
中国水稻科学 ›› 2019, Vol. 33 ›› Issue (6): 513-522.DOI: 10.16819/j.1001-7216.2019. 9090
陈专专1,2, 李先锋1, 仲敏1, 葛家奇1, 范晓磊1,2, 张昌泉1,2, 刘巧泉1,2,*( )
)
收稿日期:2019-08-07
修回日期:2019-09-13
出版日期:2019-11-10
发布日期:2019-11-10
通讯作者:
刘巧泉
基金资助:
Zhuanzhuan CHEN1,2, Xianfeng LI1, Min ZHONG1, Jiaqi GE1, Xiaolei FAN1,2, Changquan ZHANG1,2, Qiaoquan LIU1,2,*( )
)
Received:2019-08-07
Revised:2019-09-13
Online:2019-11-10
Published:2019-11-10
Contact:
Qiaoquan LIU
摘要:
目的 稻米糊化温度是影响稻米品质的重要指标,该性状受主效基因ALK/SSII-3调控,ALK基因具有多个复等位基因,本研究旨在通过RNAi技术明确籼稻亚种中两个不同ALK等位基因的效应。方法 以分别含有ALKc和ALKb等位基因的高糊化温度品种珍汕97B和低糊化温度品种龙特甫B为试验材料,使用RNAi技术构建ALK表达下调的转基因株系,通过对其稻米理化品质的测定来明确不同等位基因表达下调对稻米品质的影响。结果 对不同转基因水稻目的基因的表达分析显示本研究中转基因株系的ALK基因受到了不同程度的干扰。重点分析了不同RNAi株系稻米的糊化温度,结果表明珍汕97B的RNAi转基因稻米的糊化温度极显著降低,而在低糊化温度品种龙特甫B背景中下调表达ALK基因后对糊化温度的影响较小;转基因株系与未转化亲本相比,米粉的起始糊化温度都显著降低,表现为提前糊化;在珍汕97B背景下干扰系的峰值温度与未转化亲本相比极显著降低,而在龙特甫背景下米粉的峰值温度与未转化对照相比显著降低。对不同转基因系的理化品质分析表明,ALK下调表达植株稻米的表观直链淀粉含量显著增加,下调表达ALK后会引起米粉峰值黏度和崩解值的改变。高糊化温度品种珍汕97B干扰系与未转化对照相比胶稠度呈现极显著性差异,而低糊化温度品种龙特甫干扰系的胶稠度与未转化对照相比没有差异。结论 下调表达ALK等位基因对稻米理化品质产生显著影响,并且干扰不同等位基因的效应存在明显差异,即籼稻中的两个ALK等位基因的效应存在显著差异。
中图分类号:
陈专专, 李先锋, 仲敏, 葛家奇, 范晓磊, 张昌泉, 刘巧泉. 籼稻背景下抑制不同ALK等位基因表达对稻米品质的影响[J]. 中国水稻科学, 2019, 33(6): 513-522.
Zhuanzhuan CHEN, Xianfeng LI, Min ZHONG, Jiaqi GE, Xiaolei FAN, Changquan ZHANG, Qiaoquan LIU. Grain Quality as Affected by Down-regulation of Expression of Different ALK Alleles in indica Rice (Oryza sativa L.)[J]. Chinese Journal OF Rice Science, 2019, 33(6): 513-522.
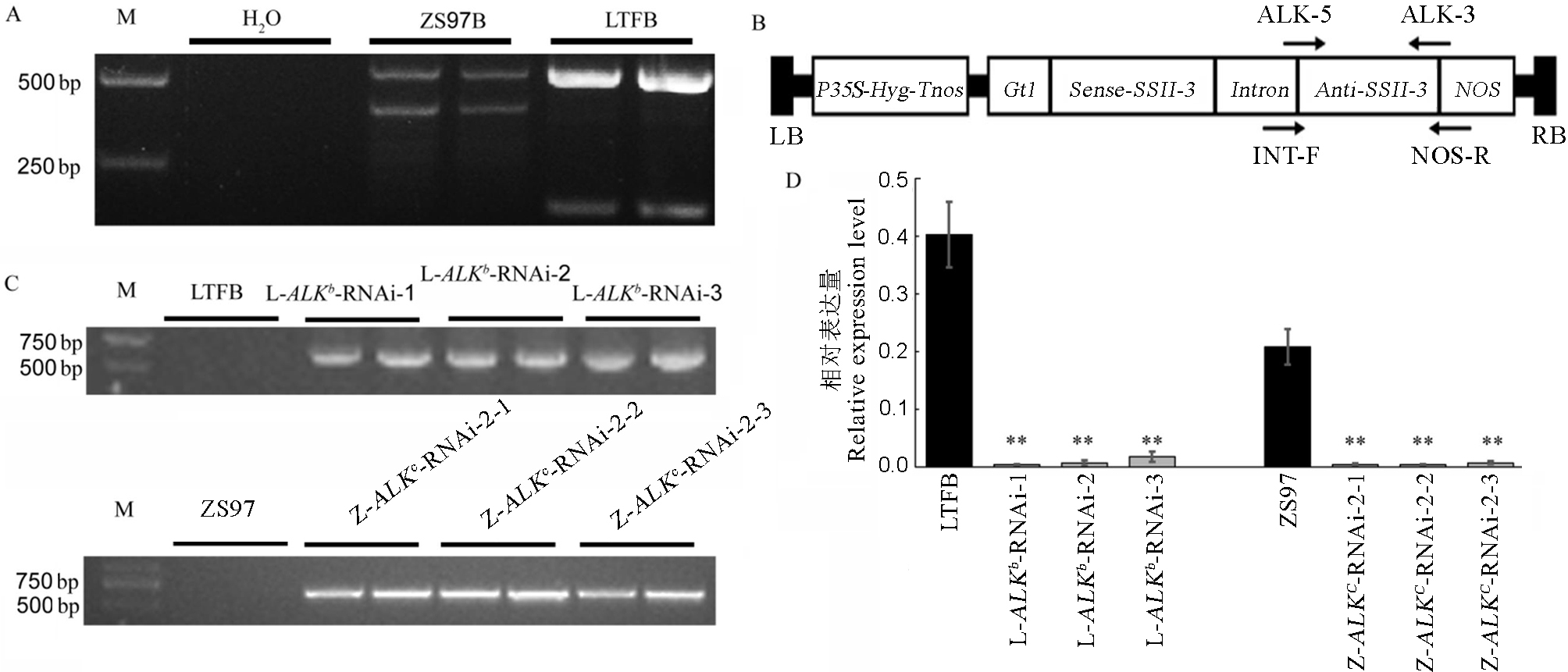
图1 ALK-RNAi载体结构及转基因水稻的鉴定 A-用引物4342(GC/TT)鉴定亲本对照龙特甫B(LTFB)和珍汕97B(ZS97B)的ALK基因型。B-含ALK-RNAi 结构双元载体的T-DNA区结构。P35S和Tnos, 分别表示CaMV35S基因的启动子和终止子区; NOS-农杆菌胭脂碱合成酶基因终止子; Hyg-潮霉素抗性基因; LB和RB分别表示T-DNA区的左右边界序列; Anti和Sense分别表示目的基因片段的反向和正向结构;图中箭头表示设计引物方向。C-ALK-RNAi转基因水稻植株的PCR鉴定。D-ALK-RNAi转基因水稻植株胚乳中ALK基因表达量分析。L-ALKb-RNAi-1、L-ALKb-RNAi-2和L-ALKb-RNAi-3为龙特甫背景下的ALKb-RNAi转基因纯合系;Z-ALKc-RNAi-2-1、Z-ALKc-RNAi-2-2和Z-ALKc-RNAi-2-3为同时携带有ALKc等位基因和ALKb-RNAi干扰结构的转基因材料纯合系。对不同背景下的转基因系和未转化对照的表达量进行方差分析,并用新复极差法进行多重比较。**表示转基因系与未转化对照之间的差异达0.01显著水平,*表示转基因系与未转化对照之间的差异达0.05显著水平(n=3)。
Fig. 1. ALK-RNAi construction and idendification of transgenic rice. A, ALK genotype in Longtefu B(LTFB) and Zhenshan 97B(ZS97B) on sites 4342 and 4343 based on primer 4342(GC/TT). B, The T-DNA structure of ALK-RNAi construct. P35S and Tnos represent the promoter and terminator region of the CaMV 35S gene respectively; NOS represents agrobacterium nopaline synthetase gene terminator; Hyg represents hygromycin resistance gene; LB and RB represent the left and right boundary sequences of the T-DNA region respectively; the anti-sense and the sense represent the reverse and forward structure of the target gene segment respectively; and the arrow in the figure indicates the direction of the designed primer. C, PCR analysis of ALK-RNAi transgenic rice plants. D, Transcription level expression of ALK Gene in RNAi Line. L-ALKb-RNAi-1, L-ALKb-RNAi-2 and L-ALKb-RNAi-3 are ALKb-RNAi transgenic homozygous lines under the background of Longtefu and Z-ALKc-RNAi-2-1, Z-ALKc-RNAi-2-2 and Z-ALKc-RNAi-2-3 are homozygous transgenic lines carrying both ALKb-RNAi interference structure and ALKc allele. Data were subjected to one-way analysis of variance (ANOVA) depending on the experiment, followed by a comparison of the means according to a Duncan’s multiple range test at P< 0.05 or P < 0.01. Double asterisks denote a highly significant difference between transgenic line and its wild type (P<0.01), single asterisk denotes a significant difference between transgenic line and it’s wild type (0.01≤ P<0.05). (n=3).
| 正向引物 Forward primer | 引物序列 Primer sequence(5′-3′) | 反向引物 Reverse primer | 引物序列 Primer sequence(5′-3′) |
|---|---|---|---|
| ALK-5 | GTCCATGGTCGACCTCAAGTAAGAACGGAG | ALK-3 | GTACTAGTCGCCTTTGGCTTC |
| 4342(GC/TT)-F1 | CAAGGAGAGCTGGAGGGGGC | 4342(GC/TT)-R1 | CATGCCGCGCACCTGGAAA |
| 4342(GC/TT)-F | TCGGCGGGCTGAGGGACAC | 4342(GC/TT)-R | TCGCATCAATGGACATAACAAACAC |
| INT-F | CCTCGTAATCAATTTTAGAT | NOS-R | GACCGCACAGGATTCAAT |
| ALK-RT-F | TGATCTGAACGAACCGGACG | ALK-RT-R | TGGGTAAAGCACCTGCAACA |
表1 本研究所用的PCR引物
Table 1 Primers used in this study.
| 正向引物 Forward primer | 引物序列 Primer sequence(5′-3′) | 反向引物 Reverse primer | 引物序列 Primer sequence(5′-3′) |
|---|---|---|---|
| ALK-5 | GTCCATGGTCGACCTCAAGTAAGAACGGAG | ALK-3 | GTACTAGTCGCCTTTGGCTTC |
| 4342(GC/TT)-F1 | CAAGGAGAGCTGGAGGGGGC | 4342(GC/TT)-R1 | CATGCCGCGCACCTGGAAA |
| 4342(GC/TT)-F | TCGGCGGGCTGAGGGACAC | 4342(GC/TT)-R | TCGCATCAATGGACATAACAAACAC |
| INT-F | CCTCGTAATCAATTTTAGAT | NOS-R | GACCGCACAGGATTCAAT |
| ALK-RT-F | TGATCTGAACGAACCGGACG | ALK-RT-R | TGGGTAAAGCACCTGCAACA |
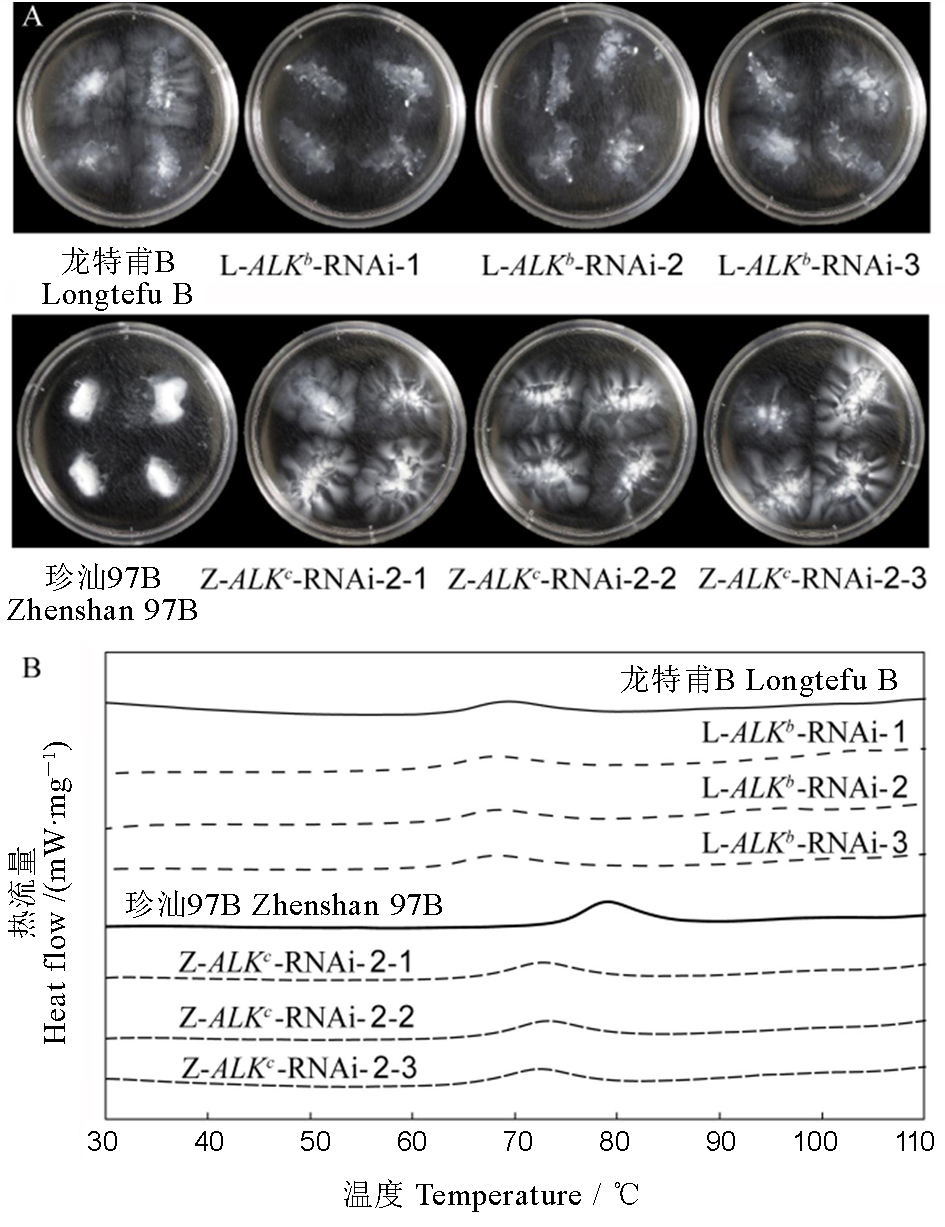
图2 ALK-RNAi转基因稻米的糊化温度 A-ALK-RNAi水稻株系精米在1.7% KOH中的碱消值; B-ALK-RNAi 水稻株系米粉的DSC曲线。
Fig 2. Gelatinization properties of ALK-RNAi transgenic rice. A, Alkali spreading value of milled rice in 1.7% KOH; B, DSC curve of rice flours of ALK-RNAi transgenic rice.
| 转基因系 | 起始温度 | 峰值温度 | 终止温度 | 热焓值 | 直链淀粉含量 | 胶稠度 | 粗蛋白含量 | |
|---|---|---|---|---|---|---|---|---|
| Transgenic line | To /℃ | Tp /℃ | Tc /℃ | △H /(J∙g-1) | AAC/% | GC/mm | Protein content/% | |
| 龙特甫B LTFB | 63.6±0.1 | 69.5±0.1 | 77.2±0.3 | 6.30±0.41 | 23.16±1.55 | 19.0±0.0 | 10.47±0.12 | |
| L-ALKb-RNAi-1 | 61.8±0.4* | 68.2±0.0** | 76.2±0.5* | 6.40±0.49 | 24.42±0.13 | 18.0±0.0 | 9.80±0.17 | |
| L-ALKb-RNAi-2 | 62.2±0.3* | 68.3±0.1** | 76.0±0.5* | 6.30±0.82 | 24.01±0.15 | 18.0±0.1 | 9.73±0.19 | |
| L-ALKb-RNAi-3 | 62.2±0.5* | 68.6±0.1** | 76.0±0.0* | 6.00±0.31 | 23.66±0.26 | 17.7±0.1 | 10.11±0.49 | |
| 珍汕97B ZS97B | 74.6±0.2 | 79.2±0.2 | 85.2±0.1 | 9.43±0.16 | 25.11±0.04 | 58.5±0.4 | 10.61±0.04 | |
| Z-ALKc-RNAi-2-1 | 67.1±0.3** | 73.4±0.1** | 80.2±0.1** | 8.20±0.08** | 27.73±0.24** | 25.5±0.1** | 9.21±0.05* | |
| Z-ALKc-RNAi-2-2 | 66.3±0.0** | 72.9±0.1** | 79.4±0.2** | 7.99±0.12** | 27.85±0.13** | 28.0±0.3** | 9.65±0.42* | |
| Z-ALKc-RNAi-2-3 | 65.6±0.1** | 72.4±0.1** | 79.0±0.0** | 7.29±0.07** | 26.69±0.01** | 45.0±0.0** | 9.03±0.46* | |
表2 ALK-RNAi转基因水稻株系米粉的热力学特性和理化特性
Table 2 Gelatinization properties and physical-chemical qualities of ALK-RNAi transgenic rice.
| 转基因系 | 起始温度 | 峰值温度 | 终止温度 | 热焓值 | 直链淀粉含量 | 胶稠度 | 粗蛋白含量 | |
|---|---|---|---|---|---|---|---|---|
| Transgenic line | To /℃ | Tp /℃ | Tc /℃ | △H /(J∙g-1) | AAC/% | GC/mm | Protein content/% | |
| 龙特甫B LTFB | 63.6±0.1 | 69.5±0.1 | 77.2±0.3 | 6.30±0.41 | 23.16±1.55 | 19.0±0.0 | 10.47±0.12 | |
| L-ALKb-RNAi-1 | 61.8±0.4* | 68.2±0.0** | 76.2±0.5* | 6.40±0.49 | 24.42±0.13 | 18.0±0.0 | 9.80±0.17 | |
| L-ALKb-RNAi-2 | 62.2±0.3* | 68.3±0.1** | 76.0±0.5* | 6.30±0.82 | 24.01±0.15 | 18.0±0.1 | 9.73±0.19 | |
| L-ALKb-RNAi-3 | 62.2±0.5* | 68.6±0.1** | 76.0±0.0* | 6.00±0.31 | 23.66±0.26 | 17.7±0.1 | 10.11±0.49 | |
| 珍汕97B ZS97B | 74.6±0.2 | 79.2±0.2 | 85.2±0.1 | 9.43±0.16 | 25.11±0.04 | 58.5±0.4 | 10.61±0.04 | |
| Z-ALKc-RNAi-2-1 | 67.1±0.3** | 73.4±0.1** | 80.2±0.1** | 8.20±0.08** | 27.73±0.24** | 25.5±0.1** | 9.21±0.05* | |
| Z-ALKc-RNAi-2-2 | 66.3±0.0** | 72.9±0.1** | 79.4±0.2** | 7.99±0.12** | 27.85±0.13** | 28.0±0.3** | 9.65±0.42* | |
| Z-ALKc-RNAi-2-3 | 65.6±0.1** | 72.4±0.1** | 79.0±0.0** | 7.29±0.07** | 26.69±0.01** | 45.0±0.0** | 9.03±0.46* | |
| 转基因系 Transgenic line | 峰值黏度 Peak viscosity /cP | 热浆黏度 Hot paste viscosity/cP | 崩解值Breakdown /cP | 冷胶黏度 Cool paste viscosity /cP | 消减值 Setback /cP | 峰值时间 Peak time /min | 起浆温度 Pasting temperature /℃ |
|---|---|---|---|---|---|---|---|
| 龙特甫B LTFB | 3699±11 | 3217±18 | 482±7 | 6385±18 | 2686±5 | 6.5 | 87.9 |
| L-ALKb-RNAi-1 | 3923±7** | 3390±10** | 533±13** | 6390±11 | 2467±8** | 6.5 | 87.0* |
| L-ALKb-RNAi-2 | 3810±13** | 3312±25** | 498±8* | 6055±7** | 2245±6** | 6.5 | 86.9* |
| L-ALKb-RNAi-3 | 3843±8** | 3137±17* | 706±10** | 6345±14* | 2502±7** | 6.5 | 87.8* |
| 珍汕97B ZS97B | 2885±12 | 2510±16 | 375±9 | 4974±15 | 2089±9 | 6.5 | 88.6 |
| Z-ALKc-RNAi-2-1 | 3608±9** | 2945±17** | 663±12** | 4749±17** | 1141±11** | 6.5 | 76.5** |
| Z-ALKc-RNAi-2-2 | 3930±11** | 3249±13** | 681±11** | 4590±16** | 660±12** | 6.6 | 77.4** |
| Z-ALKc-RNAi-2-3 | 3976±6** | 3337±14** | 639±8** | 4956±19 | 980±14** | 6.5 | 76.5** |
表3 ALK-RNAi 转基因水稻株系米粉的RVA谱特征值
Table 3 RVA parameters of mature rice flour in ALK-RNAi transgenic rice lines.
| 转基因系 Transgenic line | 峰值黏度 Peak viscosity /cP | 热浆黏度 Hot paste viscosity/cP | 崩解值Breakdown /cP | 冷胶黏度 Cool paste viscosity /cP | 消减值 Setback /cP | 峰值时间 Peak time /min | 起浆温度 Pasting temperature /℃ |
|---|---|---|---|---|---|---|---|
| 龙特甫B LTFB | 3699±11 | 3217±18 | 482±7 | 6385±18 | 2686±5 | 6.5 | 87.9 |
| L-ALKb-RNAi-1 | 3923±7** | 3390±10** | 533±13** | 6390±11 | 2467±8** | 6.5 | 87.0* |
| L-ALKb-RNAi-2 | 3810±13** | 3312±25** | 498±8* | 6055±7** | 2245±6** | 6.5 | 86.9* |
| L-ALKb-RNAi-3 | 3843±8** | 3137±17* | 706±10** | 6345±14* | 2502±7** | 6.5 | 87.8* |
| 珍汕97B ZS97B | 2885±12 | 2510±16 | 375±9 | 4974±15 | 2089±9 | 6.5 | 88.6 |
| Z-ALKc-RNAi-2-1 | 3608±9** | 2945±17** | 663±12** | 4749±17** | 1141±11** | 6.5 | 76.5** |
| Z-ALKc-RNAi-2-2 | 3930±11** | 3249±13** | 681±11** | 4590±16** | 660±12** | 6.6 | 77.4** |
| Z-ALKc-RNAi-2-3 | 3976±6** | 3337±14** | 639±8** | 4956±19 | 980±14** | 6.5 | 76.5** |
| 样品 Sample | 转基因系 Transgenic line | 综合 Integrated score | 外观 Appearance | 口感 Taste |
|---|---|---|---|---|
| 热饭 Cooked rice | 龙特甫B LTFB | 53.3±1.3 | 5.0±0.1 | 4.5±0.2 |
| L-ALKb-RNAi-1 | 48.3±1.3** | 4.3±0.1** | 3.9±0.2* | |
| L-ALKb-RNAi-2 | 51.0±0.8* | 4.7±0.1* | 4.2±0.1* | |
| L-ALKb-RNAi-3 | 50.5±1.7** | 4.6±0.2* | 4.1±0.2* | |
| 珍汕97B ZS97B | 55.0±2.6 | 5.1±0.3 | 4.8±0.3 | |
| Z-ALKc-RNAi-2-1 | 43.8±1.9** | 3.6±0.3** | 3.3±0.3** | |
| Z-ALKc-RNAi-2-2 | 42.0±0.8** | 3.5±0.0** | 3.1±0.1** | |
| Z-ALKc-RNAi-2-3 | 44.5±0.6** | 3.8±0.0** | 3.4±0.0** | |
| 冷饭 Cooled rice | 龙特甫B LTFB | 49.8±0.5 | 4.6±0.0 | 3.9±0.1 |
| L-ALKb-RNAi-1 | 44.8±0.5** | 4.1±0.1** | 3.6±0.1** | |
| L-ALKb-RNAi-2 | 48.5±1.0* | 4.3±0.1* | 3.7±0.1* | |
| L-ALKb-RNAi-3 | 46.5±0.6** | 4.3±0.1* | 3.7±0.1* | |
| 珍汕97B ZS97B | 54.0±0.8 | 5.1±0.0 | 4.7±0.1 | |
| Z-ALKc-RNAi-2-1 | 45.8±0.5** | 3.0±1.4** | 3.4±0.0** | |
| Z-ALKc-RNAi-2-2 | 44.8±1.7** | 3.4±0.2* | 3.1±0.4** | |
| Z-ALKc-RNAi-2-3 | 47.8±2.1** | 4.0±0.2 | 3.6±0.3** |
表4 ALK-RNAi转基因水稻株系稻米食味值
Table 4 Sensory evaluation of milled rice in ALK-RNAi transgenic rice lines.
| 样品 Sample | 转基因系 Transgenic line | 综合 Integrated score | 外观 Appearance | 口感 Taste |
|---|---|---|---|---|
| 热饭 Cooked rice | 龙特甫B LTFB | 53.3±1.3 | 5.0±0.1 | 4.5±0.2 |
| L-ALKb-RNAi-1 | 48.3±1.3** | 4.3±0.1** | 3.9±0.2* | |
| L-ALKb-RNAi-2 | 51.0±0.8* | 4.7±0.1* | 4.2±0.1* | |
| L-ALKb-RNAi-3 | 50.5±1.7** | 4.6±0.2* | 4.1±0.2* | |
| 珍汕97B ZS97B | 55.0±2.6 | 5.1±0.3 | 4.8±0.3 | |
| Z-ALKc-RNAi-2-1 | 43.8±1.9** | 3.6±0.3** | 3.3±0.3** | |
| Z-ALKc-RNAi-2-2 | 42.0±0.8** | 3.5±0.0** | 3.1±0.1** | |
| Z-ALKc-RNAi-2-3 | 44.5±0.6** | 3.8±0.0** | 3.4±0.0** | |
| 冷饭 Cooled rice | 龙特甫B LTFB | 49.8±0.5 | 4.6±0.0 | 3.9±0.1 |
| L-ALKb-RNAi-1 | 44.8±0.5** | 4.1±0.1** | 3.6±0.1** | |
| L-ALKb-RNAi-2 | 48.5±1.0* | 4.3±0.1* | 3.7±0.1* | |
| L-ALKb-RNAi-3 | 46.5±0.6** | 4.3±0.1* | 3.7±0.1* | |
| 珍汕97B ZS97B | 54.0±0.8 | 5.1±0.0 | 4.7±0.1 | |
| Z-ALKc-RNAi-2-1 | 45.8±0.5** | 3.0±1.4** | 3.4±0.0** | |
| Z-ALKc-RNAi-2-2 | 44.8±1.7** | 3.4±0.2* | 3.1±0.4** | |
| Z-ALKc-RNAi-2-3 | 47.8±2.1** | 4.0±0.2 | 3.6±0.3** |
| [1] | 陈静. 江苏省水稻食味改良育种研究进展. 江苏农业科学, 2015, 43(12): 77-80. |
| Chen J.A review of the research on the improvement of rice taste in Jiangsu Province.Jiangsu Agric Sci, 2015, 43(12): 77-80. (in Chinese) | |
| [2] | Tian Z X, Qian Q, Liu Q Q, Yan M X, Liu X F, Yan C J, Liu G F, Gao Z Y, Tang S Z, Zeng D L, Wang Y H, Yu J M, Gu M H, Li J Y.Allelic diversities in rice starch biosynthesis lead to diverse array of rice eating and cooking qualities.Proc Natl Acad Sci USA, 2009, 106(51): 21 760-21 765. |
| [3] | Xu C W, Mo H D.Qualitative-quantitative analysis for inheritance of gelatinization temperature in indica rice(O. sativa subsp. indica). Acta Agron Sin, 1996, 22(4): 386-389. |
| [4] | Khush G S, Paule C M, De La Cruz, N M. Rice grain quality evaluation and improvement at IRRI//The Workshop on Chemical Aspects of Rice Grain Quality, 1978. |
| [5] | Chun A, Lee H J, Hamaker B R, Janaswamy S.Effects of ripening temperature on starch structure and gelatinization, pasting, and cooking properties in rice (Oryza sativa). J Agric Food Chem, 2015, 63(12): 3085-3093. |
| [6] | Nakamura Y, Francisco J P, Hosaka Y, Sato A, Sawada T, Kubo A, Fujita N.Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol, 2005, 58(2): 213-227. |
| [7] | Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y.Mapping of a gene responsible for the difference in amylopectin structure betweenjaponica-type and indica-type rice varieties. Theor Appl Genet, 2002, 104(1): 1-8. |
| [8] | Gao Z Y, Zeng D L, Cui X, Zhou Y H, Yan M X, Huang D N, Li J Y, Qian Q.Map-based cloning of theALK gene, which controls the gelatinization temperature of rice. Sci China C Life Sci, 2003, 46(6): 661-668. |
| [9] | Bao J S, Sun M, Zhu L H, Corke H.Analysis of quantitative trait loci for some starch properties of rice (Oryza sativa L.): Thermal properties, gel texture and swelling volume. J Cereal Sci, 2004, 39(3): 379-385. |
| [10] | Fan C C, Yu X Q, Xing Y Z, Xu C G, Luo L J, Zhang Q F.The main effects, epistatic effects and environmental interactions of QTLs on the cooking and eating quality of rice in a doubled-haploid line population.Theor Appl Genet, 2005, 110(8): 1445-1452. |
| [11] | Ebadi A A, Farshadfar E, Rabiei B.Mapping QTLs controlling cooking and eating quality indicators of Iranian rice using RILs across three years.Aust J Crop Sci, 2013, 7(10): 1494-1502. |
| [12] | Zhao W G, Chung J W, Kwon S W, Lee J H, Ma K H, Park Y J.Association analysis of physicochemical traits on eating quality in rice (Oryza sativa L.). Euphytica, 2013, 191(1): 9-21. |
| [13] | Gao Z Y, Zeng D L, Cheng F M, Tian Z X, Guo L B, Su Y, Yan M X, Jiang H, Dong G J, Huang Y C, Han B, Li J Y, Qian Q.ALK, the key gene for gelatinization temperature,is a modifier gene for gel consistency in rice. J Integr Plant Biol, 2011, 53(9): 756-765. |
| [14] | Nakamura Y, Francisco P Y, Sato A, Sawada T, Kubo A, Fujita N.Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality betweenjaponica and indica rice varieties. Plant Mol Biol, 2005, 58(2): 213-227. |
| [15] | Cuevas R P, Daygon V D, Corpuz H, Nora L, Reinke R, Waters D, Fitzgerald M.Melting the secrets of gelatinisation temperature in rice.Funct Plant Biol, 2010, 37(5): 439-447. |
| [16] | Bao J S, Peng X, Hiratsuka M, Sun M, Umemoto T.Granule-bound SSIIa protein content and its relationship with amylopectin structure and gelatinization temperature of rice starch.Starch-Stärke, 2010, 61(8): 431-437. |
| [17] | Kharabianmasouleh A, Waters D L E, Reinke R F, Ward R, Henry R J. SNP in starch biosynthesis genes associated with nutritional and functional properties of rice.Sci Rep, 2012, 2: 557. |
| [18] | Waters D L E, Henry R J, Reinke R F, Fitzgerald M A. Gelatinization temperature of rice explained by polymorphisms in starch synthase.Plant Biotechnol J, 2010, 4(1): 115-122. |
| [19] | Chen M H, Bergman C J, Fjellstrom R G.SSSIIA locus genetic variation associated with alkali spreading value in international rice germplasm. Proceedings of the Plant and Animal Genome Conference, 2003, 153. |
| [20] | Bao J S, Corke H, Sun M.Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor Appl Genet, 2006, 113(7): 1171-1183. |
| [21] | Zhou Y, Zheng H Y, Wei G C, Zhou H, Han Y N, Bai X F, Xing Y Z, Han Y P.Nucleotide Diversity and Molecular Evolution of the ALK Gene in Cultivated Rice and its Wild Relatives. Plant Mol Biol Rep, 2016, 34(5): 1-8. |
| [22] | 陈秀花, 刘巧泉, 王宗阳, 王兴稳, 蔡秀玲, 张景六, 顾铭洪. 反义Wx基因导入我国籼型杂交稻重点亲本. 科学通报, 2002, 47(9): 684-688. |
| Chen X H, Liu Q Q, Wang Z Y, Wang X W, Cai X L, Zhang J L, Gu M H.Introduction of antisense Wx Gene into key parents of indica hybrid rice in China. Chin Sci Bull, 2002, 47(9): 684-688. (in Chinese) | |
| [23] | Murray M G, Thompson W F.Rapid isolation of high molecular weight plant DNA.Nucl Acids Res, 1980, 8(19): 4321-4325. |
| [24] | Zhang C Q, Zhou L H, Zhu Z B, Lu H W, Zhou X Z, Qian Y T, Li Q F, Lu Y, Gu M H, Liu Q Q.Characterization of grain quality and starch fine structure of two japonica rice(Oryza sativa) cultivars with good sensory properties at different temperatures during the filling stage. J Agric Food Chem, 2016, 64(20): 4048-4057. |
| [25] | Zhu L J, Meng X L, Shi Y C, Gu M H, Liu Q Q.High-amylose rice improves indices of animal health in normal and diabetic rats.Plant Biotechnol J, 2012, 10(3): 353-362. |
| [26] | Umemoto T, Horibata T, Aoki N, Hiratsuka M, Yano M, Inouchi N.Effects of variations in starch synthase on starch properties and eating quality of rice.Plant Prod Sci, 2008, 11(4): 472-480. |
| [27] | Miura S, Crofts N, Saito Y, Hosaka Y, Oitome N F, Watanabe T, Kumamaru T, Fujita N.Starch Synthase IIa-deficient mutant rice line produces endosperm starch with lower gelatinization temperature than japonica rice cultivars. Front Plant Sci, 2018, 9: 1-10. |
| [28] | Umemoto T, Aoki N, Lin H, Nakamura Y, Inouchi N, Sato Y.Natural variation in rice starch synthase IIa affects enzyme and starch properties.Funct Plant Biol, 2004, 31(7): 671-684. |
| [29] | Kaw R N, Cruz N M D L. Genetic analysis of amylose content, gelatinization temperature and gel consistency in rice.J Genet Breed, 1990: 103-111. |
| [30] | 李晓方, 肖昕, 刘彦卓, 刘志霞, 卢东柏, 毛兴学, 杨俊, 方正武, 李志新. 籼稻稻米品质性状遗传特点新解析. 分子植物育种, 2009, 7(6): 1077-1083. |
| Li X F, Xiao X, Liu Y Z, Liu Z X, Lu D B, Mao X X, Yang J, Fang Z W, Li Z X.Novel anlysis on genetic characters of quality traits in indica rice.Mol Plant Breed, 2009, 7(6): 1077-1083. (in Chinese with English abstract) | |
| [31] | 隋炯明, 李欣, 严松, 严长杰, 张蓉, 汤述翥, 陆驹飞, 陈宗祥, 顾铭洪. 稻米淀粉RVA谱特征与品质性状相关性研究. 中国农业科学, 2005, 38(4): 657-663. |
| Sui J M, Li X, Yan S, Yan C J, Zhang R, Tang S Z, Lu J F, Chen Z X, Gu M H.Studies on the rice RVA profile characteristics and its correlation with the quality.Sci Agric Sin, 2005, 38(4): 657-663. (in Chinese with English abstract) | |
| [32] | 曹清明, 钟海雁, 李忠海, 孙昌波. 蕨根淀粉糊化温度测定及影响因素研究. 食品与机械, 2007, 23(3): 16-19. |
| Cao Q M, Zhong H Y, Li Z H, Sun C B.Studies on the rice RVA profile characteristics and its correlation with the quality. Food & Mach, 2007, 23(3): 16-19. (in Chinese with English abstract) | |
| [33] | 张昌泉, 潘立旭, 周兴忠, 李钱峰, 刘巧泉. 不同Wx等位组合对杂交稻天优3611品质的影响 . 中国科技论文在线, 2007. |
| Zhang C Q, Pan L X, Zhou X Z, Li Q F, Liu Q Q.The combined effects of different Wx allele on grain quality of hybrid rice Tianyou 3611. Chin Sci Paper,2007. | |
| [34] | 刘鑫燕, 朱孔志, 张昌泉, 洪燃, 孙鹏, 汤述翥, 顾铭洪, 刘巧泉. 利用9311来源的粳型染色体片段代换系定位控制稻米糊化温度的微效QTL. 作物学报, 2014, 40(10): 1740-1747. |
| Liu X Y, Zhu K Z, Zhang C Q, Hong R, Sun P, Tang S Z, Gu M H, Liu Q Q.Mapping of minor QTLs for rice gelatinization temperature using chromosome segment substitution lines from indica 9311 in the japonica background. Acta Agron Sin, 2014, 40(10): 1740-1747. (in Chinese with English abstract) |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||