomica Sinica, 2022, 48(9): 2255-2264. (in Chinese with English abstract) | |
| [12] | 刘进, 姚晓云, 余丽琴, 李慧, 周慧颖, 王嘉宇, 黎毛毛. 水稻耐储藏特性三年动态鉴定与QTL分析[J]. 植物学报, 2019, 54(4): 464-473. |
| Liu J, Yao X Y, Yu L Q, Li H, Zhou H Y, Wang J Y, Li M M. Detection and analysis of dynamic quantitative trait loci at three years for seed storability in rice (Oryza sativa)[J]. Chinese Bulletin of Botany, 2019, 54(4): 464-473. (in Chinese with English abstract) | |
| [13] | Wu F X, Luo X, Wang L Q, Wei Y D, Li J G, Xie H A, Zhang J F, Xie G S. Genome-wide association study reveals the QTLs for seed storability in world rice core collections[J]. Plants (Basel), 2021, 10(4): 812. |
| [14] | Sasaki K, Fukuta Y, Sato T. Mapping of quantitative trait loci controlling seed longevity of rice (Oryza sativa L.) after various periods of seed storage[J]. Plant Breeding, 2005, 124(4): 361-366. |
| [15] | Li L F, Lin Q Y, Liu S J, Liu X, Wang W Y, Hang N T, Liu F, Zhao Z G, Jiang L, Wan J M. Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.)[J]. Plant Breeding, 2012, 131(6): 739-743. |
| [16] | Long Q Z, Zhang W W, Wang P, Shen W B, Zhou T, Liu N N, Wang R, Jiang L, Huang J X, Wang Y H, Liu Y Q, Wan J M. Molecular genetic characterization of rice seed lipoxygenase 3 and assessment of its effects on seed longevity[J]. Journal of Plant Biology, 2013, 56(4): 232-242. |
| [17] | Yuan Z Y, Fan K, Wang Y T, Tian L, Zhang C P, Sun W Q, He H Z, Yu S B. OsGRETCHENHAGEN3-2 modulates rice seed storability via accumulation of abscisic acid and protective substances[J]. Plant Physiology, 2021, 186(1): 469-482. |
| [18] | 黄娟, 梁云涛, 陈成斌, 徐志健, 梁世春, 潘英华. 普通野生稻遗传分化及水稻起源关系研究进展[J]. 南方农业学报, 2015, 46(10): 1756-1760. |
| Huang J, Liang Y T, Chen C B, Xu Z J, Liang S C, Pan Y H. Advances in origin of Oryza sativa in terms of genetic differentiation of Oryza fufipogon[J]. Journal of Southern Agriculture, 2015, 46(10): 1756-1760. (in Chinese with English abstract) | |
| [19] | 王世林, 吴婷, 周诗琪, 宋思铭, 胡标林. 结合BSA-seq和QTL分析鉴定东乡野生稻耐储性QTL[J]. 中国水稻科学, 2025, 39(6): 789-800. |
| Wang S L, Wu T, Zhou S Q, Song S M, Hu B L. Identification of QTL for seed storability in Dongxiang wild rice by integrating BSA-seq and QTL analysis[J]. Chinese Journal of Rice Science, 2025, 39(6): 789-800. (in Chinese with English abstract) | |
| [20] | Ma X, Fu Y C, Zhao X H, Jiang L Y, Zhu Z F, Gu P, Xu W Y, Su Z, Sun C Q, Tan L B. Genomic structure analysis of a set of Oryza nivara introgression lines and identification of yield-associated QTLs using whole-genome resequencing[J]. Scientific Reports, 2016, 6: 27425. |
| [21] | Meng L, Li H H, Zhang L Y, Wang J K. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations[J]. The Crop Journal, 2015, 3: 269-283. |
| [22] | McCouch S R, CGSNL (Committee on Gene Symbolization, Nomenclature and Linkage, Rice Genetics Cooperative). Gene nomenclature system for rice[J]. Rice, 2008, 1: 72-84. |
| [23] | Chen S F, Zhou Y Q, Chen Y, Chen Y R, Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): i884-i890. |
| [24] | Kim D, Langmead B, Salzberg S L. HISAT: A fast spliced aligner with low memory requirements[J]. Nature Methods, 2015, 12(4): 357-360. |
| [25] | Roberts A, Trapnell C, Donaghey J, Rinn J L, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias[J]. Genome Biology, 2011, 12(3): R22. |
| [26] | Anders S, Pyl P T, Huber W. HTSeq: A Python framework to work with high-throughput sequencing data[J]. Bioinformatics, 2015, 31(2): 166-169. |
| [27] | Love M I, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biology, 2014, 15(12): 550. |
| [28] | Dietz K-J, Mittler R, Noctor G. Recent progress in understanding the role of reactive oxygen species in plant cell signaling[J]. Plant Physiology, 2016, 171(3): 1535-1539. |
| [29] | Xue J S, Feng Y F, Zhang M Q, Xu Q L, Xu Y M, Shi J Q, Liu L F, Wu X F, Wang S, Yang Z N. The regulatory mechanism of rapid lignification for timely anther dehiscence[J]. Journal of Integrative Plant Biology, 2024, 66(8): 1788-1800. |
| [30] | 张盛春, 鞠常亮, 王小菁. 拟南芥漆酶基因AtLAC4参与生长及非生物胁迫响应[J]. 植物学报, 2012, 47(4): 357-365. |
| Zhang S C, Ju C L, Wang X Q. Arabidopsis laccase gene AtLAC4 regulates plant growth and responses to abiotic stress[J]. Chinese Bulletin of Botany, 2012, 47(4): 357-365. (in Chinese with English abstract) | |
| [31] | 徐小萍, 曹清影, 蔡柔荻, 官庆栩, 张梓浩, 陈裕坤, 徐涵, 林玉玲, 赖钟雄. 龙眼miR408与DlLAC12克隆及其在球形胚发生和非生物胁迫下的表达分析[J]. 园艺学报, 2022, 49(9): 1866-1882. |
| Xu X P, Cao Q Y, Cai R D, Guan Q X, Zhang Z H, Chen Y K, Xu H, Lin Y L, Lai Z X. Gene cloning and expression analysis of miR408 and its target DlLAC12 in globular embryo development and abiotic stress in Dimocarpus longan[J]. Acta Horticulturae Sinica, 2022, 49(9): 1866-1882. (in Chinese with English abstract) | |
| [32] | Simpson P J, Tantitadapitak C, Reed A M, Mather O C, Bunce C M, White S A, Ride J P. Characterization of two novel aldo-keto reductases from Arabidopsis: Expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress[J]. Journal of Molecular Biology, 2009, 392(2): 465-480. |
| [33] | Saito R, Shimakawa G, Nishi A, Iwamoto T, Sakamoto K, Yamamoto H, Amako K, Makino A, Miyake C. Functional analysis of the AKR4C subfamily of Arabidopsis thaliana: model structures, substrate specificity, acrolein toxicity, and responses to light and [CO2][J]. Bioscience, Biotechnology and Biochemistry, 2013, 77(10): 2038-2045. |
| [34] | Yu X H, Liu Y H, Yin L L, Peng Y B, Peng Y C, Gao Y X, Yuan B W, Zhu Q L, Cao T Y, Xie B W, Sun L Q, Chen Y, Gong Z C, Qiu Y Z, Fan X G, Li X. Radiation-promoted CDC6 protein stability contributes to radioresistance by regulating senescence and epithelial to mesenchymal transition[J]. Oncogene, 2019, 38: 549-563. |
| [35] | Abeles F B, Bosshart R P, Forrence L E, Habig W H. Preparation and purification of glucanase and chitinase from bean leaves[J]. Plant Physiology, 1971, 47(1): 129-134. |
| [1] | ZHANG Cheng, SHAO Guojun, ZHANG Xue, TIAN Shujun, SUN Chi, GUO Yanying, ZHOU Ran, HAN Yong, ZHENG Wenjing, SUN Lianping. QTL Mapping and Pyramiding Effect Analysis of Diurnal Floret Opening Time Traits in japonica Rice [J]. Chinese Journal OF Rice Science, 2026, 40(1): 106-117. |
| [2] | WANG Shilin, WU Ting, ZHOU Shiqi, SONG Siming, HU Biaolin. Identification of QTLs for Seed Storability in Dongxiang Wild Rice by Integrating BSA-Seq and QTL Analysis [J]. Chinese Journal OF Rice Science, 2025, 39(6): 789-800. |
| [3] | DING Guohua, LI Xin, CAO Liangzi, ZHOU Jinsong, LEI Lei, BAI Liangming, LUO Yu, YANG Guang, CUI Zhibo, ZHAO Minghui, SUN Shichen. Effect of Low Temperature at Booting Stage on Photosynthetic System of Different Rice Materials in Cold Region [J]. Chinese Journal OF Rice Science, 2025, 39(5): 679-689. |
| [4] | CHEN Jiale, YU Qingtao, ZHENG Chenfan, WANG Qing, TAN Yuanyuan, CHEN Baicui, LI Chengxin, JIANG Meng, SHU Qingyao. Natural Variation of OsNF-YC10 and Its Correlation with Grain Width in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(4): 552-562. |
| [5] | HUANG Fudeng, WU Chunyan, HAO Yuanyuan, HAN Yifei, ZHANG Xiaobin, SUN Huifeng, PAN Gang. Transcriptome Analysis of Top Second Leaf Sheath of Rice Under Different Nitrogen Fertilizer Levels [J]. Chinese Journal OF Rice Science, 2025, 39(4): 563-574. |
| [6] | LIANG Chuyan, ZENG Wei, WANG Jiebing, YE Jing, WU Mingming, ZHAI Rongrong, ZHANG Xiaoming, ZHANG Hengmu, YE Shenghai. Characterization and Transcriptome Analysis of a Mutant with Short Panicle and Small Grain from Zhejing 99 [J]. Chinese Journal OF Rice Science, 2025, 39(1): 67-81. |
| [7] | YANG Chuanming, WANG Lizhi, ZHANG Xijuan, YANG Xianli, WANG Yangyang, HOU Benfu, CUI Shize, LI Qingchao, LIU Kai, MA Rui, FENG Yanjiang, LAI Yongcai, LI Hongyu, JIANG Shukun. Analysis of QTL Controlling Cold Tolerance at Seedling Stage by Using a High-Density SNP Linkage Map in japonica Rice [J]. Chinese Journal OF Rice Science, 2025, 39(1): 82-91. |
| [8] | DU Yanxiu, SUN Wenyu, YUAN Zeke, ZHANG Qianqian, LI Fuhao, LI Junzhou, SUN Hongzheng. Mapping of qChalk8 Controlling Chalky Rice Rate in japonica Rice by Combining QTL-Seq with Molecular Markers [J]. Chinese Journal OF Rice Science, 2024, 38(6): 665-671. |
| [9] | FU Rongtao, CHEN Cheng, WANG Jian, ZHAO Liyu, CHEN Xuejuan, LU Daihua. Combined Transcriptome and Metabolome Analyses Reveals the Pathogenic Factors of Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(4): 375-385. |
| [10] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [11] | LIU Zhongqi, ZHANG Haiqing, HE Jiwai, GUI Jinxin. Genome-wide Association Analysis of Rice Seed Dehydration Rate at Maturity Stage [J]. Chinese Journal OF Rice Science, 2024, 38(2): 150-159. |
| [12] | HOU Benfu, YANG Chuanming, ZHANG Xijuan, YANG Xianli, WANG Lizhi, WANG Jiayu, LI Hongyu, JIANG Shukun. Mapping of Grain Shape QTLs Using RIL Population from Longdao 5/Zhongyouzao 8 [J]. Chinese Journal OF Rice Science, 2024, 38(1): 13-24. |
| [13] | HU Jiaxiao, LIU Jin, CUI Di, LE Si, ZHOU Huiying, HAN Bing, MENG Bingxin, YU Liqin, HAN Longzhi, MA Xiaoding, LI Maomao. Mapping Major QTLs for Panicle Traits Using CSSLs of Dongxiang Wild Rice (Oryza rufipogon Griff.) [J]. Chinese Journal OF Rice Science, 2023, 37(6): 597-608. |
| [14] | XIE Kaizhen, ZHANG Jianming, CHENG Can, ZHOU Jihua, NIU Fuan, SUN Bin, ZHANG Anpeng, WEN Weijun, DAI Yuting, HU Qiyan, QIU Yue, CAO Liming, CHU Huangwei. Identification and QTL Mapping of Rice Germplasm Resources with Low Amylose Content [J]. Chinese Journal OF Rice Science, 2023, 37(6): 609-616. |
| [15] | YAO Xiaoyun, CHEN Chunlian, XIONG Yunhua, HUANG Yongping, PENG Zhiqing, LIU Jin, YIN Jianhua. Identification of QTL for Milling and Appearance Quality Traits in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2023, 37(5): 507-517. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||