
中国水稻科学 ›› 2015, Vol. 29 ›› Issue (3): 241-249.DOI: 10.3969/j.issn.1001G7216.2015.03.003
董青1,2, 张迎信1,2, 张振华1,2, 周全1,2, 秦亚芝1,2,3, 王宏1,2, 程式华1,2, 曹立勇1,2,*( ), 沈希宏1,2,*(
), 沈希宏1,2,*( )
)
收稿日期:2014-11-27
修回日期:2014-12-24
出版日期:2015-05-10
发布日期:2015-05-10
通讯作者:
曹立勇,沈希宏
基金资助:
Qing DONG1,2, Ying-xin ZHANG1,2, Zhen-hua ZHANG1,2, Quan ZHOU1,2, Ya-zhi QIN1,2,3, Hong WANG1,2, Shi-hua CHENG1,2, Li-yong CAO1,2,*( ), Xi-hong SHEN1,2,*(
), Xi-hong SHEN1,2,*( )
)
Received:2014-11-27
Revised:2014-12-24
Online:2015-05-10
Published:2015-05-10
Contact:
Li-yong CAO, Xi-hong SHEN
摘要:
通过60Co-γ辐射诱变籼稻中恢8015获得了一个在全生育期叶片均呈黄绿色的突变体w390。与野生型相比,突变体中检测不到叶绿素b的存在,且叶绿素a和类胡萝卜素的含量分别降低了50.6%和44.8%;主要农艺性状调查结果显示,突变体的株高、单株有效穗数、每穗总粒数、每穗实粒数较野生型分别降低了12.0%、22.3%、18.5%和27.6%;透射电镜结果显示:突变体的类囊体数量明显减少,基粒垛叠方向异常;遗传分析表明该突变性状受一对隐性核基因控制。利用突变体与粳稻日本晴杂交构建的F2群体,将突变基因定位至水稻第10染色体长臂约71.8 kb的区域内。对该区间包含的15个ORFs进行序列分析,发现突变体中编码叶绿素酸酯氧化酶1(chlorophyllide a oxygenase 1)的基因OsCAO1的第8外显子发生了两个单碱基突变,导致第394和396位的亮氨酸和甘氨酸分别突变为组氨酸和谷氨酸,推测该突变基因是一个OsCAO1功能丧失的新等位基因。
中图分类号:
董青, 张迎信, 张振华, 周全, 秦亚芝, 王宏, 程式华, 曹立勇, 沈希宏. 水稻黄绿叶突变体w390的遗传分析和基因定位[J]. 中国水稻科学, 2015, 29(3): 241-249.
Qing DONG, Ying-xin ZHANG, Zhen-hua ZHANG, Quan ZHOU, Ya-zhi QIN, Hong WANG, Shi-hua CHENG, Li-yong CAO, Xi-hong SHEN. Genetic Analysis and Gene Mapping of a Yellow-green Leaf Mutant w390 in Rice (Oryza sativa L.)[J]. Chinese Journal OF Rice Science, 2015, 29(3): 241-249.
| 光合色素含量 Photosynthetic pigment content | 中恢8015 R8015 | w390 | P |
|---|---|---|---|
| 叶绿素a Chlorophyll a | 2.65±0.03 | 1.34±0.05 | <0.0001 |
| 叶绿素b Chlorophyll b | 0.74±0.05 | 0.00 | <0.0001 |
| 类胡萝卜素 Carotenoids | 0.66±0.04 | 0.36±0.01 | <0.0003 |
| 总含量 Total content | 4.05±0.07 | 1.70±0.06 | <0.0001 |
表1 突变体和野生型叶片的光合色素含量比较
Table 1 Comparison of photosynthetic pigment contents in leaves between the mutant and its wild-type parent.mg/g
| 光合色素含量 Photosynthetic pigment content | 中恢8015 R8015 | w390 | P |
|---|---|---|---|
| 叶绿素a Chlorophyll a | 2.65±0.03 | 1.34±0.05 | <0.0001 |
| 叶绿素b Chlorophyll b | 0.74±0.05 | 0.00 | <0.0001 |
| 类胡萝卜素 Carotenoids | 0.66±0.04 | 0.36±0.01 | <0.0003 |
| 总含量 Total content | 4.05±0.07 | 1.70±0.06 | <0.0001 |
| 性状 Trait | 中恢8015 R8015 | w390 | P |
|---|---|---|---|
| 株高 Plant height/cm | 112.3±5.2 | 98.9±5.5 | 0.0003 |
| 单株有效穗数 No. of panicles per plant | 8.1±1.4 | 6.3±1.8 | 0.0227 |
| 每穗总粒数 No. of spikelets per panicle | 118.0±9.1 | 96.2±7.3 | <0.0001 |
| 每穗实粒数 No. of grains per panicle | 71.1±6.7 | 51.5±5.9 | <0.0001 |
| 千粒重 1000-grain weight /g | 33.5±0.4 | 31.9±0.5 | 0.3067 |
| 单株产量 Grain yield per plant /g | 19.3±2.8 | 10.3±1.4 | <0.0001 |
表2 突变体和野生型的主要农艺性状比较
Table 2 Comparison of main agronomic traits between the mutant and its wild-type parent.
| 性状 Trait | 中恢8015 R8015 | w390 | P |
|---|---|---|---|
| 株高 Plant height/cm | 112.3±5.2 | 98.9±5.5 | 0.0003 |
| 单株有效穗数 No. of panicles per plant | 8.1±1.4 | 6.3±1.8 | 0.0227 |
| 每穗总粒数 No. of spikelets per panicle | 118.0±9.1 | 96.2±7.3 | <0.0001 |
| 每穗实粒数 No. of grains per panicle | 71.1±6.7 | 51.5±5.9 | <0.0001 |
| 千粒重 1000-grain weight /g | 33.5±0.4 | 31.9±0.5 | 0.3067 |
| 单株产量 Grain yield per plant /g | 19.3±2.8 | 10.3±1.4 | <0.0001 |
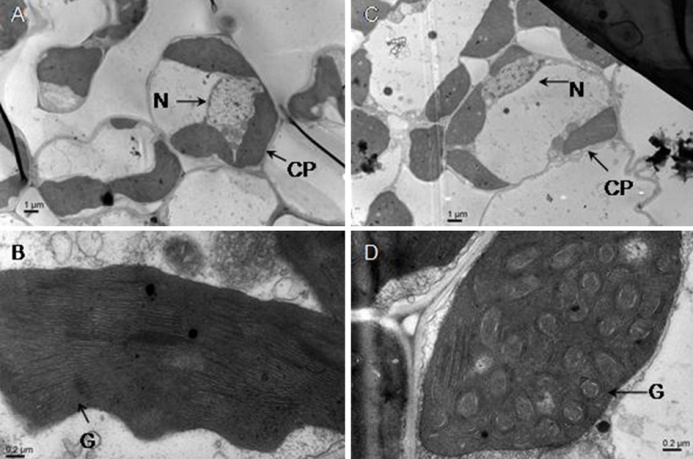
图2 野生型和突变体的叶绿体超显微结构比较(A和B-野生型的叶肉细胞和叶绿体结构; C和D-突变体的叶肉细胞和叶绿体结构; N-细胞核;CP-叶绿体; G-基粒。)
Fig. 2. Comparison of ultrastructures of chloroplasts in the mesophyll cells between the mutant and its wild-type parent.(A and B, Mesophyll cell and chloroplast of wild-type, respectively; C and D, Mesophyll cell and chloroplast of mutant, respectively. N, Nucleus; CP, Chloroplast; G, Grana.)
| 引物名称 Primer name | 上游引物 Forward primer (5'-3') | 下游引物 Reverse primer (5'-3') | 退火温度 Anealing temperature/℃ |
|---|---|---|---|
| D2 | CAGCCTCTCTAATTGACTCTC | CCAAGTAGAATGGCCAAATGT | 55 |
| D3 | GCATCAAACTATAATAACTGACT | AGGAAAGTAAACAAGGCCTTA | 55 |
| D4 | ACACATATGTAGAGTATTATCCG | GCAAGCTGTCTACACGGTT | 55 |
| D8 | AACTCTATTGTTTATGGTGG | TAATTCCGAATTTCAGCTCT | 55 |
| CAO1-1 | CATGCCTACTTGTGTCACT | ACCGCTCATGTGTACCATC | 55 |
| CAO1-2 | GGCGTATTCCAAACCTATTCG | TTAGAATAAAATACGGTGTGCT | 55 |
| CAO1-3 | ATTTCCTACTACCCGAAGCTG | GACTATAGTATGCGGTTACCTT | 55 |
| CAO1-4 | GATTACAATTGCATCCCGTGA | ATGCCATCCACAAAGATGCTC | 55 |
| CAO1-5 | CAGAAGAAGTCCATGCTCCA | CCGGTTCGATATCCAGTATTGC | 55 |
| CAO1-6 | AGATGATACTCTAGTTTCCGACA | AATTAGACAAAACCACCCTCG | 60 |
表3 精细定位和测序所用的分子标记
Table 3 Markers used for fine mapping and sequencing.
| 引物名称 Primer name | 上游引物 Forward primer (5'-3') | 下游引物 Reverse primer (5'-3') | 退火温度 Anealing temperature/℃ |
|---|---|---|---|
| D2 | CAGCCTCTCTAATTGACTCTC | CCAAGTAGAATGGCCAAATGT | 55 |
| D3 | GCATCAAACTATAATAACTGACT | AGGAAAGTAAACAAGGCCTTA | 55 |
| D4 | ACACATATGTAGAGTATTATCCG | GCAAGCTGTCTACACGGTT | 55 |
| D8 | AACTCTATTGTTTATGGTGG | TAATTCCGAATTTCAGCTCT | 55 |
| CAO1-1 | CATGCCTACTTGTGTCACT | ACCGCTCATGTGTACCATC | 55 |
| CAO1-2 | GGCGTATTCCAAACCTATTCG | TTAGAATAAAATACGGTGTGCT | 55 |
| CAO1-3 | ATTTCCTACTACCCGAAGCTG | GACTATAGTATGCGGTTACCTT | 55 |
| CAO1-4 | GATTACAATTGCATCCCGTGA | ATGCCATCCACAAAGATGCTC | 55 |
| CAO1-5 | CAGAAGAAGTCCATGCTCCA | CCGGTTCGATATCCAGTATTGC | 55 |
| CAO1-6 | AGATGATACTCTAGTTTCCGACA | AATTAGACAAAACCACCCTCG | 60 |
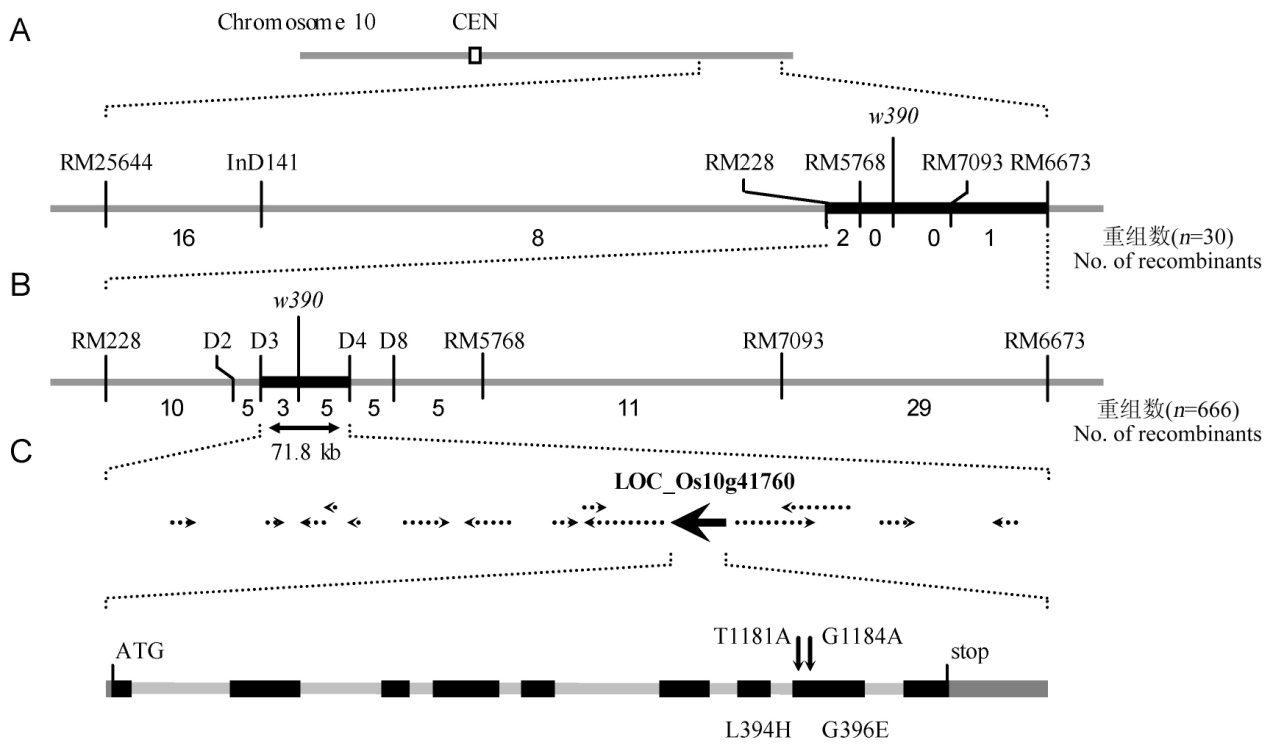
图3 突变体基因的定位与图位克隆(A-将突变体基因定位至第10染色体RM228和RM6673; B-突变体基因被界定在InDel标记D3和D4之间约71.8 kb的基因组区域内; C-序列分析发现突变体在LOC_Os10g41760的第8外显子存在两个单碱基突变,导致第394和396位的亮氨酸和甘氨酸分别突变为组氨酸和谷氨酸。)
Fig. 3. Fine mapping and positional cloning of the mutant gene.(A, The mutant locus was mapped on the chromosome 10 between RM228 and RM6673. B, The locus was narrowed to a 71.8 kb genomic region between InDel marker D3 and D4. C, Sequence analysis revealed that the mutant carried two nucleotide substitutions in the eighth exon of LOC_Os10g41760, which led to the substitution of leucine and glycine for histidine and glutamic acid, respectively.)
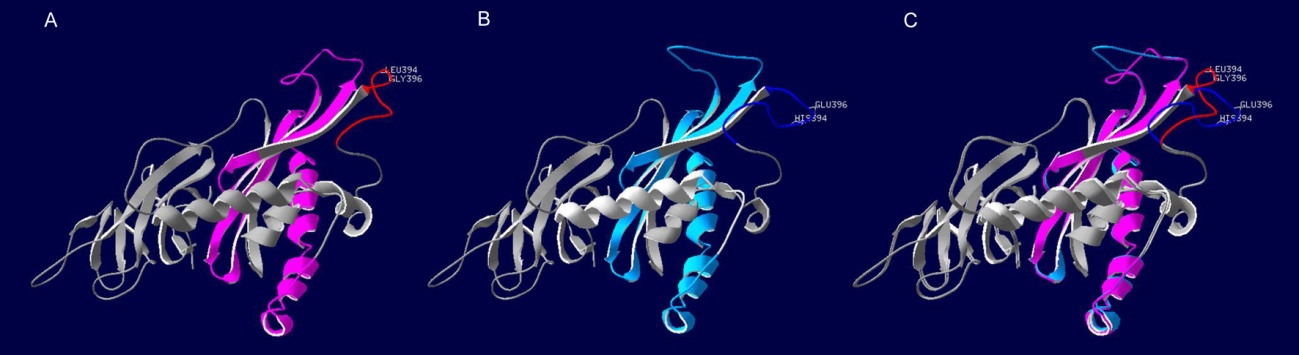
图4 野生型和突变体CAO1蛋白三维预测结构的比较(A-野生型; B-突变体; C-野生型与突变体的叠合。红色与深蓝色分别代表突变体位点在野生型与突变体中所在位置,紫色和浅蓝色分别代表野生型和突变体的PaO结构域。)
Fig. 4. Comparison of 3D protein structure model of CAO1 between wild-type and the mutant.(A, Wild-type; B, Mutant; C, Superposition of the mutant and its wild-type. The red and dark blue lines represent the structure haboring the mutations in the mutant and its wild-type, respectively. The purple and light blue regions represent the pheophorbide a oxygenase(PaO) domain in the mutant and its wild-type, respectively.)
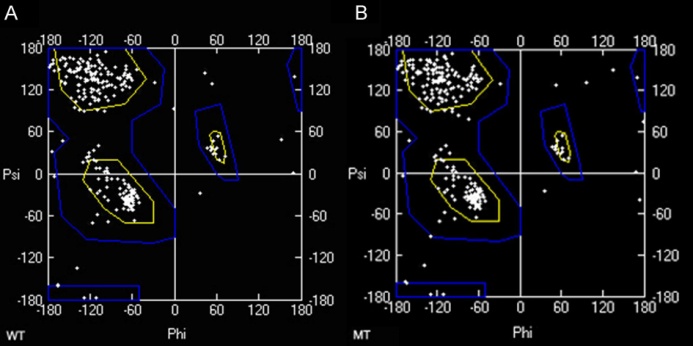
图5 野生型和突变体CAO1蛋白三维模型的拉氏构象(A-野生型; B-突变体。)
Fig. 5. Ramachandran plots of 3D protein structure models of CAO1 in the mutant and its wild-type.(A, Wild-type; B, Mutant.)
| [1] | Samuel I B.Green genes gleaned.Trends Plant Sci, 2005, 10(7): 309-312. |
| [2] | Kusaba M, Ito H, Morita R, et al.Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence.Plant Cell, 2007, 19: 1362-1375. |
| [3] | Wang P R, Gao J X, Wan C M, et al.Divinyl chlorophyll (ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice.Plant Physiol, 2010, 153: 994-1003. |
| [4] | Yamatani H, Sato Y, Masuda Y, et al.NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll-protein complexes during leaf senescence.Plant J, 2013, 74: 652-662. |
| [5] | Sugimoto H, Kusumi K, Tozawa Y, et al.The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation.Plant Cell Physiol, 2004, 45(8): 985-996. |
| [6] | Kusumi K, Sakata C, Nakamura T, et al.A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions.Plant J, 2011, 68(6): 1039-1050. |
| [7] | Su N, Hu M L, Wu D X, et al.Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production.Plant Physiol, 2012, 159(1): 227-238. |
| [8] | 邓晓娟, 张海清, 王悦, 等. 水稻叶色突变基因研究进展. 杂交水稻, 2012, 27(5): 9-14. |
| [9] | 李贤勇, 王楚桃, 李顺武, 等. 一个水稻高叶绿素含量基因的发现. 西南农业学报, 2002, 15(4): 122-123. |
| [10] | Juan M P, María C S, Kerstin K, et al.Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage.Plant Cell, 2006, 18(9): 2356-2368. |
| [11] | Miyoshi K, Ito Y, Serizawa A, et al.OsHAP3 genes regulate chloroplast biogenesis in rice.Plant J, 2003, 36(4): 532-540. |
| [12] | Dong H, Fei G L, Wu C Y, et al.A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants.Plant Physiol, 2013, 162(4): 1867-1880. |
| [13] | Zhao C F, Xu J M, Chen Y, et al.Molecular cloning and characterization of OsCHR4, a rice chromatin-remodeling factor required for early chloroplast development in adaxial mesophyll.Planta, 2012, 236(4): 1165-1176. |
| [14] | Liu W Z, Fu Y P, Hu G C, et al.Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.).Planta, 2007, 226(3): 785-795. |
| [15] | Tsugane K, Maekawa M, Takagi K, et al.An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice.Plant J, 2006, 45(1): 46-57. |
| [16] | Gothandam K M, Kim E S, Cho H, et al.OsPPR1, a pentatricopeptide repeat protein of rice is essential,for the chloroplast biogenesis.Plant Mol Biol, 2005, 58: 421-433. |
| [17] | Goh C H, Jung K H, Roberts S K, et al.Mitochondria provide the main source of cytosolic ATP for activation of outward-rectifying K+ channels in mesophyll protoplast of chlorophyll-deficient mutant rice (OsCHLH) seedlings.J Biol Chem, 2004, 279: 6874-6882. |
| [18] | Zhang H T, Li J J, Yoo J H, et al.Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development.Plant Mol Biol, 2006, 62(3): 325-337. |
| [19] | Sakuraba Y, Rahman M L, Cho S H, et al.The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions.Plant J, 2013, 74(1): 122-133. |
| [20] | Lee S, Kim J H, Yoo E S, et al.Differential regulation of chlorophyll a oxygenase genes in rice.Plant Mol Biol, 2005, 57(6): 805-818. |
| [21] | 杨海莲, 刘敏, 郭旻, 等. 一个水稻黄绿叶突变体ygl10的遗传分析和基因定位. 中国水稻科学, 2014, 28(1): 41-48. |
| [22] | Li J, Long Y, Qi G N, et al.The Os-AKT1 channel Is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex.Plant Cell, 2014, 80: 300-307. |
| [23] | Wu Z M, Zhang X, He B, et al.A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis.Plant Physiol, 2007, 145(1): 29-40. |
| [24] | Morita R, Sato Y, Masuda Y, et al.Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice.Plant J, 2009, 59(6): 940-952. |
| [25] | Jiang H W, Li M R, Liang N T, et al.Molecular cloning and function analysis of the stay green gene in rice.Plant J, 2007, 52(2): 197-209. |
| [26] | Fang J, Chai C L, Qian Q, et al.Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice.Plant J, 2008, 54(2): 177-189. |
| [27] | Li Z, Zhang Y X, Liu L, et al.Fine mapping of the lesion mimic and early senescence 1 (lmes1) in rice (Oryza sativa L.).Plant Physiol Biochem, 2014, 80: 300-307. |
| [28] | SAS Institute Inc, SAS/STAT User’s Guide. SAS Institute, Cary, NC, USA, 1999. |
| [29] | Rogers S O, Bendich A J.Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues.Plant Mol Biol, 1985, 5: 69-76. |
| [30] | Orjuela J, Garavito A, Bouniol M, et al.A universal core genetic map for rice.Theor Appl Genet, 2010, 120: 563-572. |
| [31] | Rychlik W.Oligo Primer Analysis Software Version 7.0. 2nd ed. Molecular Biology Insights, Inc, Cascade, CO, 2008. |
| [32] | 刘胜, 魏祥进, 邵高能, 等. 一个水稻“斑马叶”叶色突变体基因zebraleaf2(zl2)的图位克隆. 中国水稻科学, 2013, 27(3): 231-239. |
| [33] | Yamasato A, Nagata N, Tanaka R, et al.The N-Terminal domain of chlorophyllide a oxygenase confers protein instability in response to chlorophyll b accumulation in Arabidopsis.Plant Cell, 2005, 17: 1585-1597. |
| [34] | Harrison M A, Nemson J A, Melis A.Assembly and composition of the chlorophyll a-b light-harvesting complex of barley (Hordeum vulgare L.): Immunochemical analysis of chlorophyll b-less and chlorophyll b-deficient mutants.Photosyn Res, 1993, 38: 141-151. |
| [35] | Staehelin L A.Chloroplast structure and supramolecular organization of photosynthetic membranes// Pirson A, Zimmermann M H. Encyclopedia of Plant Physiology. Berlin: Germany:Springer-Verlag,1986: 1-84. |
| [36] | Rüdiger W.Biosynthesis of chlorophyll b and the chlorophyll cycle.Photosyn Res, 2002, 74: 187-193. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||