
中国水稻科学 ›› 2016, Vol. 30 ›› Issue (6): 577-586.DOI: 10.16819/j.1001-7216.2016.6024
杜彦修, 季新, 陈会杰, 彭廷, 张静, 李俊周, 孙红正, 赵全志*( )
)
收稿日期:2016-02-19
修回日期:2016-06-03
出版日期:2016-11-10
发布日期:2016-11-10
通讯作者:
赵全志
基金资助:
Yan-xiu DU, Xin JI, Hui-jie CHEN, Ting PENG, Jing ZHANG, Jun-zhou LI, Hong-zheng SUN, Quan-zhi ZHAO*( )
)
Received:2016-02-19
Revised:2016-06-03
Online:2016-11-10
Published:2016-11-10
Contact:
Quan-zhi ZHAO
摘要:
以水稻OsbHLH116 基因为编辑对象,根据基因编码区序列(CDS)在第一外显子区域设计长度为19 bp的sgRNA,化学合成sgRNA的寡核苷酸序列,然后与CRISPR/Cas9系统表达载体pBUN411连接,再用农杆菌介导法获得水稻转基因株系,最后利用酶切和测序相结合的方法对OsbHLH116 突变体进行了筛选鉴定和脱靶效应分析。结果表明,所构建的pBUN411-gRNA载体成功实现了对基因OsbHLH116 的定向编辑。酶切分析表明在选取的10株T0代转基因苗中得到了6个OsbHLH116 突变单株。对6个突变单株进行了TA克隆测序分析,发现了纯合突变、双等位突变和杂合突变3种类型。酶切分析表明2个潜在脱靶位点均未发生脱靶效应。
中图分类号:
杜彦修, 季新, 陈会杰, 彭廷, 张静, 李俊周, 孙红正, 赵全志. 基于CRISPR/Cas9系统的OsbHLH116基因编辑及其脱靶效应分析[J]. 中国水稻科学, 2016, 30(6): 577-586.
Yan-xiu DU, Xin JI, Hui-jie CHEN, Ting PENG, Jing ZHANG, Jun-zhou LI, Hong-zheng SUN, Quan-zhi ZHAO. CRISPR/Cas9 System-based Editing of OsbHLH116 Gene and Its Off-target Effect Analysis[J]. Chinese Journal OF Rice Science, 2016, 30(6): 577-586.
| 引物名称 Primer name | 序列 Sequence | 用途 Usage |
|---|---|---|
| sgRNA-F | 5'-GGCGCCTCCATCGGAGGAAGAGA-3' | 靶序列合成Construction of target site |
| sgRNA-R | 5'-AAACTCTCTTCCTCCGATGGAGG-3' | |
| pBUN411-VF | 5'-CCATGAAGCCTTTCAGGACATGTA-3' | 载体构建验证Verification of vector construction |
| pBUN411-VR | 5'-ACGCTGCAAACATGAGACGGAGAA-3' | |
| Basta-F | 5'-AAGCACGGTCAACTTCCGTA-3' | 除草剂基因验证Verification of Bt |
| Basta-R | 5'-GAAGTCCAGCTGCCAGAAAC-3' | |
| OsbHLH116-F | 5'-GTTGATGTGGCAAGGAGGAG-3' | 靶点两侧序列扩增Amplification of target region |
| OsbHLH116-R | 5'-TACGCACCAGACAGTTCACC-3' | |
| LOC_Os01g01380-F | 5'-ACAAGCAATGCAAATGTTGG-3' | 脱靶位点扩增Off-target amplification |
| LOC_Os01g01380-R | 5'-CTCTTCGCCCACACCATC-3' | |
| Chr4:+30193341-F | 5'-AATAGATCACGCCGTCAACC-3' | 脱靶位点扩增Off-target amplification |
| Chr4:+30193341-R | 5'-CGAGACGAATCTTTTGAGCA-3' |
表1 本研究所用引物序列
Table 1 Primer sequence used in the study.
| 引物名称 Primer name | 序列 Sequence | 用途 Usage |
|---|---|---|
| sgRNA-F | 5'-GGCGCCTCCATCGGAGGAAGAGA-3' | 靶序列合成Construction of target site |
| sgRNA-R | 5'-AAACTCTCTTCCTCCGATGGAGG-3' | |
| pBUN411-VF | 5'-CCATGAAGCCTTTCAGGACATGTA-3' | 载体构建验证Verification of vector construction |
| pBUN411-VR | 5'-ACGCTGCAAACATGAGACGGAGAA-3' | |
| Basta-F | 5'-AAGCACGGTCAACTTCCGTA-3' | 除草剂基因验证Verification of Bt |
| Basta-R | 5'-GAAGTCCAGCTGCCAGAAAC-3' | |
| OsbHLH116-F | 5'-GTTGATGTGGCAAGGAGGAG-3' | 靶点两侧序列扩增Amplification of target region |
| OsbHLH116-R | 5'-TACGCACCAGACAGTTCACC-3' | |
| LOC_Os01g01380-F | 5'-ACAAGCAATGCAAATGTTGG-3' | 脱靶位点扩增Off-target amplification |
| LOC_Os01g01380-R | 5'-CTCTTCGCCCACACCATC-3' | |
| Chr4:+30193341-F | 5'-AATAGATCACGCCGTCAACC-3' | 脱靶位点扩增Off-target amplification |
| Chr4:+30193341-R | 5'-CGAGACGAATCTTTTGAGCA-3' |
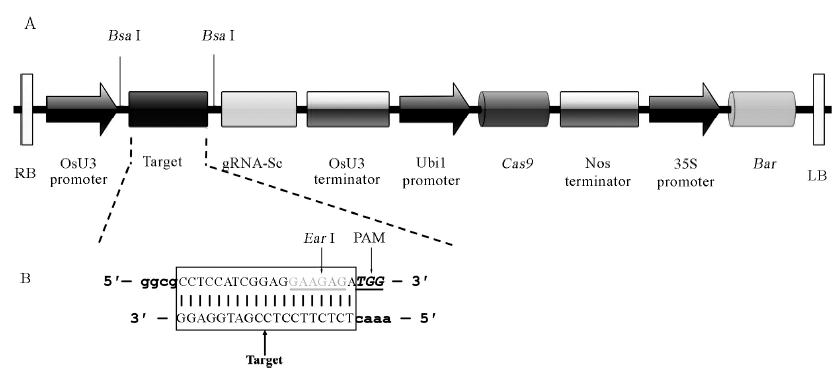
图1 pBUN411-gRNA表达载体LB和RB之间线性结构及gRNA靶序列合成 A-表达载体pBUN411-gRNA的LB和RB之间线性结构。 竖线-酶切位点Bsa Ⅰ; RB-T-DNA右边界; OsU3 promoter-水稻U3启动子;gRNA-Sc-向导RNA支架; OsU3 terminator-水稻U3终止子; Ubi1 promoter-玉米泛素基因1(Ubi1)启动子; Cas9-Cas9基因; Nos terminator-农杆菌胭脂碱合成酶基因(Nos)终止子; 35S Promoter-花椰菜花叶病毒(CaMV)35S启动子; Bar-抗除草剂基因; LB-T-DNA左边界。B-gRNA靶序列互补双链DNA。方框内为gRNA靶点序列; 灰字体为EarⅠ酶切位点; 斜体为PAM序列(不在所构建载体上);小写字体为BsaⅠ酶切后黏性互补末端。
Fig. 1. Linear structure of pBUN411-gRNA expression vectors between LB and RB and the construction of target site. A, Physical map between RB and LB of pBUN411-gRNA expression vectors. Vertical lines stand for BsaⅠ restriction sites; RB, Right border of T-DNA; OsU3 promoter, Rice U3 promoters; gRNA-Sc, Guide RNA scaffold; OsU3 terminator, Rice U3 terminators; Ubi1 promoter, Maize ubiquitin gene promoter; Cas9, Cas9 gene; Nos Terminator, Agrobacterium tumefaciens nopaline synthase gene (Nos) terminator; 35S Promoter, Cauliflower mosaic virus (CaMV) 35S promoter; Bar, Herbicide resistance Bar gene; LB, Left border of T-DNA. B, The target sites of gRNA complementary with double-stranded DNA. The target sequences are in the box; Gray letters stand for EarⅠ restriction sites; Letters with italics stand for PAM (not in the expression vectors); The lowercase stand for sticky ends of BsaⅠ.
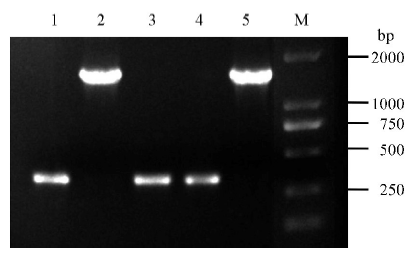
图2 pBUN411-gRNA表达载体菌落PCR 1~4-表达载体的4个菌落PCR; 5-pBUN411空载体(对照); M-DM2000 标记。
Fig. 2. Verification of pBUN411-gRNA expression vectors via colony PCR. 1-4, Four independent colonies to detect expression vectors; 5, pBUN411 empty vectors (negative contrast); M, DM2000 marker.
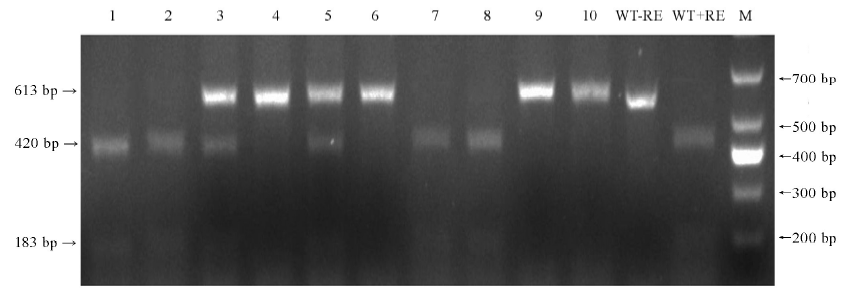
图3 10株T0代转基因植株PCR产物EarⅠ酶切分析结果 1-10—10株转基因植株; WT-野生型; M-DM1000 标记;-RE-未进行酶切;+RE-使用EarI酶切; 箭头示酶切后PCR片段。
Fig. 3. Mutation analysis of 10 T0 transgenic lines by EarⅠ digestion of PCR product. 1-10, 10 transgenic lines; WT, Wild type; M, DM1000 marker; -RE, PCR product without restriction enzyme digestion; +RE, EarⅠ digested PCR product; Arrowheads, PCR fragments after EarI digested.
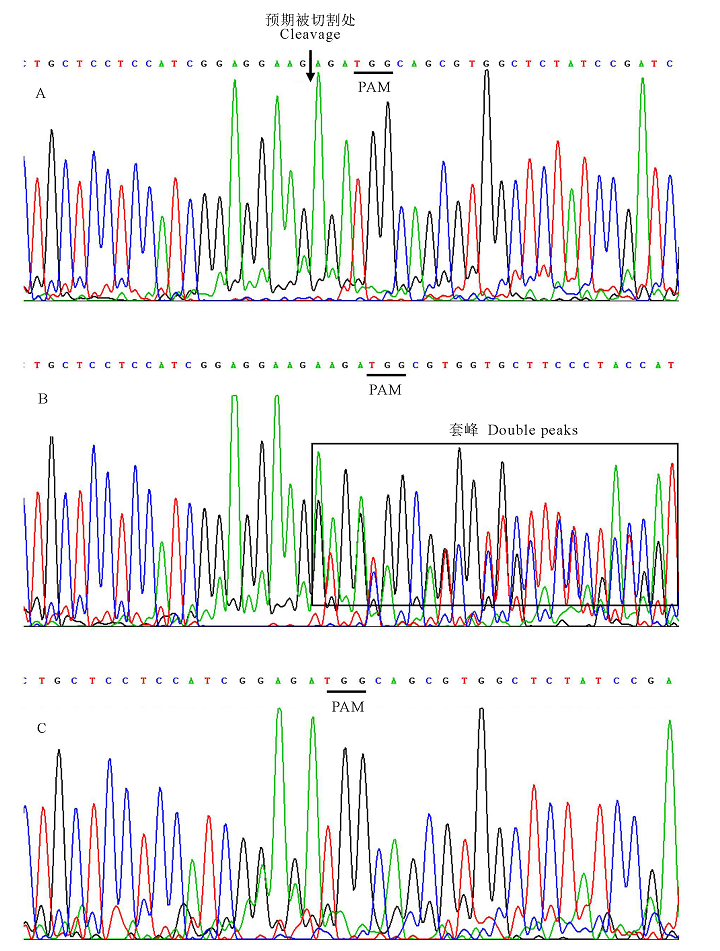
图4 4号、7号和10号单株PCR产物测序峰图 下划线-PAM序列TGG; 箭头-DNA预期被切割处; 方框—套峰。A-7号未突变单株PCR产物测序峰; B-10号双等位突变单株PCR产物测序峰; C-4号纯合突变单株PCR产物测序峰。
Fig.4. Sanger sequencing results for PCR products of number 4, 7 and 10. Underlines stand for PAM (TGG); The arrows stand for the intended cleavage site; The double peak phenomenon is in the box. A, Sanger sequencing results for PCR products of non-mutation number 7 ; B, Sanger sequencing results for PCR products of biallelic mutation number 10; C, Sanger sequencing results for PCR products of homozygous mutation number 4.

图5 野生型与6株突变体突变位点序列比对 蓝色字体-gRNA靶序列; 黄色高亮-PAM序列; 红色删除线-缺失碱基; 红色小写字体-插入碱基; -表示缺失;+表示插入;(0/0)-PCR产物测序。
Fig. 5. DNA sequence alignment of wild type with six mutant versions. The gRNA target site is shown in blue; The yellow highlighting denote PAM; Red dashes deleted bases; Insertion nucleotides are shown in red lowercases; -, Deletion; +, Insertion; (0/0)-Sequencing by using PCR product.
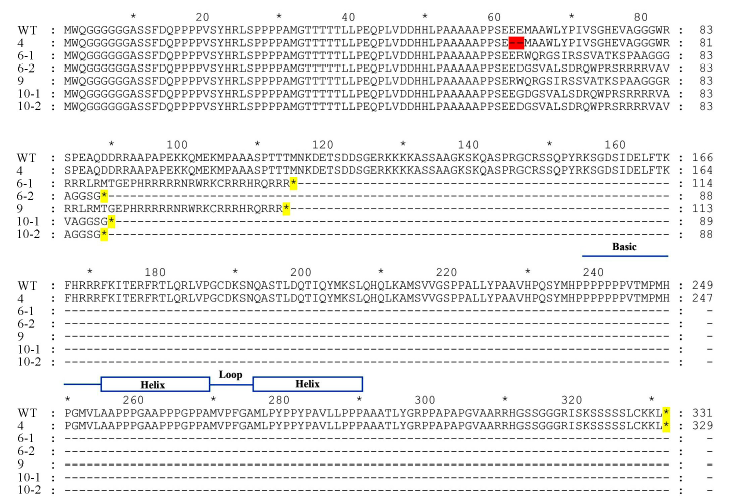
图6 野生型与株突变体氨基酸序列比对 蓝色直线和方框-碱性-螺旋-环-螺旋结构域; 红色高亮-氨基酸缺失; 黄色高亮-翻译终止。
Fig.6. Comparison of the amino acid sequences between wild type and OsbHLH116 mutants. Blue lines and boxes denote basic-helix-loop-helix domain; The red highlighting denote amino acid deletion; The yellow highlighting denote translation termination.
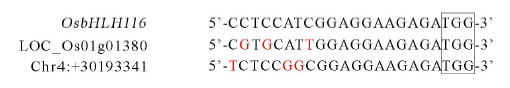
图7 推定的脱靶位点序列与靶位点序列比对分析 方框内为PAM (NGG) 序列,红色字体为与靶序列比对不匹配碱基。
Fig. 7. Sequences comparative analysis between the putative off-target sites and target sites.
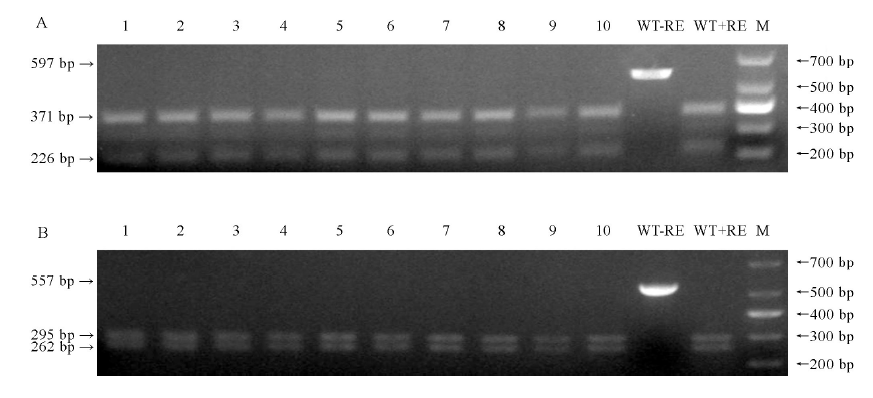
图8 10株T0代转基因植株脱靶位点酶切验证 1~10-10株转基因植株; WT-野生型; M-DM1000 标记; -RE-未进行酶切; +RE-使用EarI酶切; 箭头-酶切后PCR片段。A-基因LOC_Os01g01380脱靶位点酶切鉴定; B-Chr4:+30193341脱靶位点酶切鉴定。
Fig. 8. PCR/ restriction enzyme (PCR/RE) assay to detect off-target of 10 plants of T0 transgenic lines. 1-10, 10 transgenic lines; WT, Wild type; M, DM1000 Marker; -RE, PCR product without restriction enzyme digestion; +RE, EarI digested PCR product; Arrowheads, PCR fragments after EarI digestion. A, PCR/RE assay to detect off-target of LOC_Os01g01380; B, PCR/RE assay to detect off-target of Chr4: +30 193 341.
| [1] | Wiedenheft B, Sternberg S H, Doudna J A.RNA-guided genetic silencing systems in bacteria and archaea.Nature, 2012, 482(7385): 331-338. |
| [2] | Cong L, Ran F A, Cox D, et al.Multiplex genome engineering using CRISPR/Cas systems.Science, 2013, 339(6121): 819-823. |
| [3] | Miao J, Guo D, Zhang J, et al.Targeted mutagenesis in rice using CRISPR-Cas system.Cell Res, 2013, 23(10): 1233. |
| [4] | Shan Q, Wang Y, Li J, et al.Targeted genome modification of crop plants using a CRISPR-Cas system.Nat Biotechnol, 2013, 31(8): 686-688. |
| [5] | Atchley W R, Terhalle W, Dress A.Positional dependence, cliques, and predictive motifs in the bHLH protein domain.J Mol Evol, 1999, 48(5): 501-516. |
| [6] | Nesi N, Debeaujon I, Jond C, et al.The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell, 2000, 12(10): 1863-1878. |
| [7] | Martı'nez-Garcı'A J F, Huq E, Quail P H. Direct targeting of light signals to a promoter element-bound transcription factor.Science, 2000, 288(5467): 859-863. |
| [8] | Massari M E, Murre C.Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms.Mol Cell Biol, 2000, 20(2): 429-440. |
| [9] | Pires N, Dolan L.Origin and diversification of basic-helix-loop-helix proteins in plants.Mol Biol Evol, 2010, 27(4):862-874. |
| [10] | Komatsu K, Maekawa M, Ujiie S, et al.LAX and SPA: Major regulators of shoot branching in rice.PNAS, 2003, 100(20): 11765-11770. |
| [11] | Sakamoto W, Ohmori T, Kageyama K, et al.The Purple leaf (Pl) locus of rice: The Plw allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis.Plant Cell Physiol, 2001, 42(9): 982-991. |
| [12] | Seo J S, Joo J, Kim M J, et al.OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice.Plant J, 2011, 65(6): 907-921. |
| [13] | Xing H, Dong L, Wang Z, et al.A CRISPR/Cas9 toolkit for multiplex genome editing in plants.BMC Plant Biol, 2014, 14(1): 327. |
| [14] | Nishimura A, Aichi I, Matsuoka M.A protocol for Agrobacterium-mediated transformation in rice. Nat Protoc, 2006, 1(6): 2796-2802. |
| [15] | Hall T A.BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.Nucl Acids Sympos Ser, 1999, 41: 95-98. |
| [16] | Nicholas K B, Nicholas H, Deerfield D W.GeneDoc: Analysis and visualization of genetic variation.Embnew News, 1997, 4: 1-4. |
| [17] | Li X, Duan X, Jiang H, et al.Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol, 2006, 141(4): 1167-1184. |
| [18] | Carroll D, Morton J J, Beumer K J, et al.Design, construction and in vitro testing of zinc finger nucleases.Nat Protoc, 2006, 1(3): 1329-1341. |
| [19] | Li T, Liu B, Spalding M H, et al.High-efficiency TALEN-based gene editing produces disease-resistant rice.Nat Biotechnol, 2012, 30(5): 390-392. |
| [20] | Mussolino C, Cathomen T.RNA guides genome engineering.Nat Biotechnol, 2013, 31(3): 208-209. |
| [21] | Mao Y, Zhang H, Xu N, et al.Application of the CRISPR-Cas system for efficient genome engineering in plants.Mol Plant, 2013, 6(6): 2008. |
| [22] | Jinek M, Chylinski K, Fonfara I, et al.A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.Science, 2012, 337(6096): 816-821. |
| [23] | Ran F A, Hsu P D, Lin C, et al.Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity.Cell, 2013, 154(6): 1380-1389. |
| [24] | Zhou H, Liu B, Weeks D P, et al.Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice.Nucl Acids Res, 2014, 42(17): 10903-10914. |
| [25] | Feng Z, Mao Y, Xu N, et al.Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications inArabidopsis. Proc Natl Acad Sci, 2014, 111(12): 4632-4637. |
| [26] | Zhang H, Zhang J, Wei P, et al.The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation.Plant Biotechnol J, 2014, 12(6): 797-807. |
| [27] | Li X, Duan X, Jiang H, et al.Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol, 2006, 141(4): 1167-1184. |
| [28] | Gu X, Liu T, Feng J, et al.The qSD12 underlying gene promotes abscisic acid accumulation in early developing seeds to induce primary dormancy in rice.Plant Mol Biol, 2010, 73(1-2): 97-104. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||