
Chinese Journal OF Rice Science ›› 2021, Vol. 35 ›› Issue (6): 629-638.DOI: 10.16819/j.1001-7216.2021.210206
• 研究报告 • Previous Articles
Yudong CAO, Xiangyi XIAO, Naizhong YE, Xiaowen DING, Xiaoxuan YI, Jinling LIU*( ), Yinghui XIAO*(
), Yinghui XIAO*( )
)
Received:2021-02-19
Revised:2021-03-16
Online:2021-11-10
Published:2021-11-10
Contact:
Jinling LIU, Yinghui XIAO
曹煜东, 肖湘谊, 叶乃忠, 丁晓雯, 易晓璇, 刘金灵*( ), 肖应辉*(
), 肖应辉*( )
)
通讯作者:
刘金灵,肖应辉
基金资助:Yudong CAO, Xiangyi XIAO, Naizhong YE, Xiaowen DING, Xiaoxuan YI, Jinling LIU, Yinghui XIAO. Auxin Regulator OsGRF4 Simultaneously Regulates Rice Grain Shape and Blast Resistance[J]. Chinese Journal OF Rice Science, 2021, 35(6): 629-638.
曹煜东, 肖湘谊, 叶乃忠, 丁晓雯, 易晓璇, 刘金灵, 肖应辉. 生长素调控因子OsGRF4协同调控水稻粒形和稻瘟病抗性[J]. 中国水稻科学, 2021, 35(6): 629-638.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2021.210206
| 引物用途 Primer usage | 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|---|
| CRISPR/Cas9载体构建 Constriction of CRISPR/Cas9 vector | CRS-GRF4-F | GCCGCAGCTCCTCGTACTGCGCCG |
| CRS-GRF4-R | AAACCGGCGCAGTACGAGGAGCTG | |
| 敲除转基因植株鉴定 Identification of knockout transgenic plants | GRF4-CRSJD-F | AACCCATTTTCTTGGCTC |
| GRF4-CRSJD-R | CGCCTGATCGGAATAAAC |
Table 1 Primers for construction and identification of CRISPR/Cas9 vectors.
| 引物用途 Primer usage | 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|---|
| CRISPR/Cas9载体构建 Constriction of CRISPR/Cas9 vector | CRS-GRF4-F | GCCGCAGCTCCTCGTACTGCGCCG |
| CRS-GRF4-R | AAACCGGCGCAGTACGAGGAGCTG | |
| 敲除转基因植株鉴定 Identification of knockout transgenic plants | GRF4-CRSJD-F | AACCCATTTTCTTGGCTC |
| GRF4-CRSJD-R | CGCCTGATCGGAATAAAC |
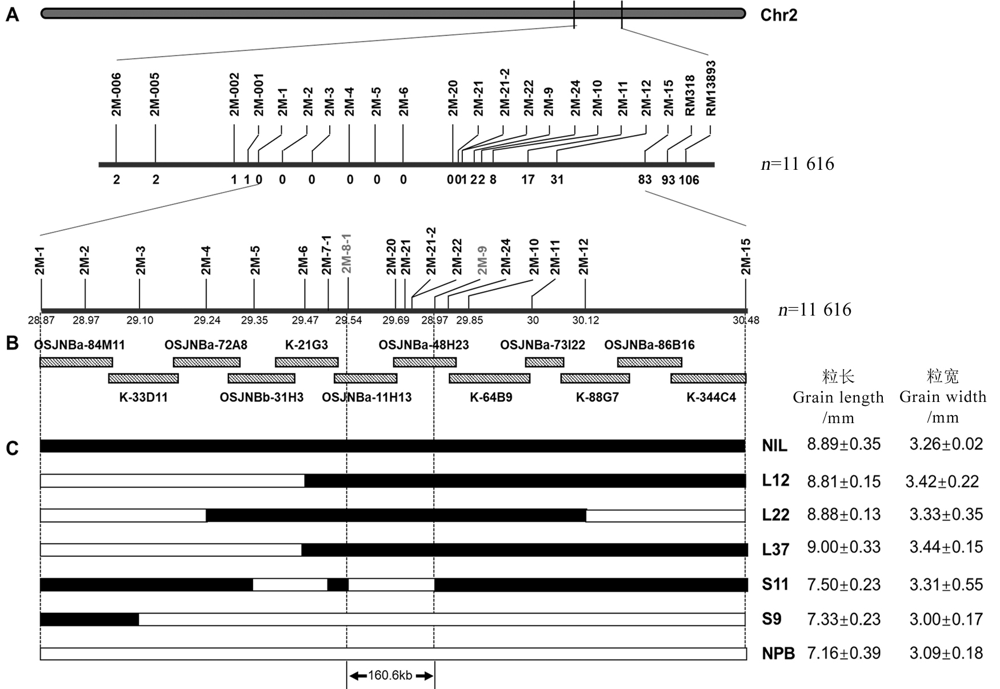
Fig. 1. Construction of genetic map and physical map of GS2.2. A, Genetic map of GS2.2; B, BAC clone contig and physical map of GS2.2 locus; C, The GS2.2 physical interval was determined by overlapping recombinant lines. NIL, Near-isogenic line with large grains; NPB, Nipponbare; “L+Number” represents a recombinant strain with large grains, and the “S+Number” represents a recombinant strain with small grains. The phenotype of grain length and width for NPB, NIL and recombinant lines was presented on the right.
| 候选基因 Candidate gene | 蛋白编码结构 Protein coding structure | |
|---|---|---|
| ORF1 | Probable WRKY transcription factor 14 | |
| ORF2 | Protein NRT1/ PTR FAMILY 8.3 | |
| ORF3 | ADP-ribosylation factor | |
| ORF4 | Uncharacterized LOC4330423 | |
| ORF5 | Putative surface protein SACOL0050 | |
| ORF6 | 60S ribosomal protein L12-1 | |
| ORF7 | DNA topoisomerase 2 | |
| ORF8 | Protein-coding | |
| ORF9 | Os02g0699800 | |
| ORF10 | Transcription initiation factor TFIID subunit 8 | |
| ORF11 | Uncharacterized protein At1g51745 | |
| ORF12 | Cell division cycle 20.2, cofactor of APC complex | |
| ORF13 | Myb family transcription factor APL | |
| ORF14 | Ubiquinol oxidase 2, mitochondrial | |
| ORF15 | Polyamine transporter PUT1 | |
| ORF16 | Cyclin-dependent kinase F-4 | |
| ORF17 | Protein Brevis radix-like 2 | |
| ORF18 | Growth-regulating factor 4 | |
Table 2 Prediction and analysis of GS2.2 candidate genes.
| 候选基因 Candidate gene | 蛋白编码结构 Protein coding structure | |
|---|---|---|
| ORF1 | Probable WRKY transcription factor 14 | |
| ORF2 | Protein NRT1/ PTR FAMILY 8.3 | |
| ORF3 | ADP-ribosylation factor | |
| ORF4 | Uncharacterized LOC4330423 | |
| ORF5 | Putative surface protein SACOL0050 | |
| ORF6 | 60S ribosomal protein L12-1 | |
| ORF7 | DNA topoisomerase 2 | |
| ORF8 | Protein-coding | |
| ORF9 | Os02g0699800 | |
| ORF10 | Transcription initiation factor TFIID subunit 8 | |
| ORF11 | Uncharacterized protein At1g51745 | |
| ORF12 | Cell division cycle 20.2, cofactor of APC complex | |
| ORF13 | Myb family transcription factor APL | |
| ORF14 | Ubiquinol oxidase 2, mitochondrial | |
| ORF15 | Polyamine transporter PUT1 | |
| ORF16 | Cyclin-dependent kinase F-4 | |
| ORF17 | Protein Brevis radix-like 2 | |
| ORF18 | Growth-regulating factor 4 | |
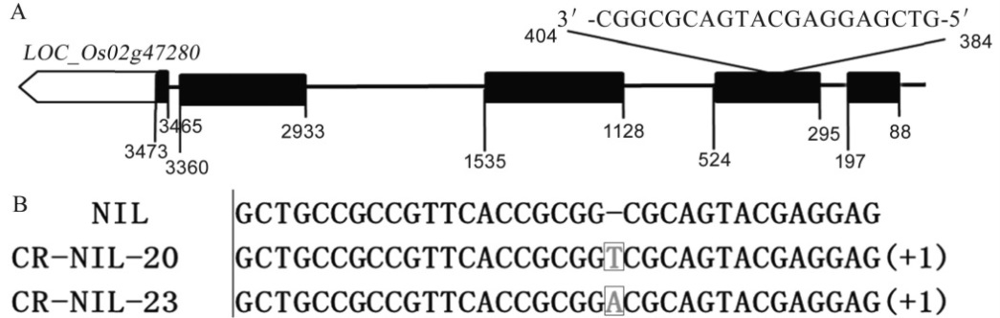
Fig. 3. Sequence of target site for OsGRF4 CRISPR / Cas9 gene editing and genotype of OsGRF4 CRISPR / Cas9 gene editing lines in NIL background. A, Sequence of target site for OsGRF4 CRISPR / Cas9 gene editing; B, Genotype of OsGRF4 CRISPR / Cas9 gene editing lines in NIL.
| 转基因株系 Transgenic lines | 基因型 Genotype | 基因编辑类型 Gene editing type |
|---|---|---|
| Reference | GCTGCCGCCGTTCACCGCGGCGCAGTACGAGGAGC | |
| CR-NIL-15 | GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAGC | +1 |
| CR-NIL-16 | GCTGCCGCCGTTCACCGCGGACGCAGTACGAGGAG | +1 |
| GCTGCCGCCGTTCACCGCGGCCGCAGTACGAGGAG | +1 | |
| CR-NIL-17 | AGGCTGCCGCCGTTCACCGC-(46 bp del)CTGGTGGCAGGCGTG | -46 |
| AGGCTGCCGCCGTTCACCGCGGGCGCAGTACGAGG | +1 | |
| CR-NIL-19 | GGCTGCCGCCGTTCACCGCGGCGCAGTACGAGGAG (WT) | +0 |
| GGCTGCCGCCGTTCACCGCGTACGCAGTACGAGGA | +1 | |
| CR-NIL-20 | GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAG | +1 |
| CR-NIL-23 | GCTGCCGCCGTTCACCGCGGACGCAGTACGAGGAG | +1 |
| CR-NIL-22 | GCTGCCGCCGTTCACCGCGGGCGCAGTACGAGGAG | +1 |
| GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAG | +1 |
Table 3 OsGRF4 CRISPR / Cas9 gene editing knockout mutation types.
| 转基因株系 Transgenic lines | 基因型 Genotype | 基因编辑类型 Gene editing type |
|---|---|---|
| Reference | GCTGCCGCCGTTCACCGCGGCGCAGTACGAGGAGC | |
| CR-NIL-15 | GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAGC | +1 |
| CR-NIL-16 | GCTGCCGCCGTTCACCGCGGACGCAGTACGAGGAG | +1 |
| GCTGCCGCCGTTCACCGCGGCCGCAGTACGAGGAG | +1 | |
| CR-NIL-17 | AGGCTGCCGCCGTTCACCGC-(46 bp del)CTGGTGGCAGGCGTG | -46 |
| AGGCTGCCGCCGTTCACCGCGGGCGCAGTACGAGG | +1 | |
| CR-NIL-19 | GGCTGCCGCCGTTCACCGCGGCGCAGTACGAGGAG (WT) | +0 |
| GGCTGCCGCCGTTCACCGCGTACGCAGTACGAGGA | +1 | |
| CR-NIL-20 | GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAG | +1 |
| CR-NIL-23 | GCTGCCGCCGTTCACCGCGGACGCAGTACGAGGAG | +1 |
| CR-NIL-22 | GCTGCCGCCGTTCACCGCGGGCGCAGTACGAGGAG | +1 |
| GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAG | +1 |
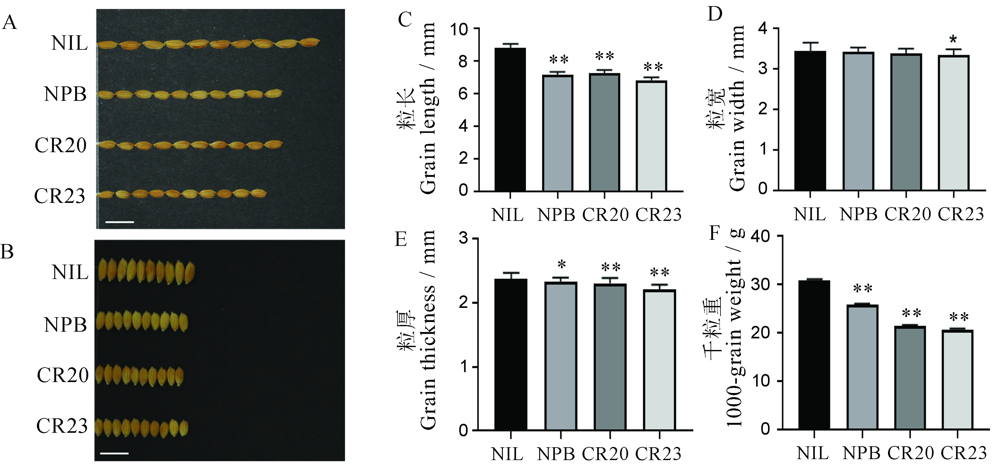
Fig. 4. Grain shape of gene editing lines in GS2.2-NIL background. A and B, Phenotype of grain length and width. C-F, Statistic data of grain length, width, thickness and 1000-grain weight. *P<0.05; **P<0.01(t-test). Error bar indicated the SD value (n=30). NIL, GS2.2-NIL; CR20, CR-NIL-20; CR23, CR-NIL-23.
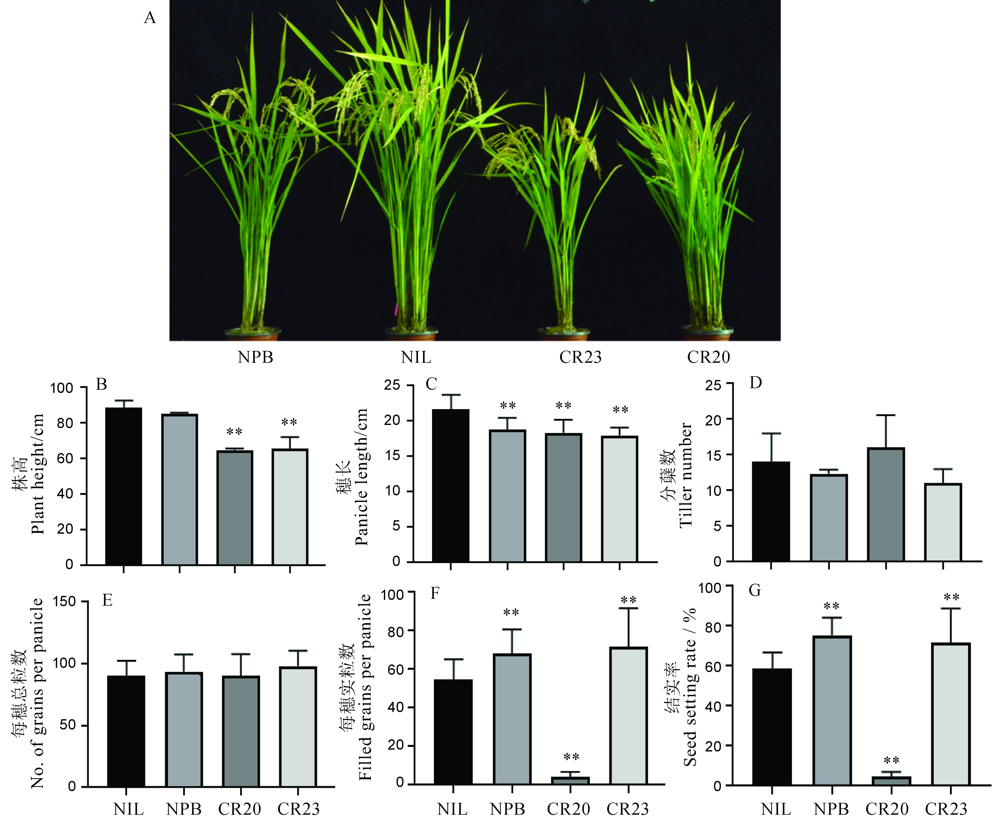
Fig. 5. Phenotype of agronomic traits of OsGRF4 gene editing lines in NIL background. A, Plant architecture for gene editing lines. B-G, Plant height, panicle length, tiller number, total grains per panicle, number of full grains per panicle and seed setting rate for gene editing lines. The t-test was used for significance analysis. Error bar indicated the SD value (n=3). **P<0.01.NIL, GS2.2-NIL; CR20, CR-NIL-20; CR23, CR-NIL-23.
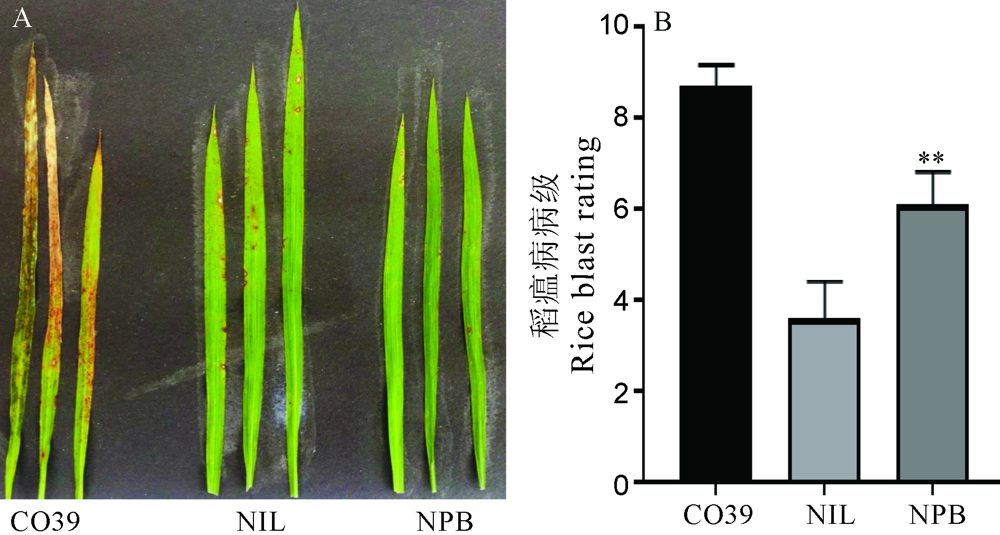
Fig. 6. Symptoms of GS2.2-NIL and Nipponbare(NPB) after infected by rice blast fungal in natural disease nurse. A, Symptoms of leaf after rice blast fungal infection in the natural disease nurse; B, Leaf blast rating after rice blast fungal infection in the natural disease nurse. The t-test was used for significance analysis. Error bar indicated the SD(n=10). **P<0.01。
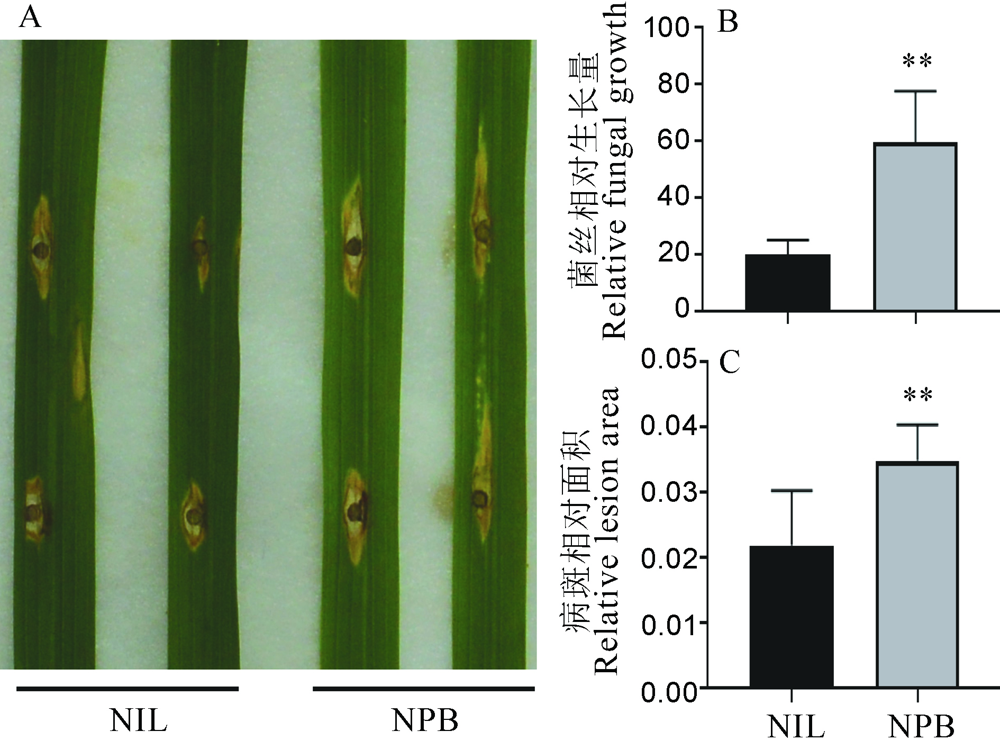
Fig. 7. Symptoms of GS2.2-NIL and Nipponbare(NPB) after in vitro inoculation with rice blast fungus. A, Leaf disease phenotype; B, Relative blast fungal biomass in the infected leaf; C, Area of leaf infected by blast fungal. The t-test was used for significance analysis, error bar indicated the SD value(n=3). **P<0.01(t-test).
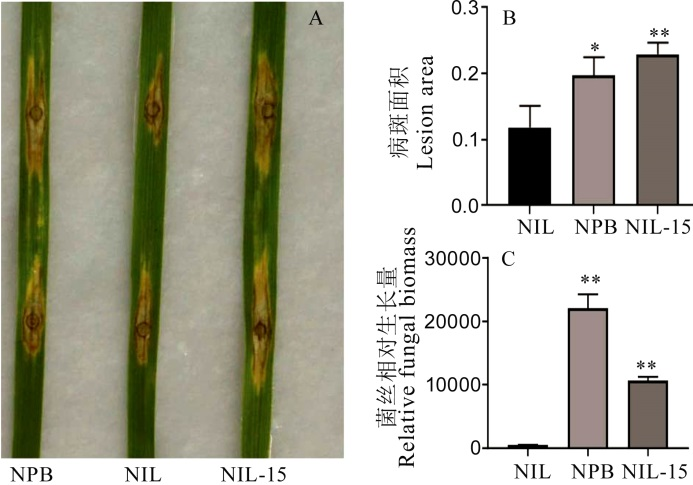
Fig. 8. Symptoms of GS2.2-NIL, Nipponbare(NPB) and CRISPR/Cas9 editing lines after in vitro inoculation with rice blast fungus. A, Phenotype of infection; B, Infected leaf area; C, The relative fungal biomass. The t-test was used for significance analysis, error bar indicated the SD value (n=3). *P<0.05; **P<0.01(t-test). NIL-15, CR-NIL-15.
| [1] | 康艺维, 陈玉宇, 张迎信. 水稻粒型基因克隆研究进展及育种应用展望[J]. 中国水稻科学, 2020, 34(6): 479-490. |
| Kang Y W, Chen Y Y, Zhang Y X.Research progress and breeding prospects of grain size associated genes in rice[J]. Chinese Journal of Rice Science, 2020, 34(6): 479-490. (in Chinese with English abstract) | |
| [2] | 杨维丰, 詹鹏麟, 林少俊, 苟亚军, 张桂权, 王少奎. 水稻粒形的遗传研究进展[J]. 华南农业大学学报, 2019, 40(5): 203-210. |
| Yang W F, Zhan P L, Lin S J, Gou Y J, Zhang G Q, Wang S K.Research progress of grain shape genetics in rice[J]. Journal of South China Agricultural University, 2019, 40(5): 203-210. (in Chinese with English abstract) | |
| [3] | 刘喜, 牟昌铃, 周春雷, 程治军, 江玲, 万建民. 水稻粒型基因克隆和调控机制研究进展[J]. 中国水稻科学, 2018, 32(1): 1-11. |
| Liu X, Mou C L, Zhou C L, Cheng Z J, Jiang L, Wan J M.Research progress on cloning and regulation mechanism of rice grain shape genes[J]. Chinese Journal of Rice Science, 2018, 32(1): 1-11. (in Chinese with English abstract) | |
| [4] | Huang R, Jiang L, Zheng J, Wang T, Wang H, Huang Y, Hong Z.Genetic bases of rice grain shape: So many genes, so little known[J]. Trends in Plant Science, 2013, 18(4): 218-226. |
| [5] | 尉鑫, 曾智锋, 杨维丰, 韩婧, 柯善文. 水稻粒形遗传调控研究进展[J]. 安徽农业科学, 2019, 47(5): 21-28. |
| Wei X, Zeng Z F, Yang W F, Han J, Ke S W.Advances in studies on genetic regulation of rice grain shape.Journal of Anhui Agricultural Science, 2019, 47(5): 21-28. (in Chinese with English abstract) | |
| [6] | 马超, 原佳乐, 张苏, 贾琦石, 冯雅岚. GRF转录因子对植物生长发育及胁迫响应调控的分子机制[J]. 核农学报, 2017, 31(11): 2145-2153. |
| Ma C, Yuan J L, Zhang S, Jia Q S, Feng Y L.The molecular mechanisms of growth-regulating factors (GRFs) in plant growth, development and stress response[J]. Journal of Nuclear Agricultural Sciences, 2017, 31(11): 2145-2153. (in Chinese with English abstract) | |
| [7] | 袁岐, 张春利, 赵婷婷, 许向阳. 植物中GRF转录因子的研究进展[J]. 基因组学与应用生物学, 2017, 36(8): 3145-3151. |
| Yuan Q, Zhang C L, Zhao T T, Xu X Y.Research advances of GRF transcription factor in plant[J]. Genomics and Applied Biology, 2017, 36(8): 3145-3151. (in Chinese with English abstract) | |
| [8] | Szczygieł-Sommer A, Gaj M D.The miR396-GRF regulatory module controls the embryogenic response in Arabidopsis via an auxin-related pathway[J]. Inter- national Journal of Molecular Sciences, 2019, 20: 5221. |
| [9] | Liang G, He H, Li Y, Wang F, Yu D.Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis[J]. Plant Physiology, 2014, 164(1): 249-258. |
| [10] | van Knaap E, Kim J H, Kende H. A novel gibberellin- induced gene from rice and its potential regulatory role in stem growth[J]. Plant Physiology, 2000, 122(3): 695. |
| [11] | Chandran V, Wang H, Gao F, Cao X L, Chen Y P, Li G B, Zhu Yong, Yang X M, Zhang L L, Zhao Z X, Zhao J H, Wang Y G, Li S C, Fan J, Li Y, Zhao J Q, Li S Q, Wang W M. miR396-OsGRFs module balances growth and rice blast disease-resistance[J]. Frontiers in Plant Science, 2019, 9: 1999. |
| [12] | Dai Z, Tan J, Zhou C, Yang X, Yang F, Zhang S, Sun S, Miao X, Shi Z.The OsmiR396-OsGRF8-OsF3H- flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa)[J]. Plant Biotechnology Journal, 2019, 17: 1657-1669. |
| [13] | Sun P, Zhang W, Wang Y, He Q, Shu F, Liu H, Wang J, Wang J, Yuan L, Deng H.OsGRF4 controls grain shape, panicle length and seed shattering in rice[J]. Journal of Integrative Plant Biology, 2016, 58(10): 836-847. |
| [14] | Li S, Gao F, Xie K, Zeng X, Cao Y, Zeng J, He Z, Ren Y, Li W, Deng Q, Wang S, Zheng A, Zhu J, Liu H, Wang L, Li P.The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice[J]. Plant Biotechnology Journal, 2016, 14(11): 2134-2146. |
| [15] | Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y, Harberd N P, Fu X.Modulating plant growth-metabolism coordination for sustainable agriculture[J]. Nature, 2018, 560(7720): 595-600. |
| [16] | Chen X, Jiang L, Zheng J, Chen F, Wang T, Wang M, Tao Y, Wang H, Hong Z, Huang Y, Huang R.A missense mutation in Large Grain Size 1 increases grain size and enhances cold tolerance in rice[J]. Journal of Experimental Botany, 2019, 70(15): 3851-3866. |
| [17] | 叶乃忠. 水稻粒形基因GS2.2的精细定位[D]. 长沙: 湖南农业大学, 2016. |
| Ye N Z.Fine mapping of rice grain shape gene GS2.2[D]. Changsha: Hunan Agricultural University, 2016. (in Chinese with English abstract) | |
| [18] | 楼巧君, 陈亮, 罗利军. 三种水稻基因组DNA快速提取方法的比较[J]. 分子植物育种, 2005, 13(5): 749-752. |
| Lou Q J, Chen L, Luo L J.Comparison of three rapid rice DNA extraction methods[J]. Molecular Plant Breeding, 2005, 13(5): 749-752. (in Chinese with English abstract) | |
| [19] | 张会军. 水稻Pi21和OsBadh2基因编辑改良空育131的稻瘟病抗性及香味品质[D]. 武汉: 华中农业大学, 2016. |
| Zhang H J.Rice blast resistance and aroma quality improved by editing rice Pi21 and OsBadh2 genes [D]. Wuhan: Huazhong Agricultural University, 2016. (in Chinese with English abstract) | |
| [20] | 王文文, 孙红正, 李俊周, 彭廷, 杜彦修, 张静, 赵全志. 水稻miR395d基因过表达载体的构建及功能初步分析[J]. 分子植物育种, 2019, 17(9): 2876-2881. |
| Wang W W, Sun H Z, Li J Z, Peng T, Du Y X, Zhang J, Zhao Q Z.Construction and preliminary functional analysis of over-expression vector of mir395d gene in rice[J]. Molecular Plant Breeding, 2019, 17(9): 2876-2881. (in Chinese with English abstract) | |
| [21] | 崔华威, 杨艳丽, 黎敬涛, 罗文富, 苗爱敏, 胡振兴, 韩小女. 一种基于Photoshop的叶片相对病斑面积快速测定方法[J]. 安徽农业科学, 2009, 37(22): 10760-10762, 10805. |
| Cui H W, Yang Y L, Li J T, Luo W F, Miao A M, Hu Z X, Han X N.A faster method for measuring relative lesion area on leaves based on software photoshop[J]. Journal of Anhui Agricultural Science, 2009, 37(22): 10760-10762, 10805. (in Chinese with English abstract) | |
| [22] | International Rice Research Institute. Standard Evaluation System for Rice (SES)[M]. Los Baños, Philippines: International Rice Research Institute, 1996: 17-18. |
| [23] | 孙平勇, 张武汉, 张莉, 舒服, 何强, 彭志荣, 邓华凤. 水稻氮高效、耐冷基因OsGRF4功能标记的开发及其利用[J]. 作物学报, 2021, 47(4): 684-690. |
| Sun P Y, Zhang W H, Zhang L, Shu F, He Q, Peng Z R, Deng H F.Development and application of functional marker for high nitrogen use efficiency and chilling tolerance gene OsGRF4 in rice. Acta Agronomica Sinica, 2021, 47(4): 684-690. (in Chinese with English abstract) | |
| [24] | 刘学英, 李姗, 吴昆, 刘倩, 高秀华, 傅向东. 提高农作物氮肥利用效率的关键基因发掘与应用[J]. 科学通报, 2019, 64(25): 2633-2640. |
| Liu X Y, Li S, Wu K, Liu Q, Gao X H, Fu X D.Sustainable crop yields from the coordinated modulation of plant growth and nitrogen metabolism[J]. Chinese Science Bulletin, 2019, 64(25): 2633-2640. (in Chinese with English summary) | |
| [25] | Li W, Zhu Z, Chen M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J, Zhou X, Zhu X, Chen Z, Wang J, Zhao W, Ma B, Qin P, Chen W, Wang Y, Liu J, Wu X, Li P, Wang J, Zhu L, Li S, Chen X. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance [J]. Cell, 2017, 170(1): 114-126.e15. |
| [26] | Zhou X, Liao H, Chen M, Yin J, Chen Yi, Wang J, Zhu X, Chen Z, Yuan C, Wang W, Wu X, Li P, Zhu L, Li S, Ronald P C, Chen X.Loss of function of a rice TPR-domain RNA-binding protein confers broad- spectrum disease resistance[J]. Proceedings of the National Academy of Sciences of the Unite States of America, 2018, 115: 3174-3179. |
| [27] | Wang J, Zhou L, Shi H, Chen M, Yu H, Yi H, He M, Yin J, Zhu X, Li Y, Li W, Liu J, Wang J, Chen X, Qing H, Wang Y, Liu G, Wang W, Li P, Wu X, Zhu L, Zhou J, Ronald P C, Li S, Li J, Chen X,.A single transcription factor promotes both yield and immunity in rice[J]. Science, 2018, 361: 1026-1028. |
| [1] |
WANG Yichen, ZHU Benshun, ZHOU Lei, ZHU Jun, YANG Zhongnan.
Sterility Mechanism of Photoperiod/Thermo-sensitive Genic Male Sterile Lines and Development and Prospects of Two-line Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(5): 463-474. |
| [2] |
XU Yongqiang XU Jun, FENG Baohua, XIAO Jingjing, WANG Danying, ZENG Yuxiang, FU Guanfu.
Research Progress of Pollen Tube Growth in Pistil of Rice and Its Response to Abiotic stress [J]. Chinese Journal OF Rice Science, 2024, 38(5): 495-506. |
| [3] |
HE Yong, LIU Yaowei, XIONG Xiang, ZHU Danchen, WANG Aiqun, MA Lana, WANG Tingbao, ZHANG Jian, LI Jianxiong, TIAN Zhihong.
Creation of Rice Grain Size Mutants by Editing OsOFP30 via CRISPR/Cas9 System [J]. Chinese Journal OF Rice Science, 2024, 38(5): 507-515. |
| [4] |
LÜ Yang, LIU Congcong, YANG Longbo, CAO Xinglan, WANG Yueying, TONG Yi, Mohamed Hazman, QIAN Qian, SHANG Lianguang, GUO Longbiao.
Identification of Candidate Genes for Rice Nitrogen Use Efficiency by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(5): 516-524. |
| [5] |
YANG Hao, HUANG Yanyan, WANG Jian, YI Chunlin, SHI Jun, TAN Chutian, REN Wenrui, WANG Wenming.
Development and Application of Specific Molecular Markers for Eight Rice Blast Resistance Genes in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(5): 525-534. |
| [6] |
JIANG Peng, ZHANG Lin, ZHOU Xingbing, GUO Xiaoyi, ZHU Yongchuan, LIU Mao, GUO Chanchun, XIONG Hong, XU Fuxian.
Yield Formation Characteristics of Ratooning Hybrid Rice Under Simplified Cultivation Practices in Winter Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(5): 544-554. |
| [7] |
YANG Mingyu, CHEN Zhicheng, PAN Meiqing, ZHANG Bianhong, PAN Ruixin, YOU Lindong, CHEN Xiaoyan, TANG Lina, HUANG Jinwen.
Effects of Nitrogen Reduction Combined with Biochar Application on Stem and Sheath Assimilate Translocation and Yield Formation in Rice Under Tobacco-rice Rotation [J]. Chinese Journal OF Rice Science, 2024, 38(5): 555-566. |
| [8] |
XIONG Jiahuan, ZHANG Yikai, XIANG Jing, CHEN Huizhe, XU Yicheng, WANG Yaliang, WANG Zhigang, YAO Jian, ZHANG Yuping .
Effect of Biochar-based Fertilizer Application on Rice Yield and Nitrogen Utilization in Film- mulched PaddyFields [J]. Chinese Journal OF Rice Science, 2024, 38(5): 567-576. |
| [9] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [10] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [11] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [12] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [13] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [14] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [15] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||