
中国水稻科学 ›› 2022, Vol. 36 ›› Issue (1): 27-34.DOI: 10.16819/j.1001-7216.2022.210302
杨晋宇1,#, 白琛2,3,#, 丁小惠2,3, 申红芳1, 王磊1, 应杰政1,*( ), 鄂志国1,*(
), 鄂志国1,*( )
)
收稿日期:2021-03-03
修回日期:2021-04-19
出版日期:2022-01-10
发布日期:2022-01-10
通讯作者:
应杰政,鄂志国
作者简介:第一联系人:#共同第一作者;
基金资助:
YANG Jinyu1,#, BAI Chen2,3,#, DING Xiaohui2,3, SHEN Hongfang1, WANG Lei1, YING Jiezheng1,*( ), E Zhiguo1,*(
), E Zhiguo1,*( )
)
Received:2021-03-03
Revised:2021-04-19
Online:2022-01-10
Published:2022-01-10
Contact:
YING Jiezheng, E Zhiguo
About author:First author contact:#These authors contributed equally to the work;
摘要:
【目的】通过对水稻雄性不育突变体的研究,可以鉴定更多与育性或花粉发育相关的基因,有助于解析水稻雄性生殖发育的整个调控网络。【方法】常规种植条件下,突变体ms7 (male sterile 7)与对照种植于浙江富阳和海南陵水,比较它们的育性及主要农艺性状差异,利用混池关联分析和图位克隆方法进行目标基因定位。【结果】整个生育期,突变体ms7生长速率与野生型一致,成熟期的株高、分蘖数、叶数、叶大小、穗长和每穗颖花数等性状与野生型相比也没有显著差异,但ms7结实率为0,表现为完全雄性不育,花药瘦小且颜色发白,半薄切片显示绒毡层降解推迟,花粉镜检呈染败。遗传分析表明花粉败育受单个隐性基因控制,定位于第7染色体上BSA11与YD7045之间1.17 Mb的范围内。【结论】本研究为水稻雄性不育基因ms7的克隆和功能研究打下了基础。
杨晋宇, 白琛, 丁小惠, 申红芳, 王磊, 应杰政, 鄂志国. 水稻雄性核不育突变体ms7的遗传分析及基因定位[J]. 中国水稻科学, 2022, 36(1): 27-34.
YANG Jinyu, BAI Chen, DING Xiaohui, SHEN Hongfang, WANG Lei, YING Jiezheng, E Zhiguo. Genetic Analysis and Gene Mapping of a Male Sterile Mutant ms7 in Rice[J]. Chinese Journal OF Rice Science, 2022, 36(1): 27-34.

图1 突变体ms7及其野生型中恢161的表型 A―灌浆期的植株形态;B―ms7和野生型的结实情况;C―ms7和野生型的颖花形态;D―ms7和野生型的花粉碘化钾染色。
Fig. 1. Phenotype of ms7 and its wild type Zhonghui 161. A, ms7 and its wild type Zhonghui 161 (WT) at grain filling stage; B, Panicle of ms7 and WT at the harvest stage; C, Spikelet of ms7 and WT; D, I2-KI staining of ms7 and WT pollens.
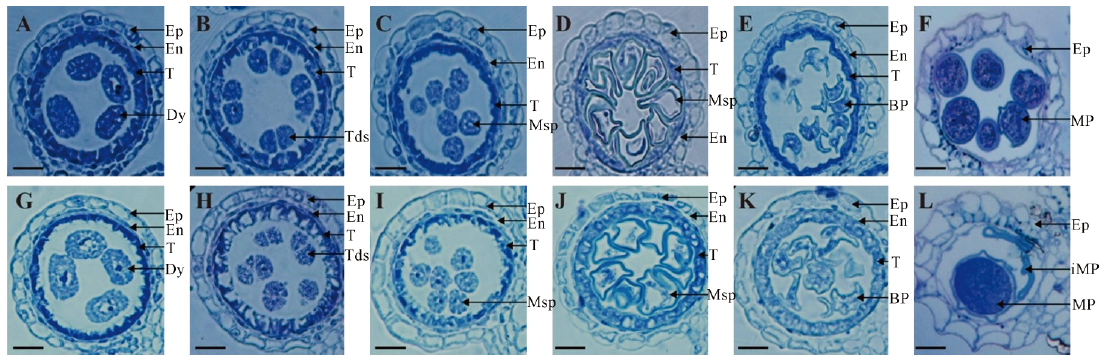
图2 野生型中恢161和突变体ms7不同发育时期花药半薄切片观察 A~F为野生型花药在8a至12时期的切片;G~L为ms7突变体花药在8a至12时期的切片。 Ep―表皮;En―内壁;T―绒毡层;Dy―二分体;Tds―四分体;Msp―小孢子;BP―二胞花粉;MP―成熟花粉粒;iMP―未成熟花粉粒。标尺为20 μm。
Fig. 2. Contrast of slice of anther between the ms7 mutant and its wild type. A-F, Wild-type anthers from stages 8a to 12; G-L, ms7 anthers from stages 8a to 12. Ep, Epidermis; En, Endothecium; T, Tapetum; Dy, Dyad cell; Tds, Tetrads; Msp, Microspore; BP, Biceullar pollen; MP, Mature pollen; iMP, Immature pollen. Bar=20 μm.
| 群体Group | 可育植株数No. of fertile plants | 不育植株数 No. of sterile plants | 卡方值 χ2 |
|---|---|---|---|
| F2-32 | 240 | 72 | 0.6154 |
| F2-33 | 232 | 73 | 0.1847 |
| F2-34 | 313 | 106 | 0.0199 |
| F2-35 | 273 | 76 | 1.9341 |
表1 ms7中雄性不育性状的遗传分析
Table 1 Genetic analysis of the male-sterile trait in ms7.
| 群体Group | 可育植株数No. of fertile plants | 不育植株数 No. of sterile plants | 卡方值 χ2 |
|---|---|---|---|
| F2-32 | 240 | 72 | 0.6154 |
| F2-33 | 232 | 73 | 0.1847 |
| F2-34 | 313 | 106 | 0.0199 |
| F2-35 | 273 | 76 | 1.9341 |
| 样品 Sample | SNP位点数 SNP number | 转换的SNP数 Transition number | 颠换的SNP数 Transversion number | Ti/Tv | 杂合SNP数 Heterozygosity number | 纯合SNP数 Homozygosity number |
|---|---|---|---|---|---|---|
| 16F4Mix | 3 383 944 | 2 408 546 | 969 917 | 2.48 | 2 098 068 | 1 285 876 |
| 16S4 | 3 383 383 | 2 408 846 | 968 983 | 2.49 | 2 069 857 | 1 313 526 |
| IR9667 | 2 714 614 | 1 929 831 | 781 109 | 2.47 | 1 517 729 | 1 196 885 |
| ms7 | 3 014 721 | 2 147 993 | 863 049 | 2.49 | 549 661 | 2 465 060 |
表2 SNP数据统计表
Table 2. Statistics of SNP data.
| 样品 Sample | SNP位点数 SNP number | 转换的SNP数 Transition number | 颠换的SNP数 Transversion number | Ti/Tv | 杂合SNP数 Heterozygosity number | 纯合SNP数 Homozygosity number |
|---|---|---|---|---|---|---|
| 16F4Mix | 3 383 944 | 2 408 546 | 969 917 | 2.48 | 2 098 068 | 1 285 876 |
| 16S4 | 3 383 383 | 2 408 846 | 968 983 | 2.49 | 2 069 857 | 1 313 526 |
| IR9667 | 2 714 614 | 1 929 831 | 781 109 | 2.47 | 1 517 729 | 1 196 885 |
| ms7 | 3 014 721 | 2 147 993 | 863 049 | 2.49 | 549 661 | 2 465 060 |
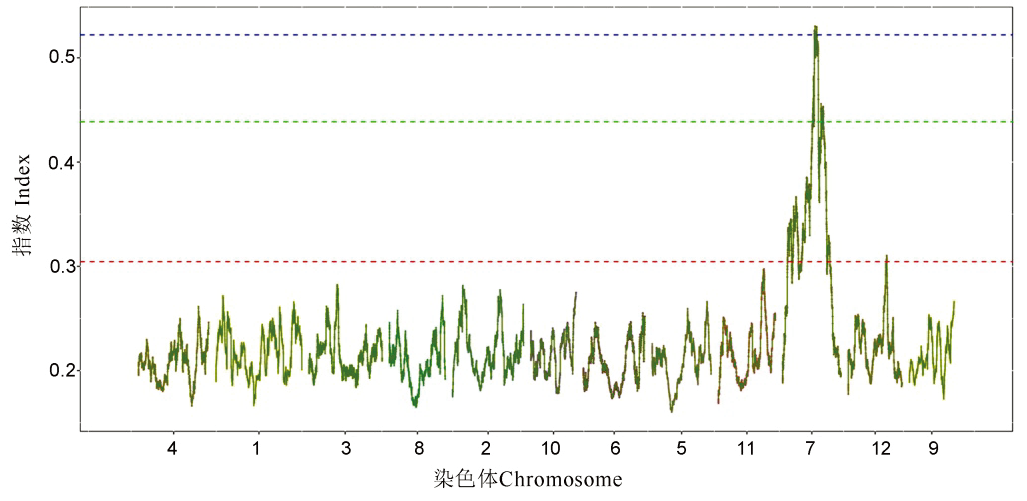
图3 SNP指数在染色体上的分布 曲线是变异位点在染色体上所对应的Index值;红、绿和蓝三根线分别是95%、99%和99.9%置信区间阈值线。
Fig. 3. Distribution of SNP-index on the chromosomes. The curve is the index value corresponding to the variation site on the chromosomes; the confidence interval thresholds from bottom to top are 95%, 99% and 99.9%, respectively.
| 引物名称 Primer name | 差异位点 Position | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|---|
| YD7036 | 16 913 833 | CCAGGGGATCTAAACAGG | TTGGACCATCACATAAGC |
| BSA11 | 16 969 331 | TGCCAATGTCGATTCCAAGAG | TCCAGCCTCTGTATACTTGCA |
| BSA6 | 17 002 663 | CCCTCACCTTCTGCAAGCTA | CGGTCTTTGAGGAGGTATCGA |
| BSA36 | 17 512 757 | AACAGAAAGCAACCAACCGC | TTCTTGGAGGTCAGGCAAGA |
| YD7013 | 17 777 239 | TGGTCTGCACTGCCTTAA | GGATGGACGGATGGGATA |
| YD7017 | 17 995 826 | ATCTTTCCCGATTGAGCT | CAGGCTTCATTCAAGACAA |
| YD7045 | 18 137 137 | AGGCTAAGGCGAGGAGAA | GCATACCAGCAAGGACGA |
| BSA47 | 18 400 602 | TTCATTCTTCTACCTTTGCCATCGG | TCTCTCTCACACTGGTCTCCTCCTC |
表3 8个InDel标记的引物信息
Table 3. Sequences and physical position of the eight InDel markers.
| 引物名称 Primer name | 差异位点 Position | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|---|
| YD7036 | 16 913 833 | CCAGGGGATCTAAACAGG | TTGGACCATCACATAAGC |
| BSA11 | 16 969 331 | TGCCAATGTCGATTCCAAGAG | TCCAGCCTCTGTATACTTGCA |
| BSA6 | 17 002 663 | CCCTCACCTTCTGCAAGCTA | CGGTCTTTGAGGAGGTATCGA |
| BSA36 | 17 512 757 | AACAGAAAGCAACCAACCGC | TTCTTGGAGGTCAGGCAAGA |
| YD7013 | 17 777 239 | TGGTCTGCACTGCCTTAA | GGATGGACGGATGGGATA |
| YD7017 | 17 995 826 | ATCTTTCCCGATTGAGCT | CAGGCTTCATTCAAGACAA |
| YD7045 | 18 137 137 | AGGCTAAGGCGAGGAGAA | GCATACCAGCAAGGACGA |
| BSA47 | 18 400 602 | TTCATTCTTCTACCTTTGCCATCGG | TCTCTCTCACACTGGTCTCCTCCTC |
| [1] | 鄂志国, 程本义, 孙红伟, 汪玉军, 朱练峰, 林海, 王磊, 童汉华, 陈红旗. 近40年我国水稻育成品种分析[J]. 中国水稻科学, 2019,33(6):523-531. |
| E Z G, Cheng B, Sun H, Wang Y, Zhu L, Lin Hai, Wang L, Tong H, Chen H. Analysis on Chinese improved rice varieties in recent four decades[J]. Chinese Journal of Rice Science, 2019,33(6):523-531. (in Chinese with English abstract) | |
| [2] | 范优荣, 曹晓风, 张启发. 光温敏雄性不育水稻的研究进展[J]. 科学通报, 2016,61(35):3822-3832. |
| Fan Y R, Cao X F, Zhang Q F. Progress on photoperiod thermo-sensitive genic male sterile rice[J]. Chinese Science Bulletin, 2016,61(35):3822-3832. (in Chinese with English abstract) | |
| [3] | Fan Y, Yang J, Mathioni S M, Yu J, Shen J, Yang X, Wang L, Zhang Q, Cai Z, Xu C, Li X, Xiao J, Meyers B C, Zhang Q,. PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016,113(52):15144. |
| [4] | Zhou H, Liu Q, Li J, Jiang D, Zhou L, Wu P, Lu S, Li F, Zhu L, Liu Z, Chen L, Liu Y G, Zhuang C. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA[J]. Cell Research, 2012,22(4):649-660. |
| [5] | Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice[J]. Proceedings of the National Academy of Sciences, 2012,109(7):2654-2659 |
| [6] | Zhou H, Zhou M, Yang Y, Li J, Zhu L, Jiang D, Dong J, Liu Q, Gu L, Zhou L, Feng M, Qin P, Hu X, Song C, Shi J, Song X, Ni E, Wu X, Deng Q, Liu Z, Chen M, Liu Y G, Cao X, Zhuang C. RNase Z S1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice [J]. Nature Communications, 2014,5:4884. |
| [7] | Pitnjam K, Chakhonkaen S, Toojinda T, Muangprom A. Identification of a deletion in tms2 and development of gene-based markers for selection[J]. Planta, 2008,228(5):813-822. |
| [8] | Yu B, Liu L, Wang T. Deficiency of very long chain alkanes biosynjournal causes humidity-sensitive male sterility via affecting pollen adhesion and hydration in rice[J]. Plant, Cell & Environment, 2019,42(12):3340. |
| [9] | Xue Z, Xu X, Zhou Y, Wang X, Zhang Y, Liu D, Zhao B, Duan L, Qi X. Deficiency of a triterpene pathway results in humidity-sensitive genic male sterility in rice[J]. Nature Communications, 2018,9:604. |
| [10] | Chen H, Zhang Z, Ni E, Lin J, Peng G, Huang J, Zhu L, Deng L, Yang F, Luo Q, Sun W, Liu Z, Zhuang C, Liu YG, Zhou H. HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynjournal and the humidity-sensitive genic male sterility in rice (Oryza sativa)[J]. New Phytologist, 2020,225(5):2077-2093. |
| [11] | 王多祥, 祝万万, 袁政, 张大兵. 水稻雄性发育功能基因的发掘及应用[J]. 生命科学, 2016,28(10):1180-1188. |
| Wang DX, Zhu WW, Yuan Z, Zhang DB. Functional research of rice male reproduction and its utilization in breeding[J]. Chinese Bulletin of Life Sciences, 2016,28(10):1180-1188. (in Chinese with English abstract) | |
| [12] | Li L, Li Y, Song S, Deng H, Li N, Fu X, Chen G, Yuan L. An anther development F-box (ADF) protein regulated by tapetum degeneration retardation (TDR) controls rice anther development[J]. Planta, 2015,241(1):157-166. |
| [13] | Lee S, Jung K H, An G, Chung Y Y. Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system[J]. Plant Molecular Biology, 2004,54(5):755-765. |
| [14] | Niu N, Liang W, Yang X, Jin W, Wilson Z A, Hu J, Zhang D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice[J]. Nature Communications, 2013,4 : 1445. |
| [15] | Shi J, Tan H, Yu X H, Liu Y, Liang W, Ranathunge K, Franke R B, Schreiber L, Wang Y, Kai G, Shanklin J, Ma H, Zhang D. Defective Pollen Wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase[J]. Plant Cell, 2011,23(6):2225-2246. |
| [16] | Mondol P C, Xu D, Duan L, Shi J, Wang C, Chen X, Chen M, Hu J, Liang W, Zhang D. Defective Pollen Wall 3 (DPW3), a novel alpha integrin-like protein, is required for pollen wall formation in rice[J]. New Phytologist, 2020,225(2):807-822. |
| [17] | Yang X, Liang W, Chen M, Zhang D, Zhao X, Shi J. Rice fatty acyl-CoA synthetase OsACOS12 is required for tapetum programmed cell death and male fertility[J]. Planta, 2017,246(1):105-122. |
| [18] | Xu D, Shi J, Rautengarten C, Yang L, Qian X, Uzair M, Zhu L, Luo Q, An G, Waßmann F, Schreiber L, Heazlewood J L, Scheller H V, Hu J, Zhang D, Liang W. Defective Pollen Wall 2 (DPW2) encodes an acyl transferase required for rice pollen development[J]. Plant Physiology, 2017,173(1):240-255. |
| [19] | Men X, Shi J, Liang W, Zhang Q, Lian G, Quan S, Zhu L, Luo Z, Chen M, Zhang D. Glycerol-3-phosphate acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice[J]. Journal of Experimental Botany, 2017,68(3):513-526. |
| [20] | Yu J, Meng Z, Liang W, Behera S, Kudla J, Tucker MR, Luo Z, Chen M, Xu D, Zhao G, Wang J, Zhang S, Kim YJ, Zhang D. A rice Ca2+ binding protein is required for tapetum function and pollen formation [J]. Plant Physiology, 2016,172(3):1772-1786. |
| [21] | Zheng S, Li J, Ma L, Wang H, Zhou H, Ni E, Jiang D, Liu Z, Zhuang C. OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers[J]. Proceedings of the National Academy of Sciences of USA, 2019,116(15):7549-7558. |
| [22] | Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB[J]. Plant Cell, 2009,21(5):1453. |
| [23] | Liu Z, Bao W, Liang W, Yin J, Zhang D. Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development[J]. Journal of Integrative Plant Biology, 2010,52(7):670-678. |
| [24] | Xiang X J, Sun L P, Yu P, Yang Z F, Zhang P P, Zhang Y X, Wu W X, Chen D B, Zhan X D, Khan R M, Abbas A, Cheng S H, Cao L Y. The MYB transcription factor Baymax1 plays a critical role in rice male fertility[J]. Theoretical and Applied Genetics, 2021,134(2):453-471. |
| [25] | Jung K H, Han M J, Lee Y S, Kim Y W, Hwang I, Kim M J, Kim Y K, Nahm B H, An G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development[J]. Plant Cell, 2005,17(10):2705-2722. |
| [26] | Ko S S, Li M J, Ku M S B, Ho Y C, Lin Y J, Chuang M H, Hsing H X, Lien Y C, Yang H T, Chang H C, Chan M T. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice[J]. Plant Cell, 2014,26(6):2486-2504. |
| [27] | Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W. The rice basic Helix-Loop-Helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell, 2014,26(4):1512-1524. |
| [28] | Li N, Zhang D S, Liu H S, Yin C S, Li X X, Liang W Q, Yuan Z, Xu B, Chu H W, Wang J, Wen T Q, Huang H, Luo D, Ma H, Zhang D B. The rice Tapetum Degeneration Retardation gene is required for tapetum degradation and anther development[J]. Plant Cell, 2006,18(11):2999-3014. |
| [29] | Ji C, Li H, Chen L, Xie M, Wang F, Chen Y, Liu YG. A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development[J]. Molecular Plant, 2013,6(5):1715-1718. |
| [30] | Cao H, Li X, Wang Z, Ding M, Sun Y, Dong F, Chen F, Liu L, Doughty J, Li Y, Liu Y X. Histone H2B monoubiquitination mediated by HISTONE MONOUBI-QUITINATION1 and HISTONE MONOUBIQUITI-NATION2 is involved in anther development by regulating tapetum degradation-related genes in rice[J]. Plant Physiology, 2015,168(4):1389. |
| [31] | Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson Z A, Zhang D. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice[J]. Plant Physiology, 2011,156(2):615-630. |
| [32] | Yang Z, Liu L, Sun L, Yu P, Zhang P, Abbas A, Xiang X, Wu W, Zhang Y, Cao L, Cheng S. OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice[J]. Plant Molecular Biology, 2019,99(1-2):175-191. |
| [33] | Yang Z, Sun L, Zhang P, Zhang Y, Yu P, Liu L, Abbas A, Xiang X, Wu W, Zhan X, Cao L, Cheng S. TDR INTERACTING PROTEIN 3, encoding a PHD-finger transcription factor, regulates Ubisch bodies and pollen wall formation in rice[J]. Plant Journal, 2019,99(5):844-861. |
| [34] | Li X, Gao X, Wei Y, Deng L, Ouyang Y, Chen G, Li X, Zhang Q, Wu C. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5’-triphosphate-dependent RNA helicases regulates tapetum degeneration[J]. Plant Cell, 2011,23(4):1416-1434. |
| [35] | Bai W, Wang P, Hong J, Kong W, Xiao Y, Yu X, Zheng H, You S, Lu J, Lei D, Wang C, Wang Q, Liu S, Liu X, Tian Y, Chen L, Jiang L, Zhao Z, Wu C, Wan J. Earlier degraded tapetum1 (EDT1) encodes an ATP-citrate lyase required for tapetum programmed cell death[J]. Plant Physiology, 2019,181(3):1223-1238. |
| [36] | Zhang P, Zhang Y, Sun L, Sinumporn S, Yang Z, Sun B, Xuan D, Li Z, Yu P, Wu W, Wang K, Cao L, Cheng S. The rice AAA-ATPase OsFIGNL1 is essential for male meiosis[J]. Frontiers in Plant Science, 2017,8:1639. |
| [37] | Tan H, Liang W, Hu J, Zhang D. MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice[J]. Developmental Cell, 2012,22(6):1127-1137. |
| [38] | Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009,25(14), 1754-1760. |
| [39] | McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo M A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome research, 2010,20(9):1297-1303. |
| [40] | Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009,25(16):2078-2079. |
| [41] | Zhang D, Luo X, Zhu L. Cytological analysis and genetic control of rice anther development[J]. Journal of Genetics and Genomics, 2011,38(9):379-390. |
| [42] | Wang C, Wang Y, Cheng Z, Zhao Z, Chen J, Sheng P, Yu Y, Ma W, Duan E, Wu F, Liu L, Qin R, Zhang X, Guo X, Wang J, Jiang L, Wan J. The role of OsMSH4 in male and female gamete development in rice meiosis[J]. Journal of Experimental Botany, 2016,67(5):1447-1459. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||