
Chinese Journal OF Rice Science ›› 2025, Vol. 39 ›› Issue (6): 751-759.DOI: 10.16819/j.1001-7216.2025.240709
• Research Papers • Previous Articles Next Articles
TAO Shibo1,2, XU Na1, XU Zhengjin1, LIU Chang1,*( ), XU Quan1,*(
), XU Quan1,*( )
)
Received:2024-07-13
Revised:2024-08-20
Online:2025-11-10
Published:2025-11-19
Contact:
LIU Chang, XU Quan
陶士博1,2, 许娜1, 徐正进1, 刘畅1,*( ), 徐铨1,*(
), 徐铨1,*( )
)
通讯作者:
刘畅,徐铨
基金资助:TAO Shibo, XU Na, XU Zhengjin, LIU Chang, XU Quan. Cloning of Cold6 Conferring Cold Tolerance in Rice[J]. Chinese Journal OF Rice Science, 2025, 39(6): 751-759.
陶士博, 许娜, 徐正进, 刘畅, 徐铨. 水稻发芽期耐冷基因Cold6的克隆[J]. 中国水稻科学, 2025, 39(6): 751-759.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2025.240709
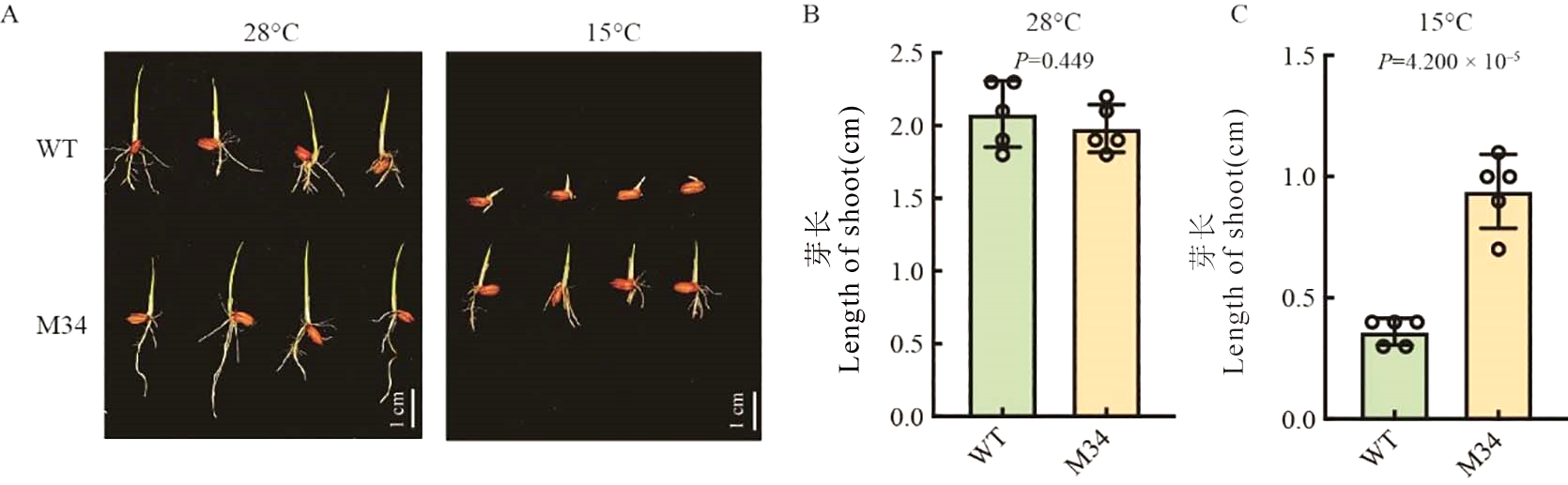
Fig. 1. Phenotype of WT and M34 under 28 ℃ and 15℃ A, WT and M34 5 d after sowing under 28℃ and 15 ℃, bar = 1 cm. B, Length of shoot 5 d after sowing under 28 ℃. C, Length of shoot 5 d after sowing under 15 ℃. Data are mean ± SE (n = 5).
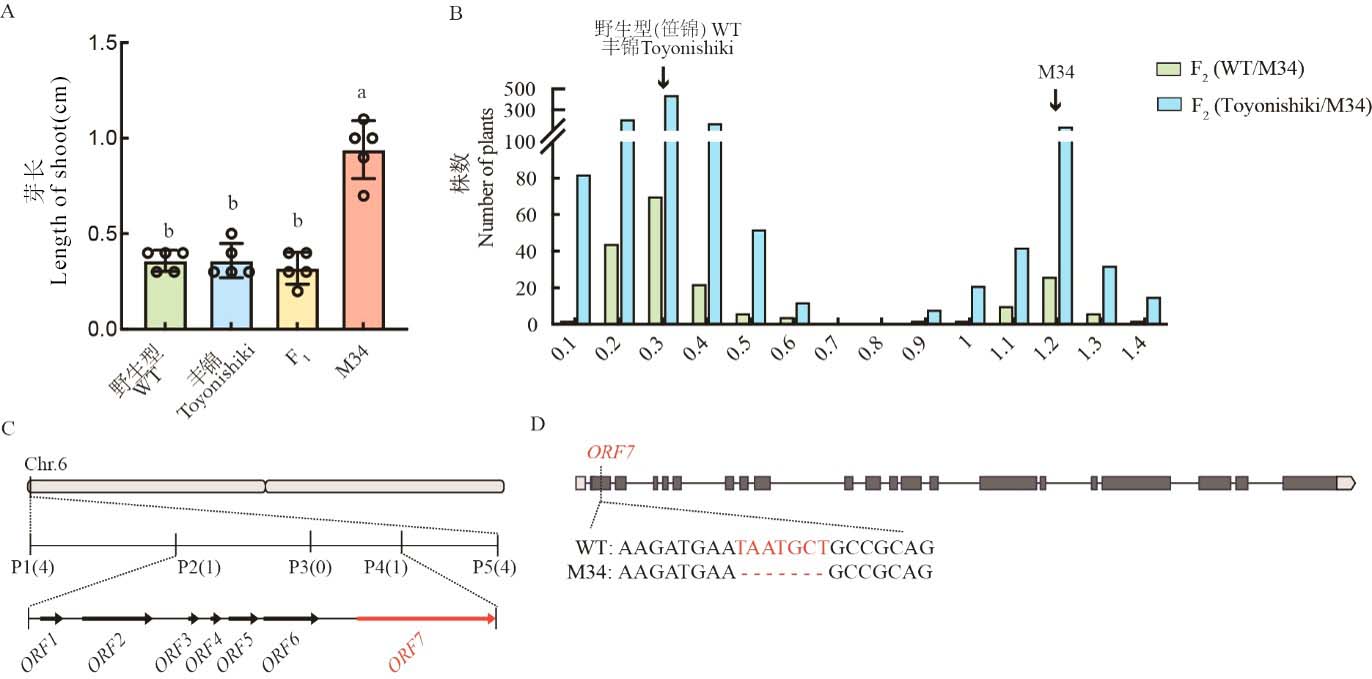
Fig. 2. Map-based cloning of candidate gene Cold6 A, Length of shoot of WT, M34, and F1 plants 5 d after sowing under 15℃. Data are mean ± SE (n = 5). Different letters denote significant differences (P < 0.05) by Duncan’s multiple range test; B, Distribution of shoot length of F2 population under 15℃ 5 d after sowing; C, Fine mapping of candidate gene; D, Sequence difference between WT and M34 in ORF7.
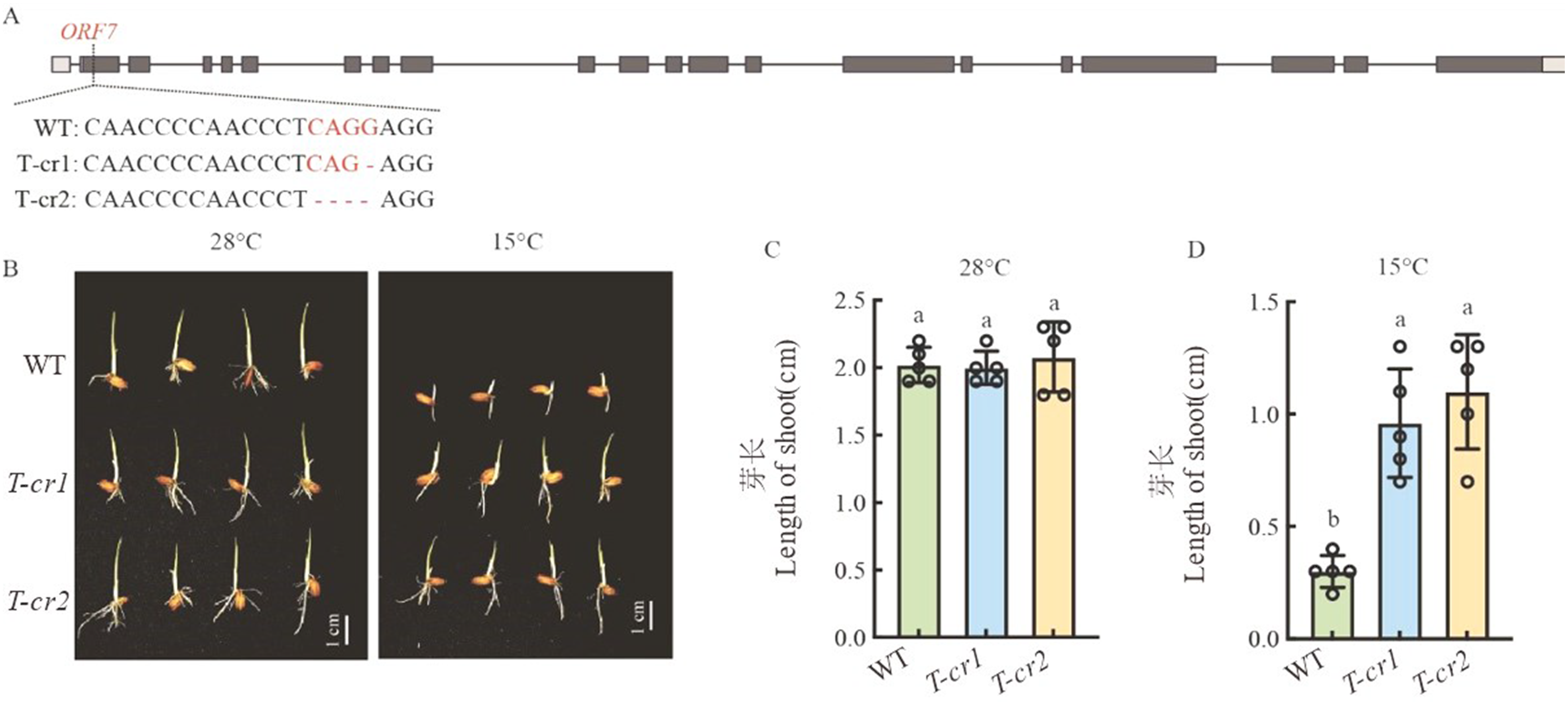
Fig. 3. CRISPR/Cas9 gene editing of Cold6 and phenotypic investigation A, Target sites and mutation sequences of Cold6 gene; B, WT and gene edited plants under 28 ℃ and 15℃ 5 d after sowing, scale bar = 1 cm; C, Shoot length of WT and gene edited plants under 28 ℃ 5 d after sowing; D, Shoot length of WT and gene edited plants under 15 ℃ 5 d after sowing. Data are mean ± SE (n = 5). Different letters denote significant differences (P < 0.05) by Duncan’s multiple range test.
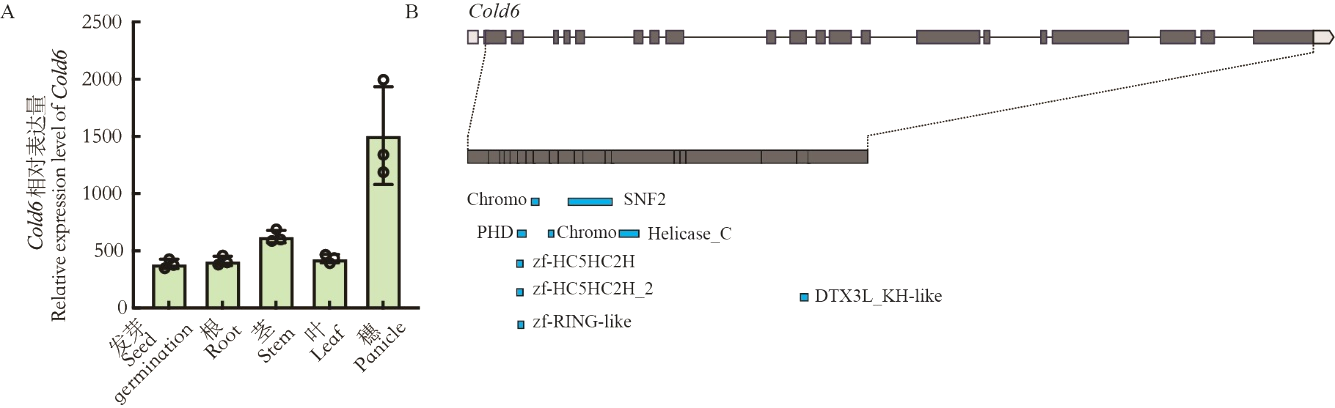
Fig. 4. Expression pattern and motif domain prediction of Cold6 A, Relative expression of Cold6 in different organs of plants, data are mean ± SE (n = 3); B, Prediction of motif in Cold6.
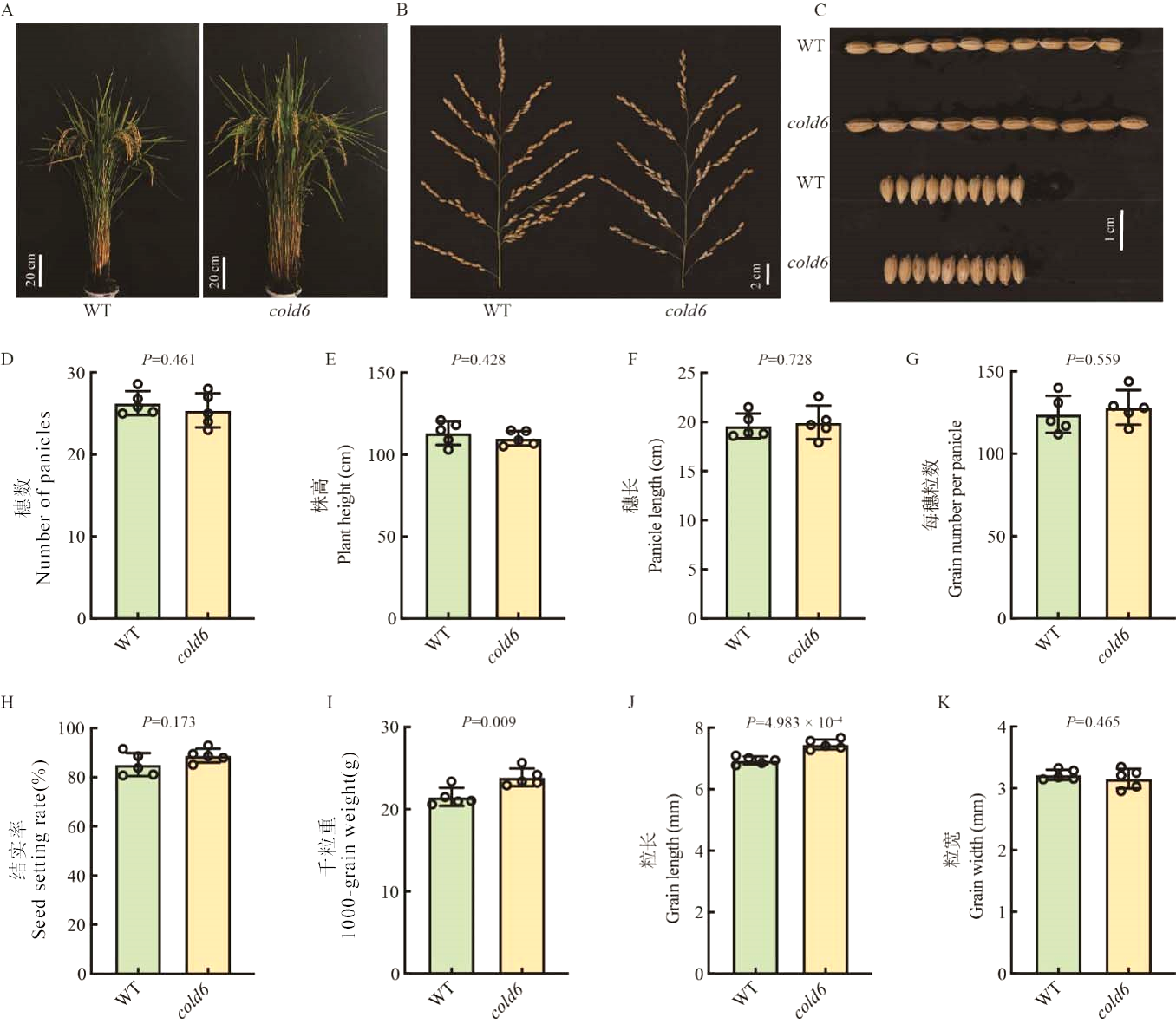
Fig. 5. Effect of Cold6 on agronomic traits in rice A, Plant architecture of WT and cold6 mutant, scale bar = 20 cm; B, Panicle architecture of WT and cold6 mutant, scale bar = 2 cm; C, Grain shape of WT and cold6 mutant, scale bar = 1 cm; D-K, Number of panicles, plant height, panicle length, grain number per panicle, seed setting rate, 1,000-grain weight, grain length, and grain width of WT and cold6 mutant. Data are mean ± SE (n = 5).
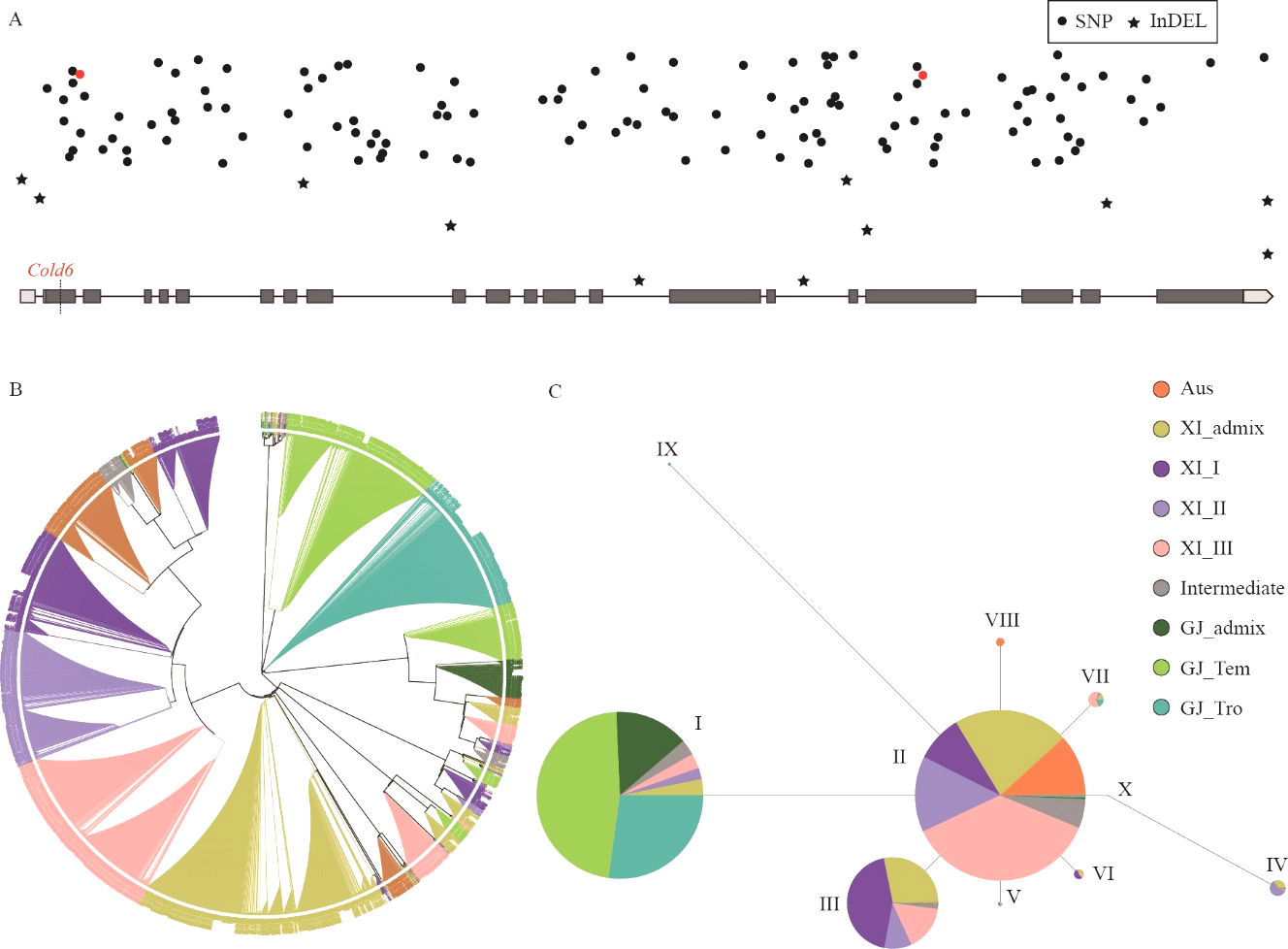
Fig. 6. Phylogenetic and haplotype analysis of Cold6 A, Distribution of SNP and InDel on Cold6. Red color indicates the SNP causing the change of protein function predicted by PloyPhen-2; B, Phylogenetic analysis of Cold6; C, Haplotype analysis of Cold6.
| [1] | Qian Q. Genomics-assisted germplasm improvement[J]. Journal of Integrative Plant Biology, 2018, 60(2): 82-84. |
| [2] | Zhang Z, Li J, Pan Y, Zhou L, Shi H, Zeng Y, Guo H, Yang S, Zheng W, Yu J, Sun X, Li G, Ding Y, Ma L, Shen S, Dai L, Zhang H, Guo Y, Li Z. Natural variation in CTB4a enhances rice adaptation to cold habitats[J]. Nature Communications, 2017, 8: 14788. |
| [3] | Lu G, Wu F Q, Wu W, Wang H T, Zheng X M, Zhang Y, Chen X, Zhou K, Jin M, Cheng Z, Li X, Jiang L, Wang H, Wan J. Rice LTG1 is involved in adaptive growth and fitness under low ambient temperature[J]. The Plant Journal, 2014, 78: 468-480. |
| [4] | Liu C T, Wang W, Mao B G, Chu C C. Cold stress tolerance in rice: Physiological changes, molecular mechanism, and future prospects[J]. Yi Chuan, 2018, 40: 171-185. |
| [5] | Yonemaru J I, Yamamoto T, Fukuoka S, Uga Y, Hori K, Yano M J R. Q-TARO: QTL Annotation Rice Online Database[J]. Rice, 2010, 3: 194-203. |
| [6] | Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice [J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105: 12623-12628. |
| [7] | Wang X, Zou B, Shao Q, Cui Y, Lu S, Zhang Y, Huang Q, Huang J, Hua J. Natural variation reveals that OsSAP16 controls low-temperature germination in rice[J]. Journal of Experimental Botany, 2018, 69: 413-421. |
| [8] | Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D. COLD1 confers chilling tolerance in rice[J]. Cell, 2015, 160: 1209-1221. |
| [9] | Zhao J, Zhang S, Dong J, Yang T, Mao X, Liu Q, Wang X, Liu B. A novel functional gene associated with cold tolerance at the seedling stage in rice[J]. Plant Biotechnology Journal, 2017, 15: 1141-1148. |
| [10] | Liu C, Schläppi M R, Mao B, Wang W, Wang A, Chu C. The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage[J]. Plant Biotechnology Journal, 2019, 17: 1834-1849. |
| [11] | Liu C, Ou S, Mao B, Tang J, Wang W, Wang H, Cao S, Schläppi M R, Zhao B, Xiao G, Wang X, Chu C. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates[J]. Nature Communications, 2018, 9: 3302. |
| [12] | Mao D, Xin Y, Tan Y, Hu X, Bai J, Liu Z, Yu Y, Li L, Peng C, Fan T, Zhu Y, Guo Y, Wang S, Lu D, Xing Y, Yuan L, Chen C. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116: 3494-3501. |
| [13] | Saito K, Hayano-Saito Y, Kuroki M, Sato Y J P. Map-based cloning of the rice cold tolerance gene Ctb1[J]. Plant Science, 2010, 179: 97-102. |
| [14] | Gu S, Zhang Z, Li J, Sun J, Cui Z, Li F, Zhuang J, Chen W, Su C, Wu L, Wang X, Guo Z, Xu H, Zhao M, Ma D, Chen W. Natural variation in OsSEC13 HOMOLOG 1 modulates redox homeostasis to confer cold tolerance in rice[J]. Plant Physiology, 2023, 193: 2180-2196. |
| [15] | Wu J, Liu H, Zhang Y, Zhang Y, Li D, Liu S, Lu S, Wei L, Hua J, Zou B. A major gene for chilling tolerance variation in indica rice codes for a kinase OsCTK1 that phosphorylates multiple substrates under cold[J]. The New Phytologist, 2024, 242: 2077-2092. |
| [16] | Zu X, Luo L, Wang Z, Gong J, Yang C, Wang Y, Xu C, Qiao X, Deng X, Song X, Chen C, Tan B C, Cao X. A mitochondrial pentatricopeptide repeat protein enhances cold tolerance by modulating mitochondrial superoxide in rice[J]. Nature Communications, 2023, 14: 6789. |
| [17] | Zhong Y, Luo Y, Sun J, Qin X, Gan P, Zhou Z, Qian Y, Zhao R, Zhao Z, Cai W, Luo J, Chen L L, Song J M. Pan-transcriptomic analysis reveals alternative splicing control of cold tolerance in rice[J]. The Plant Cell, 2024, 36: 2117-2139. |
| [18] | Li R, Song Y, Wang X, Zheng C, Liu B, Zhang H, Ke J, Wu X, Wu L, Yang R, Jiang M. OsNAC5 orchestrates OsABI5 to fine-tune cold tolerance in rice[J]. Journal of Integrative Plant Biology, 2024, 66: 660-682. |
| [19] | Shen Y, Cai X, Wang Y, Li W, Li D, Wu H, Dong W, Jia B, Sun M, Sun X Y. MIR1868 negatively regulates rice cold tolerance at both the seedling and booting stages[J]. The Crop Journal, 2024, 12: 375-383. |
| [20] | Jiang S, Yang C, Xu Q, Wang L, Yang X, Song X, Wang J, Zhang X, Li B, Li H, Li Z, Li W. Genetic dissection of germinability under low temperature by building a resequencing linkage map in japonica rice[J]. International Journal of Molecular Sciences, 2020, 21(4): 1284. |
| [21] | Wang Y, Li F, Zhang F, Wu L, Xu N, Sun Q, Chen H, Yu Z, Lu J, Jiang K, Wang X, Wen S, Zhou Y, Zhao H, Jiang Q, Wang J, Jia R, Sun J, Tang L, Xu H, Hu W, Xu Z, Chen W, Guo A, Xu Q. Time-ordering japonica/geng genomes analysis indicates the importance of large structural variants in rice breeding[J]. Plant Biotechnology Journal, 2023, 21: 202-218. |
| [22] | Adzhubei I A, Schmidt S, Peshkin L, Ramensky V E, Gerasimova A, Bork P, Kondrashov A S, Sunyaev S R. A method and server for predicting damaging missense mutations[J]. Nature Methods, 2010, 7: 248-249. |
| [23] | Saito K, Miura K, Nagano K, Hayano-Saito Y, Araki H, Kato A. Identification of two closely linked quantitative trait loci for cold tolerance on chromosome 4 of rice and their association with anther length[J]. Theoretical and Applied Genetics, 2001, 103: 862-868. |
| [24] | Jiang N, Shi S, Shi H, Khanzada H, Wassan G, Zhu C, Peng X, Yu Q, Chen X, He X, Fu J, Hu L, Xu J, Ouyang L, Sun X, Zhou D, He H, Bian J. Mapping QTL for seed germinability under low temperature using a new high-density genetic map of rice[J]. Frontiers in Plant Science, 2017, 8: 1223. |
| [25] | Lee J, Lee W, Kwon S W. A quantitative shotgun proteomics analysis of germinated rice embryos and coleoptiles under low-temperature conditions[J]. Proteome Science, 2015, 13: 27. |
| [26] | Yang L M, Liu H L, Zhao H W, Wang J G, Sun J, Zheng H L, Lei L, Zou D T. Mapping quantitative trait loci and meta-analysis for cold tolerance in rice at booting stage[J]. Euphytica, 2019, 215 (5): 89 |
| [27] | 吴爱婷, 宋佳谕, 胡涛, 刘思彤, 高继平, 黄丽湘, 高银隆, 赵明辉. 超级稻沈农265苗期耐冷性QTL定位[J]. 核农学报, 2018, 32(8): 1477-1482. |
| Wu A T, Song J Y, Hu T, Liu S T, Gao J P, Huang L X, Gao Y L, Zhao M H. QTLs mapping for cold tolerance at seedling stage in super rice variety Shennong 265[J]. Journal of Nuclear Agricultural Sciences, 2018, 32(8): 1477-1482. (in Chinese with English abstract) | |
| [28] | Hu Y, Zhu N, Wang X, Yi Q, Zhu D, Lai Y, Zhao Y. Analysis of rice Snf2 family proteins and their potential roles in epigenetic regulation[J]. Plant Physiology and Biochemistry, 2013, 70: 33-42. |
| [29] | Hu Y, Liu D, Zhong X, Zhang C, Zhang Q, Zhou D X. CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109: 5773-5778. |
| [1] | SONG Anqi, WU Songquan, MA Qiuyue, BAN Wanning, LIU Xiangguo, JIN Yongmei. Plant Prime Editing: A New Direction in Crop Breeding [J]. Chinese Journal OF Rice Science, 2025, 39(6): 711-730. |
| [2] | CHEN Ling, LIN Wenying, LIANG Limei, OUYANG Younan, YE Shenghai, JI Zhijuan. Flowering Habits of Rice and Its Application in Breeding japonica Cytoplasmic Male Sterile Lines [J]. Chinese Journal OF Rice Science, 2025, 39(6): 731-743. |
| [3] | WANG Juan, WU Lijuan, HONG Haibo, YAO Zhiwen, WANG Lei, E Zhiguo. Research Progress on Biological Functions of Ubiquitin-conjugating Enzymes in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 744-750. |
| [4] | CHEN Wei, YE Yuanmei, ZHAO Jianhua, FENG Zhiming, CHEN Zongxiang, HU Keming, ZUO Shimin. Modifying Heading Date of Nanjing 46 via CRISPR/Cas9-mediated Genome Editing [J]. Chinese Journal OF Rice Science, 2025, 39(6): 760-770. |
| [5] | HOU Guihua, ZHOU Liguo, LEI Jianguo, CHEN Hong, NIE Yuanyuan. Preliminary Analysis of Function and Mechanism of OsRDR5 Gene in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 779-788. |
| [6] | WANG Shilin, WU Ting, ZHOU Shiqi, SONG Siming, HU Biaolin. Identification of QTLs for Seed Storability in Dongxiang Wild Rice by Integrating BSA-Seq and QTL Analysis [J]. Chinese Journal OF Rice Science, 2025, 39(6): 789-800. |
| [7] | LU Shuai, TAO Tao, LIU Ran, ZHOU Wenyu, CAO Lei, YANG Qingqing, ZHANG Mingqiu, REN Xinzhe, YANG Zhidi, XU Fuxiang, HUAN Haidong, GONG Yuanhang, ZHANG Haocheng, JIN Sukui, CAI Xiuling, GAO Jiping, LENG Yujia. Identification and Gene Cloning of a Long Sterile Lemma and Small Grain Mutant lsg8 in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2025, 39(6): 813-824. |
| [8] | DENG Huan, LIU Yapei, WANG Chunlian, GUO Wei, CHEN Xifeng, JI Zhiyuan. Mapping Analysis of a New Bacterial Blight Resistance Gene Xa49(t) in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 825-831. |
| [9] | ZHANG Lanlan, LIU Dilin, MA Xiaozhi, HUO Xing, KONG Le, LI Jinhua, FU Chongyun, LIAO Yilong, ZHU Manshan, ZENG Xueqin, LIU Wuge, WANG Feng. Quality Traits Affecting Eating Quality in indica Rice Under Different Nitrogen Application Levels in Early and Late Seasons in South China [J]. Chinese Journal OF Rice Science, 2025, 39(6): 832-846. |
| [10] | BIAN Jinlong, REN Gaolei, QIU Shi, XU Fangfu, HU Zhonglei, ZHANG Hongcheng, WEI Haiyan. Effect of Application Methods of Mixed Controlled-release Nitrogen Fertilizer on Yield and Nitrogen Utilization of Good Taste Quality japonica Rice Under Different Mechanical Transplanting Methods in the Huaibei Region [J]. Chinese Journal OF Rice Science, 2025, 39(6): 847-862. |
| [11] | LI Xing, ZHANG Ruichun, CHEN Ge, XIE Jiaxin, XIAO Zhengwu, CAO Fangbo, CHEN Jiana, HUANG Min. Yield Formation and Photosynthetic Characteristics of Machine-transplanted Late-season Rice with Short Growth Duration [J]. Chinese Journal OF Rice Science, 2025, 39(6): 863-872. |
| [12] | LUO Zizi, ZHANG Dejun, CHEN Dongdong, BI Miao, ZHU Yuhan, HAN Xu, WU Qiang, LI Yuechen. Characteristics of Spatiotemporal Evolution of Thermal Resources in Ratoon Rice at Heading and Grain-filling Stages in Sichuan Basin in 1981−2022 [J]. Chinese Journal OF Rice Science, 2025, 39(6): 873-886. |
| [13] | HAO Wenqian, CAI Xingjing, YANG Haidong, WU Yuyang, TENG Xuan, XUE Chao, GONG Zhiyun. Advances in Roles of Different Types of Histone Modifications in Responses of Rice to Abiotic Stresses [J]. Chinese Journal OF Rice Science, 2025, 39(5): 575-585. |
| [14] | LU Tingting, YAN Wenhui, SU Xinquan, ZENG Luohua, HUA Liqin, CHEN Jianghua, FENG Baohua, WANG Yuexing, HU Jiang, FU Guanfu. Research Progress on Physiological and Ecological Mechanisms and Regulation Pathways of Yield, Quality and Stress Resistance Response in Perennial Rice [J]. Chinese Journal OF Rice Science, 2025, 39(5): 586-600. |
| [15] | WU Wanting, XU Qian, LIU Dantong, ZHU Changjin, DU Haotian, JU Haoran, HUO Zhongyang, DAI Qigen, LI Guohui, XU Ke. Research Progress in Regulation of Anthocyanin Accumulation in Colored Rice [J]. Chinese Journal OF Rice Science, 2025, 39(5): 601-614. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||