
中国水稻科学 ›› 2015, Vol. 29 ›› Issue (5): 457-466.DOI: 10.3969/j.issn.1001G7216.2015.05.002
孙廉平1,2, 张迎信2, 张沛沛2, 杨正福2, 占小登2, 沈希宏2, 张振华2, 胡霞2, 轩丹丹2, 吴玮勋2, 曹立勇2,*( ), 程式华1,2,*(
), 程式华1,2,*( )
)
收稿日期:2015-02-04
修回日期:2015-05-06
出版日期:2015-09-10
发布日期:2015-09-10
通讯作者:
曹立勇,程式华
作者简介:*通讯录作者:E-mail:caoliyong@cass.cn;shcheng@mail.hz.zj.cn
基金资助:
Lian-ping SUN1,2, Ying-xin ZHANG2, Pei-pei ZHANG2, Zheng-fu YANG2, Xiao-deng ZHAN2, Xi-hong SHEN2, Zhen-hua ZHANG2, Xia HU2, Dan-dan XUAN2, Wei-xun WU2, Li-yong CAO2,*( ), Shi-hua CHENG1,2,*(
), Shi-hua CHENG1,2,*( )
)
Received:2015-02-04
Revised:2015-05-06
Online:2015-09-10
Published:2015-09-10
Contact:
Li-yong CAO, Shi-hua CHENG
About author:*Corresponding author:E-mailcaoliyong@cass.cn;shcheng@mail.hz.zj.cn
摘要:
在60Co-γ射线辐射诱变籼稻中恢8015的突变体库内发现了一个花器官发育突变体,暂命名为开颖不育突变体ohms1(open hull and male sterile 1)。ohms1突变体表现为颖花开裂,在雄蕊和柱头之间形成类似内外稃的结构,使得突变体的颖花形成类似“三齿稃”状的三个颖壳,小穗完全不育,花粉育性为60%~70%,但自交不结实。遗传分析和基因定位结果表明,ohms1受一对隐性单基因控制,位于第3染色体短臂KY2和KY29标记之间,物理距离约42 kb,该区域包含4个开放阅读框ORFs。进一步序列分析发现,突变体中一个编码MADS盒的基因LOC_Os03g11614的第5内含子末位碱基由A突变为G。酶切实验和cDNA测序证实,该基因的第5内含子未被剪切,致使该基因的第6外显子所编码的14个氨基酸完整缺失,但并未造成该蛋白MADS结构域的改变或移码。qRT-PCR结果显示,突变体中OsMADS1 基因的表达水平显著降低,水稻开花调控因子和内外稃发育调控基因的表达量也发生了显著变化。说明该基因对水稻花器官发育尤其是内外稃发育和小花原基的分化具有重要作用。
孙廉平, 张迎信, 张沛沛, 杨正福, 占小登, 沈希宏, 张振华, 胡霞, 轩丹丹, 吴玮勋, 曹立勇, 程式华. 一个由可变剪接造成的水稻开颖不育突变体ohms1 的鉴定及基因定位[J]. 中国水稻科学, 2015, 29(5): 457-466.
Lian-ping SUN, Ying-xin ZHANG, Pei-pei ZHANG, Zheng-fu YANG, Xiao-deng ZHAN, Xi-hong SHEN, Zhen-hua ZHANG, Xia HU, Dan-dan XUAN, Wei-xun WU, Li-yong CAO, Shi-hua CHENG. Characterization and Gene Mapping of an Open Hull Male Sterile Mutant ohms1 Caused by Alternative Splicing in Rice[J]. Chinese Journal OF Rice Science, 2015, 29(5): 457-466.
| 引物名称 Primer name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') | 实验目的 Purpose |
|---|---|---|---|
| InD44 | GGAATCCCTCCCTTCTTGTC | GGTCGGTAAAGACGGTGAAA | Fine mapping |
| KY2 | GTGGGAAGAAGAACATCAACTG | GCACACAAGATAAACCCAATCAGC | Fine mapping |
| KY12 | ACCACGAGGGTGACCGTAGA | GCGAGGGTTGATGAGATAGCA | Fine mapping |
| KY17 | CGAGAGGCGAAGGAAATAGAACG | CTCCTCCTCCTCCTGGTTCTCC | Fine mapping |
| KY25 | CCATGGTCGCCATTGACACG | CCTGCTATAACACTCGCACAGATGC | Fine mapping |
| KY26 | GGTGGTGAGCCAAGAACTGACC | CCTCAAGGAATCCTCGTAAGTCG | Fine mapping |
| KY29 | CCAAGTGTGTCCGAGCTTAGTGC | TGAGTCAAAGCGAAAGTCAACAGG | Fine mapping |
| RM7576 | CTGCCCTGCCTTTTGTACAC | GCGAGCATTCTTTCTTCCAC | Fine mapping |
| InD45 | CCAGGGATCTTCTCATCCAA | CCTGGCTAGCATACCACACA | Fine mapping |
| InD46 | GCCATTGATCTTCTGCAGGT | TTTGTTGTCAATGCCCTGTT | Fine mapping |
| RD0304 | GGCGTCACTGCTCGTA | GCCTGAAGCGTCCACA | Fine mapping |
| CAPS1 | GCCATCGATCACCCTGAAAGTC | CTGATCAGCAAGAACAGTGC | ohms1 site detection |
| CKY | AGCCAAACCACACCACCATAAAG | AGGACACTGTTTGCATTGGCT | cDNA sequencing |
表1 本研究所用的引物
Table 1 Primers used in this study.
| 引物名称 Primer name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') | 实验目的 Purpose |
|---|---|---|---|
| InD44 | GGAATCCCTCCCTTCTTGTC | GGTCGGTAAAGACGGTGAAA | Fine mapping |
| KY2 | GTGGGAAGAAGAACATCAACTG | GCACACAAGATAAACCCAATCAGC | Fine mapping |
| KY12 | ACCACGAGGGTGACCGTAGA | GCGAGGGTTGATGAGATAGCA | Fine mapping |
| KY17 | CGAGAGGCGAAGGAAATAGAACG | CTCCTCCTCCTCCTGGTTCTCC | Fine mapping |
| KY25 | CCATGGTCGCCATTGACACG | CCTGCTATAACACTCGCACAGATGC | Fine mapping |
| KY26 | GGTGGTGAGCCAAGAACTGACC | CCTCAAGGAATCCTCGTAAGTCG | Fine mapping |
| KY29 | CCAAGTGTGTCCGAGCTTAGTGC | TGAGTCAAAGCGAAAGTCAACAGG | Fine mapping |
| RM7576 | CTGCCCTGCCTTTTGTACAC | GCGAGCATTCTTTCTTCCAC | Fine mapping |
| InD45 | CCAGGGATCTTCTCATCCAA | CCTGGCTAGCATACCACACA | Fine mapping |
| InD46 | GCCATTGATCTTCTGCAGGT | TTTGTTGTCAATGCCCTGTT | Fine mapping |
| RD0304 | GGCGTCACTGCTCGTA | GCCTGAAGCGTCCACA | Fine mapping |
| CAPS1 | GCCATCGATCACCCTGAAAGTC | CTGATCAGCAAGAACAGTGC | ohms1 site detection |
| CKY | AGCCAAACCACACCACCATAAAG | AGGACACTGTTTGCATTGGCT | cDNA sequencing |

图1 中恢8015野生型(左)与突变体(右)在抽穗期的植株及花器官发育形态
Fig. 1. Phenotype of plant and floral organ of wild type Zhonghui 8015(left)and ohms1 mutant(right)at heading stage.
| 组合 Combination | F1结实率 Seed-setting rate of F1/% | F2 | |||
|---|---|---|---|---|---|
| 野生型植株数 No. of wild-type plants | 突变表型植株数 No. of mutant-type plants | ||||
| ohms1/中花11 ohms1/Zhonghua 11 | 85.22 | 374 | 113 | 0.84 | 3.84 |
| ohms1/02428 | 88.76 | 4658 | 1496 | 1.57 | |
表2 突变体基因ohms1的遗传分析
Table 2 Genetic analysis of the mutant ohms1.
| 组合 Combination | F1结实率 Seed-setting rate of F1/% | F2 | |||
|---|---|---|---|---|---|
| 野生型植株数 No. of wild-type plants | 突变表型植株数 No. of mutant-type plants | ||||
| ohms1/中花11 ohms1/Zhonghua 11 | 85.22 | 374 | 113 | 0.84 | 3.84 |
| ohms1/02428 | 88.76 | 4658 | 1496 | 1.57 | |
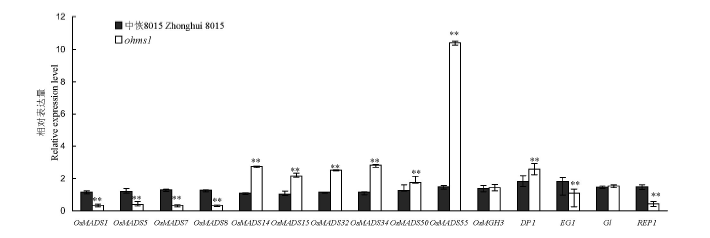
图5 实时荧光定量PCR分析野生型与突变体中水稻花器官发育相关基因的表达
Fig. 5. Expression analysis of rice floral-organ-development-associated genes between the wild-type and the mutant by qRT-PCR.
| [1] | Coen E S, Meyerowitz E M.The war of the whorls:Genetic interactions controlling flower development.Nature, 1991, 353(6339):31-37. |
| [2] | Colombo L, Franken J, Koetje E, et al.The petunia MADS box gene FBP11 determines ovule identity.Plant Cell, 1995, 7(11):1859-1868. |
| [3] | Ferrario S, Immink R G H, Shchennikova A, et al. The MADS box gene FBP2 is required for SEPALLATA function in petunia.Plant Cell, 2003, 15(4):914-925. |
| [4] | Ditta G, Pinyopich A, Robles P, et al.The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity.Curr Biol, 2004, 14(21):1935-1940. |
| [5] | Mandel M A, Yanofsky M F.The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristemsand is negatively regulated byAPETALA1. Plant Cell, 1995, 7(11):1763-1771. |
| [6] | Lohmann J U, Weigel D.Building beauty: The genetic control of floral patterning.Dev Cell, 2002, 2(2):135-142. |
| [7] | Lamb R S, Irish V F.Functional divergence with in the APETAL3/PISTILLATA floral homeotic gene lineages.Proc Natl Acad Sci USA, 2003, 100(11):6558-6563. |
| [8] | Jeon J, Lee S, Jung K H, et al.Production of transgenic rice plants show in greduced heading date and plant height by ectopic expression of rice MADS-box genes.Mol Breeding, 2000, 6(6):581-592. |
| [9] | Wang K J, Tang D, Hong L L, et al.DEP and AFO regulate reproductive habit in rice.PLoS Genet, 2010, 6(1):1-9. |
| [10] | Lu S J, Wei H, Wang Y, et al.Overexpression of a transcription factor OsMADS15 modifies plant architecture and flowering time in rice (Oryza sativa L.).Plant Mol Biol Rep, 2012, 30(6):1461-1469. |
| [11] | Fornara F, Parenicova L, Falasca G, et al.Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes.Plant Physiol, 2004, 135(4):2207-2219. |
| [12] | Kobayashi K, Yasuno N, Sato Y, et al.Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-Like MADS box genes and PAP2, a SEPALLATA MADS Box gene.Plant Cell, 2012, 24(5):1848-1859. |
| [13] | Sentoku N, Kato H, Kitano H, et al.OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy.Mol Gene Genomics, 2005, 273(1):1-9. |
| [14] | Xiao H, Wang Y, Liu D F, et al.Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference.Plant Mol Biol, 2003, 52(5):957-966. |
| [15] | Nagasawa N, MiyOshi M, Sano Y, et al. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice.Development, 2003, 130(4):705-718. |
| [16] | Arora R, Agarwal P, Ray S, et al.MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress.BMC Genom, 2007, 8:242-262. |
| [17] | Yadav S R, Prasad P, Vijayraghavan U.Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ.Genetics, 2007, 176(1):283-294. |
| [18] | Yamaguchi T, Lee D Y, Miyao A, et al.Functional diversification of the two C-class MADS box genes OsMADS3 and OsMADS58 inOryza sativa. Plant Cell, 2006, 18(1):15-28. |
| [19] | Dreni L, Pilatone A, Yun D P, et al.Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy.Plant Cell, 2011, 23(8):2850-2863. |
| [20] | Yamaguchi T, Nagasawa N, Kawasaki D, et al.The YABBY gene DROOPING LEAF regulates carpel specification and midrib development inOryza sativa. Plant Cell, 2004, 16(2):500-509. |
| [21] | Dreni L, Jacchia S, Fornara F, et al.The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice.Plant J, 2007, 52(4):690-699. |
| [22] | Yin L L, Xue H W.The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development.Plant Cell, 2012, 24(3):1049-1065. |
| [23] | Jeon J S, Jang S, Lee S, et al.An leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development.Plant Cell, 2000, 12(6):871-884. |
| [24] | Prasad K, Parameswaran S, Vijayraghavan U.OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs.Plant J, 2005, 43(6):915-928. |
| [25] | Khanday I, Yadav R S, Vijayraghavan U.Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways.Plant Physiol, 2013, 161(4):1970-1983. |
| [26] | Cui R F, Han J K, Zhao S Z, et al.Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa).Plant J, 2010, 61(5):767-781. |
| [27] | Moon S, Jung K H, Lee D E, et al.The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size.Mol Cells, 2006, 21(1):147-152. |
| [28] | Suzaki T, Ohneda M, Toriba T, et al.FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice.PLoS Genet, 2009, 5(10):1-9. |
| [29] | Chu H W, Qian Q, Liang W Q, et al.The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice.Plant Physiol, 2006, 142(3):1039-1052. |
| [30] | Xiao H, Tang J F, Li Y F, et al.STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice.Plant J, 2009, 59(5):789-801. |
| [31] | Jang S, Lee B, Kim C, et al.The OsFOR1 gene encodes a polygalacturonase-inhibiting protein (PGIP) that regulates floral organ number in rice.Plant Mol Biol, 2003, 53(3):357-372. |
| [32] | Ren D Y, Li Y F, Zhao F M, et al.MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice.Plant Physiol, 2013, 162(2):872-884. |
| [33] | Yoshida A, Suzaki T, Tanaka W, et al.The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet.Proc Natl Acad Sci USA, 2009, 106(47):20103-20108. |
| [34] | 卢扬江, 郑康乐. 提取水稻DNA 的一种简易方法. 中国水稻科学, 1992, 6(1):47-48. |
| [35] | 李云峰. 水稻小穗不确定性基因LHS1-3和雄蕊雌蕊化基因PS的图位克隆与功能分析. 重庆:西南大学. 2008. |
| [36] | Agrawal G K, Abe K, Yamazaki M, et al.Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene.Plant Mol Biol, 2005, 59(1):125-135. |
| [37] | Chen J J, Ding J H, Ouyang Y D, et al.A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice.Proc Natl Acad Sci USA, 2008, 105(32):11436-11441. |
| [38] | Yang J Y, Zhao X B, Cheng K, et al.A killer-protector system regulates both hybrid sterility and segregation distortion in rice.Science, 2012, 337(6100):1336-1340. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||