
中国水稻科学 ›› 2022, Vol. 36 ›› Issue (5): 447-458.DOI: 10.16819/j.1001-7216.2022.210714
伏荣桃1,2, 王剑1,2, 陈诚1,2, 赵黎宇1, 陈雪娟1, 卢代华1,2( )
)
收稿日期:2021-09-06
修回日期:2021-11-10
出版日期:2022-09-10
发布日期:2022-09-09
通讯作者:
卢代华
基金资助:
FU Rongtao1,2, WANG Jian1,2, CHEN Cheng1,2, ZHAO Liyu1, CHEN Xuejuan1, LU Daihua1,2( )
)
Received:2021-09-06
Revised:2021-11-10
Online:2022-09-10
Published:2022-09-09
Contact:
LU Daihua
摘要:
【目的】由稻曲病菌引起的稻曲病不仅造成水稻减产,而且还会产生对动物和植物有毒的真菌毒素。探明水稻幼穗对稻曲病菌毒素胁迫响应的分子机制,可为发掘水稻抗稻曲病基因以及抗病分子育种开辟新的思路。【方法】用稻曲病菌毒素处理水稻幼穗,采用转录组测序技术对水稻幼穗进行转录组测序,以水稻9311基因组作为参考基因组进行对比,利用TPM法计算基因表达量,设定参数(差异倍数的绝对值不小于2,且q值不大于0.05)筛选差异表达基因。结合基因差异表达分析、富集功能分析,鉴定水稻响应胁迫的关键基因,并利用实时荧光定量PCR技术对差异表达基因进行验证。【结果】在稻曲病菌毒素胁迫12 h后,水稻幼穗出现2526个差异表达基因(DEG);通过GO富集、KEGG代谢途经和KOG功能分析,将差异基因划分为GO功能下的64个条目、32个代谢途径和KOG功能下23个类别,包括淀粉和蔗糖代谢、苯丙类生物合成、碳代谢、糖酵解/糖异生、氨基糖和核苷酸糖代谢等生物学过程。DEG中有66个植物转录因子,分属7种植物转录因子家族,包括WRKY和Myb两大转录因子。分析二萜类生物合成与淀粉和蔗糖代谢途径相关基因发现,OsCPS2、OsKSL4和细胞色素P450等基因表达量上调,而淀粉酶、β-呋喃果糖苷酶和UDP-焦磷酸化酶等基因表达量下调,推测这些基因在水稻响应稻曲病菌毒素胁迫时发挥重要的作用。【结论】稻曲病菌毒素作为非生物胁迫因素对水稻幼穗具有毒性;通过干扰淀粉和蔗糖代谢等途径而影响种子营养物质的合成,降低水稻抵抗病原菌侵染水稻的能力。
伏荣桃, 王剑, 陈诚, 赵黎宇, 陈雪娟, 卢代华. 水稻幼穗响应稻曲病菌毒素胁迫早期的转录组分析[J]. 中国水稻科学, 2022, 36(5): 447-458.
FU Rongtao, WANG Jian, CHEN Cheng, ZHAO Liyu, CHEN Xuejuan, LU Daihua. Transcriptome Analysis of Young Rice Panicles in Early Response to Exposure to Mycotoxin of Ustilaginoidea virens[J]. Chinese Journal OF Rice Science, 2022, 36(5): 447-458.
| 样品名 Sample | 过滤前数据 Total raw reads / bp | 过滤后数据 Total clean reads / bp | 过滤后数据Clean bases(G) / bp | Q30/% | N/% | GC/% | 总对比数据 Total mapped reads /% |
|---|---|---|---|---|---|---|---|
| CK1-1 | 43 418 126 | 41 710 168 | 5.82 | 90.75 | 0.01 | 50.07 | 36 532 652(94.56%) |
| CK1-2 | 38 359 472 | 35 721 996 | 4.98 | 87.46 | 0.01 | 52.25 | 32 694 216(94.92%) |
| CK1-3 | 49 946 750 | 47 116 642 | 6.53 | 86.91 | 0.01 | 52.26 | 42 564 507(94.74%) |
| CL1-1 | 44 766 290 | 43 134 338 | 6.11 | 90.88 | 0.01 | 49.21 | 38 396 232(95.14%) |
| CL1-2 | 43 990 022 | 42 300 572 | 5.84 | 89.18 | 0.01 | 48.86 | 35 673 451(93.65%) |
| CL1-3 | 44 322 824 | 42 304 834 | 5.97 | 89.59 | 0.01 | 49.82 | 39 456 359(94.76%) |
表1 样本测序数据质量控制数据统计
Table 1. Statistics of transcriptone sequencing data.
| 样品名 Sample | 过滤前数据 Total raw reads / bp | 过滤后数据 Total clean reads / bp | 过滤后数据Clean bases(G) / bp | Q30/% | N/% | GC/% | 总对比数据 Total mapped reads /% |
|---|---|---|---|---|---|---|---|
| CK1-1 | 43 418 126 | 41 710 168 | 5.82 | 90.75 | 0.01 | 50.07 | 36 532 652(94.56%) |
| CK1-2 | 38 359 472 | 35 721 996 | 4.98 | 87.46 | 0.01 | 52.25 | 32 694 216(94.92%) |
| CK1-3 | 49 946 750 | 47 116 642 | 6.53 | 86.91 | 0.01 | 52.26 | 42 564 507(94.74%) |
| CL1-1 | 44 766 290 | 43 134 338 | 6.11 | 90.88 | 0.01 | 49.21 | 38 396 232(95.14%) |
| CL1-2 | 43 990 022 | 42 300 572 | 5.84 | 89.18 | 0.01 | 48.86 | 35 673 451(93.65%) |
| CL1-3 | 44 322 824 | 42 304 834 | 5.97 | 89.59 | 0.01 | 49.82 | 39 456 359(94.76%) |
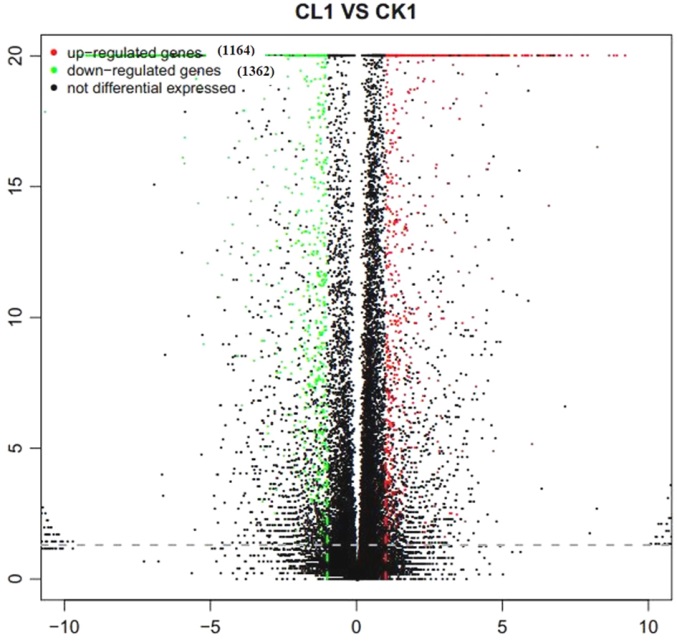
图2 差异表达基因的火山图分布 横轴为基因在不同组样本间的差异表达倍数(Log2 B/A);纵轴为基因表达量变化的统计学显著程度(P值);图中每个点代表一个基因,其中红色表示上调基因,绿色表示下调基因,黑色表示非差异基因。
Fig. 2. Volcano-plot distribution of DEGs. Horizontal axis represents the multiple value of differetially expressed gene between different groups of samples; Vertical axis represents statistically significant degree(P value) of gene expression changes; Red, Up-regulated DEGs; Blue, Down-regulated DEGs; Black, Non-DEGs.
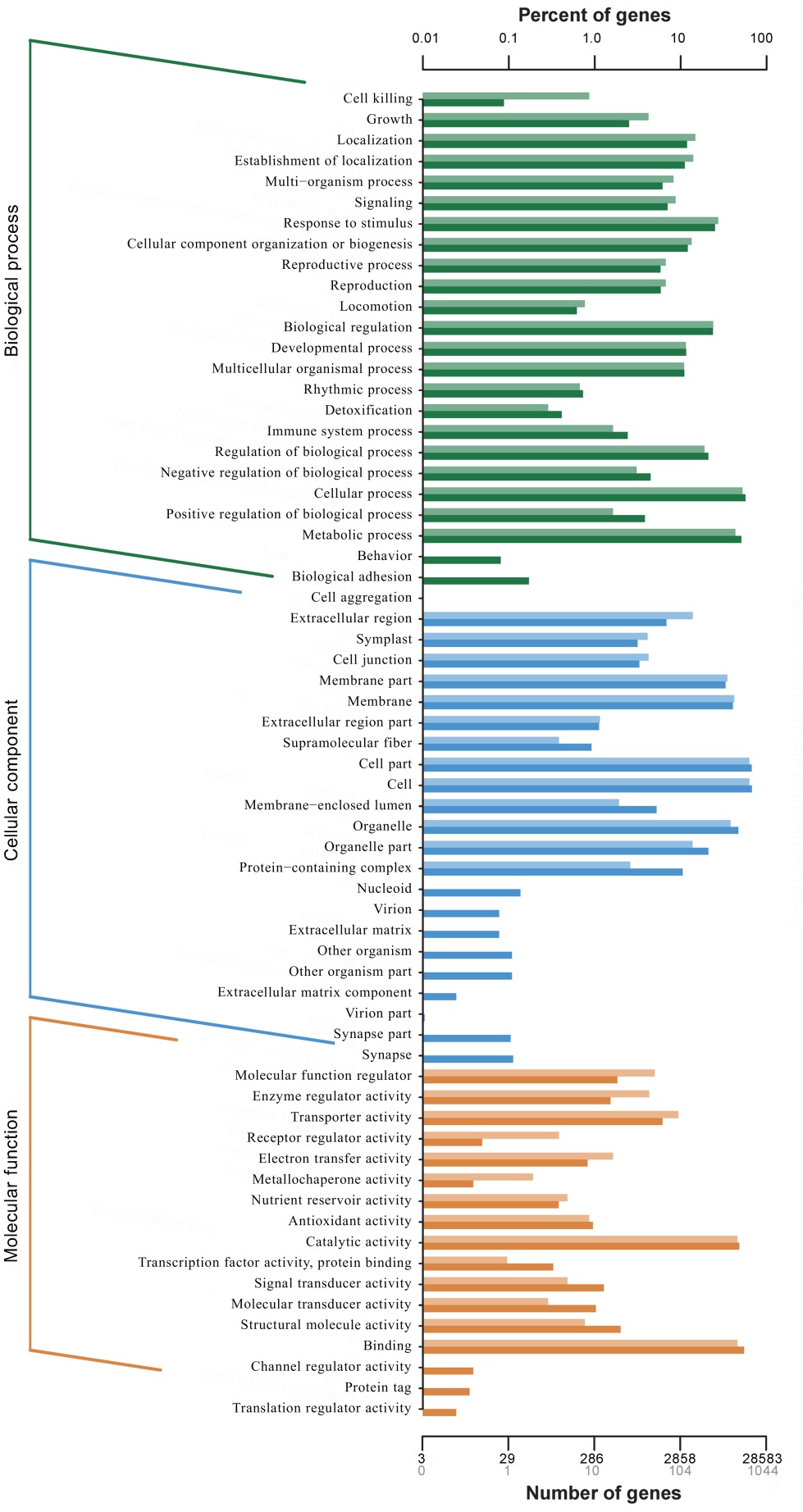
图3 差异表达基因的GO功能分类 纵轴为功能分类; 横轴为该分类内基因个数(下)及其占被注释上基因总数的百分比(上)。不同颜色代表不同的分类。柱状图和坐标轴上浅色代表差异基因,深色代表所有基因。
Fig. 3. GO function classification of DEGs. Vertical axis is function classification; Horizontal axis is the number of genes in the classification (bottom) and their percentage in the total number of annotated genes (top). Light colors represent differentially expressed genes and dark colors represent all genes.
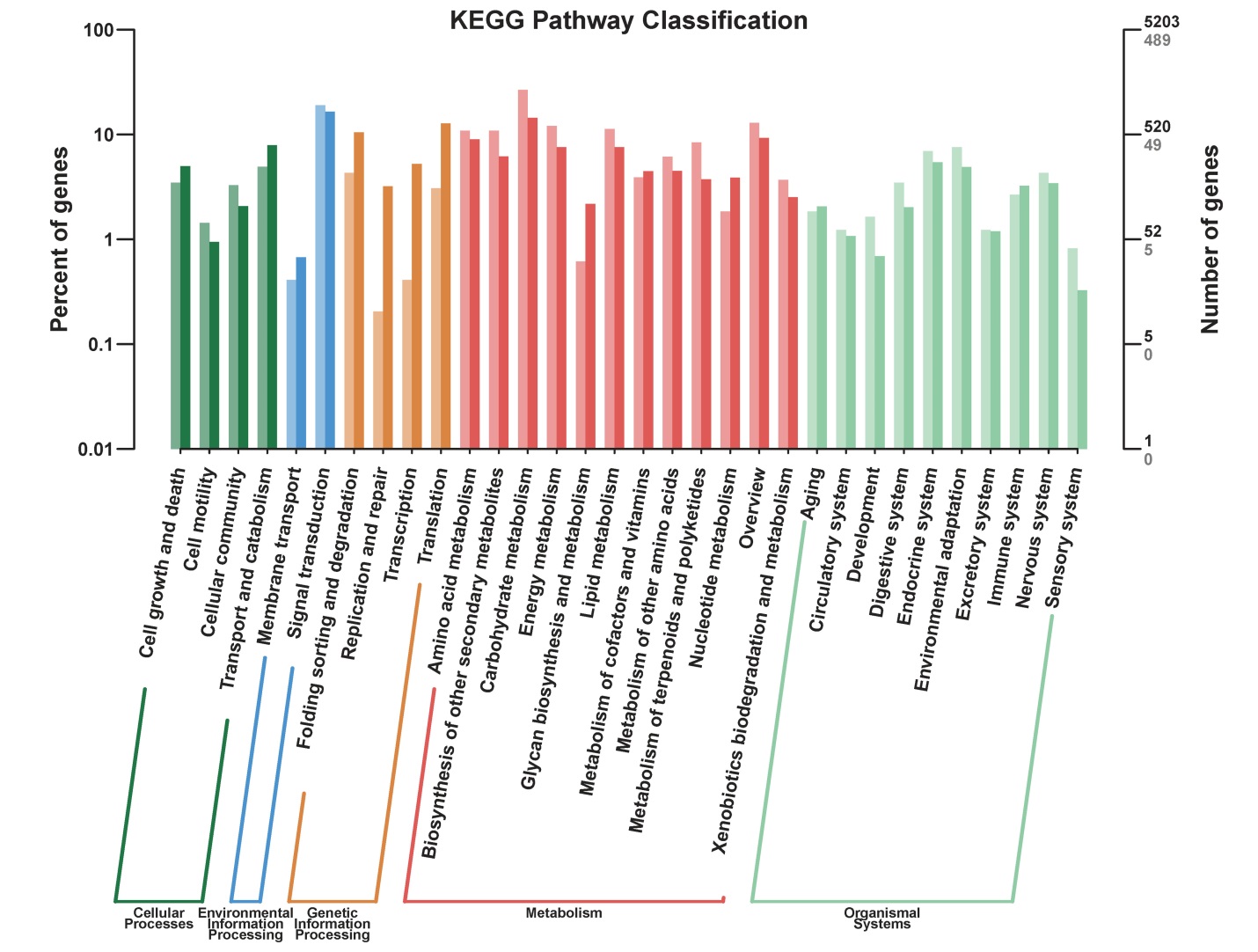
图4 差异表达基因KEEG代谢途径分类 横轴代表的是不同的通路,左侧纵轴代表该通路内基因个数占被注释上基因总数的百分比(左),右侧纵轴代表基因数目(浅色字体代表差异基因,深色字体代表所有基因)。
Fig. 4. KEGG pathway classification of DEGs. The horizontal axis represents different pathways; The left vertical axis represents the percentage of the number of genes in this pathway in the total number of annotated genes (left), the right vertical axis represents the number of genes (light color represents differentially expressed genes, dark color represents all genes).
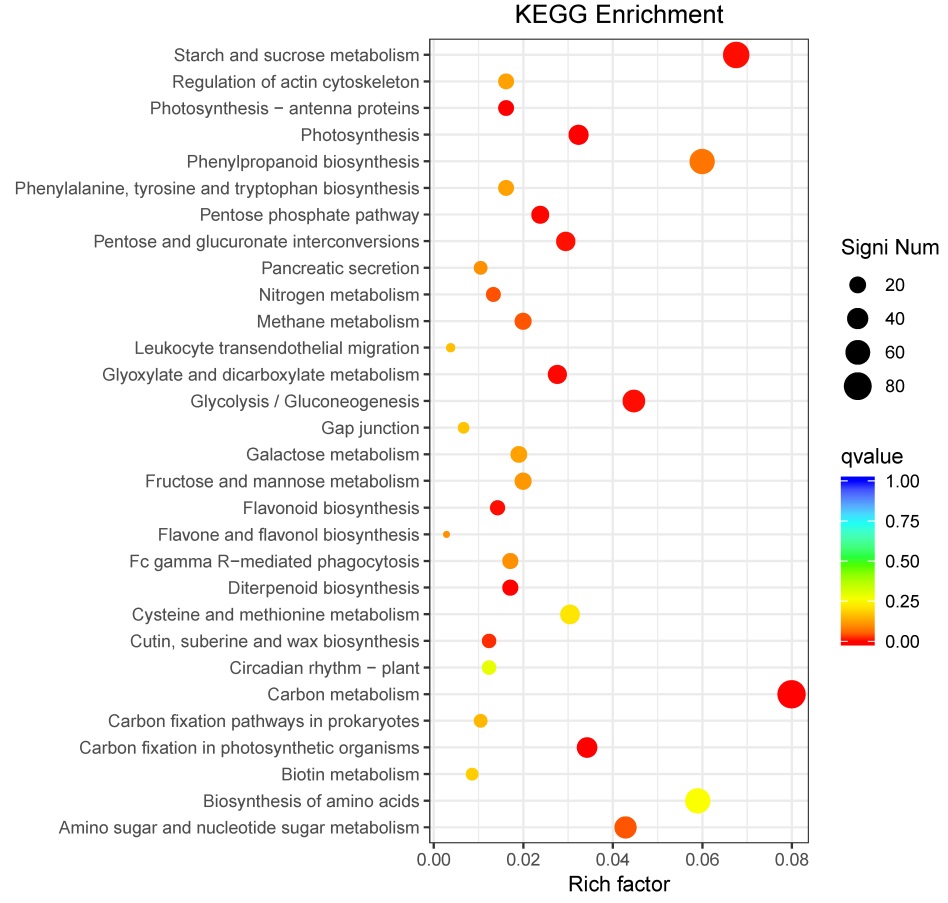
图5 差异基因通路富集散点图 纵轴表示功能注释信息,横轴表示通路对应的富集因子(Rich factor), 富集因子越大,表示富集的程度越高。点的颜色表示q值的大小,q值越小则颜色越接近红色。点的大小表示每个条目下包含的差异基因的个数,点越大则基因越多(富集程度最高的前30个通路)。
Fig. 5. Scatter plot of enriched KEGG pathways for DEGs. Vertical axis represents functional information; Horizontal axis shows the rich factor corresponding to pathway, and the greater the rich factor, the greater the enrichment degree. The size of the dots represents the number of DEGs under the term, and the larger the dots, the more genes (the top 30 pathways with the highest enrichment).
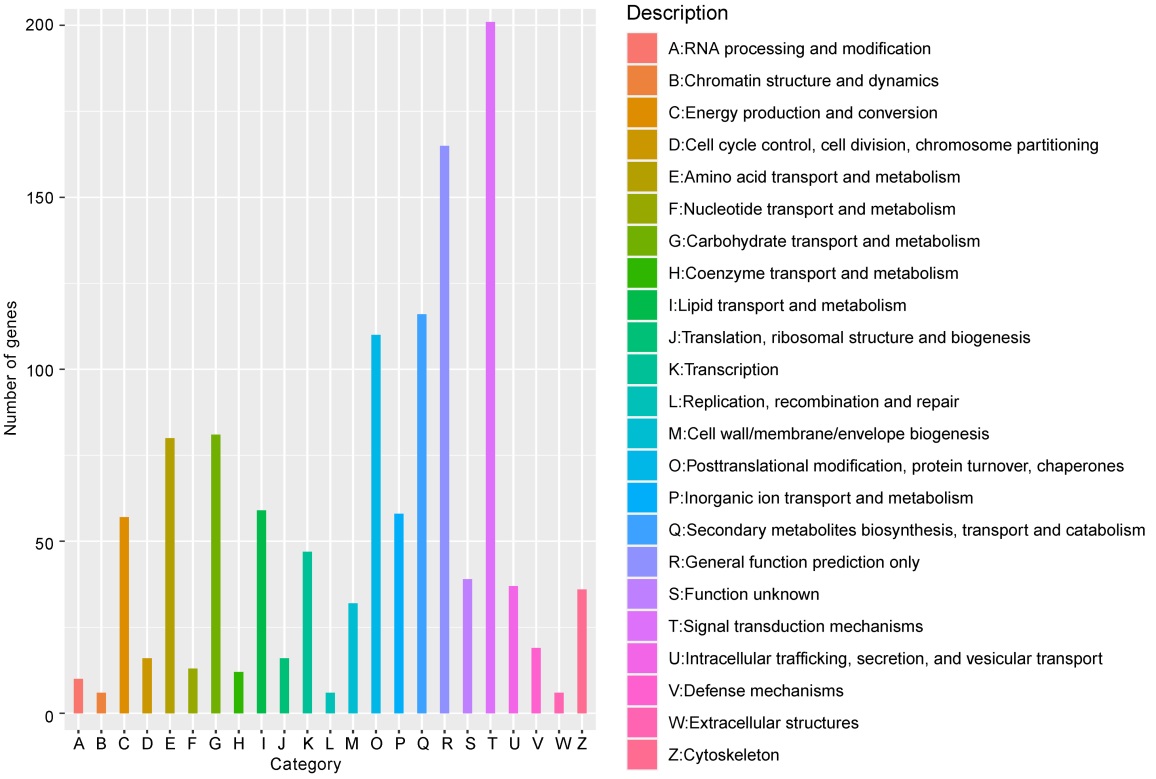
图6 差异基因KOG功能分类 横轴每种颜色代表一个KOG的功能分类;纵轴为注释到该分类下的基因数目。
Fig. 6. KOG function classification of DEGs. Each color on the horizontal axis represents a functional classification of KOG; Vertical axis is number of genes annotated under the classification.
| 转录因子家族 Transcription factor family | 受影响的转录因子数量 Number of affected transcription factors | 主要的植物调控功能 Main regulatory functions in plants | ||
|---|---|---|---|---|
| 上调 Up-regulated | 下调 Down-regulated | 总数 Total | ||
| Myb | 8 | 7 | 15 | 植物生长发育,抵御生物、非生物胁迫a, b, c |
| WRKY | 2 | 0 | 2 | 植物生长发育,抵御生物、非生物胁迫a, b, c |
| HSF(Heat shock transcription factor) | 6 | 3 | 9 | 抵御生物、非生物胁迫b, c |
| bHLH(basic Helix loop helix) | 1 | 3 | 4 | 植物生长发育,抵御生物、非生物胁迫a, b, c |
| MADS-box | 3 | 9 | 12 | 植物生长发育,抵御非生物胁迫a, c |
| GATA | 2 | 7 | 9 | 植物生长发育,抵御非生物胁迫a, c |
| HOX | 6 | 9 | 15 | 植物生长发育,抵御非生物胁迫a, c |
| 总数Total | 28 | 38 | 66 | |
表2 稻曲菌毒素影响的植物转录因子
Table 2. Plant transcription factors affected by mycotoxin from U. virens.
| 转录因子家族 Transcription factor family | 受影响的转录因子数量 Number of affected transcription factors | 主要的植物调控功能 Main regulatory functions in plants | ||
|---|---|---|---|---|
| 上调 Up-regulated | 下调 Down-regulated | 总数 Total | ||
| Myb | 8 | 7 | 15 | 植物生长发育,抵御生物、非生物胁迫a, b, c |
| WRKY | 2 | 0 | 2 | 植物生长发育,抵御生物、非生物胁迫a, b, c |
| HSF(Heat shock transcription factor) | 6 | 3 | 9 | 抵御生物、非生物胁迫b, c |
| bHLH(basic Helix loop helix) | 1 | 3 | 4 | 植物生长发育,抵御生物、非生物胁迫a, b, c |
| MADS-box | 3 | 9 | 12 | 植物生长发育,抵御非生物胁迫a, c |
| GATA | 2 | 7 | 9 | 植物生长发育,抵御非生物胁迫a, c |
| HOX | 6 | 9 | 15 | 植物生长发育,抵御非生物胁迫a, c |
| 总数Total | 28 | 38 | 66 | |
| 序号No. | 基因名称 Gene name | 基因登录号 Gene ID | 引物序列 Primer sequence (5′-3′) |
|---|---|---|---|
| 1 | WRKY71 | BGIOSGA007670 | F: TGCAGGTGGTGAAAGATGGG |
| R: GAAACAGGCTCGAATTTGCACT | |||
| 2 | WRKY24 | BGIOSGA004722 | F: AGAGATGGTGAGTGCCCTGT |
| R: CGATGTCGCTCATGGTTTGG | |||
| 3 | Myb | BGIOSGA004072 | F: CATCTCGCAAAACGGCGAAG |
| R: GTTCGGACATCACATAGTCGC | |||
| 4 | Myb | BGIOSGA006005 | F: TGCCTTGGATACGCGAAAGA |
| R: CCGCTTCTTGAGGTGAGTGT | |||
| 5 | UGPase | BGIOSGA007245 | F: TCTGGGATGGCCATAGAAAAACA |
| R: GTACTGGACCAACAACGTGC | |||
| 6 | Chitinase | BGIOSGA006087 | F: GTACTGGACCAACAACGTGC |
| R: TTAGCAGGTGAGGTTGCTGC | |||
| 7 | Peroxidase | BGIOSGA037333 | F: GGGATTTGCCATAAGCGAAACA |
| R:CCACATTCTCGGTTGTTGCC | |||
| 8 | SBEII | BGIOSGA018719 | F: GAGTCGAGCTGGAATTGTGTTG |
| R: GCAGAGTGCCCACATTCATC | |||
| 9 | NADPH oxidase | BGIOSGA037531 | F: AGATGAATTCGCTCCAATGATTTGT |
| R: GGCCAAGCTGAAATTGTGGC | |||
| UBI | F: CGCAAGAAGAAGTGTGGTCA | ||
| R: ACGATTGATTTAACCAGTCCATGA |
表3 qRT-PCR验证引物
Table 3. Primers used for qRT-PCR verifying.
| 序号No. | 基因名称 Gene name | 基因登录号 Gene ID | 引物序列 Primer sequence (5′-3′) |
|---|---|---|---|
| 1 | WRKY71 | BGIOSGA007670 | F: TGCAGGTGGTGAAAGATGGG |
| R: GAAACAGGCTCGAATTTGCACT | |||
| 2 | WRKY24 | BGIOSGA004722 | F: AGAGATGGTGAGTGCCCTGT |
| R: CGATGTCGCTCATGGTTTGG | |||
| 3 | Myb | BGIOSGA004072 | F: CATCTCGCAAAACGGCGAAG |
| R: GTTCGGACATCACATAGTCGC | |||
| 4 | Myb | BGIOSGA006005 | F: TGCCTTGGATACGCGAAAGA |
| R: CCGCTTCTTGAGGTGAGTGT | |||
| 5 | UGPase | BGIOSGA007245 | F: TCTGGGATGGCCATAGAAAAACA |
| R: GTACTGGACCAACAACGTGC | |||
| 6 | Chitinase | BGIOSGA006087 | F: GTACTGGACCAACAACGTGC |
| R: TTAGCAGGTGAGGTTGCTGC | |||
| 7 | Peroxidase | BGIOSGA037333 | F: GGGATTTGCCATAAGCGAAACA |
| R:CCACATTCTCGGTTGTTGCC | |||
| 8 | SBEII | BGIOSGA018719 | F: GAGTCGAGCTGGAATTGTGTTG |
| R: GCAGAGTGCCCACATTCATC | |||
| 9 | NADPH oxidase | BGIOSGA037531 | F: AGATGAATTCGCTCCAATGATTTGT |
| R: GGCCAAGCTGAAATTGTGGC | |||
| UBI | F: CGCAAGAAGAAGTGTGGTCA | ||
| R: ACGATTGATTTAACCAGTCCATGA |
| [1] | Tanaka E, Ashizawa T, Sonoda R, Sonoda R, Tanka C. Villosiclava virens gen. nov., comb. nov., teleomorph of Ustilaginoidea virens,the causal agent of rice false smut[J]. Mycotaxon, 2008, 106(1): 491-501. |
| [2] | Fan J, Yang J, Wang Y Q, Li G B, Li Y, Huang F, Wang W M. Current understanding on Villosiclava virens, a unique flower-infecting fungus causing rice false smut disease[J]. Molecular Plant Pathology, 2016, 17(9): 1321-1330. |
| [3] | 伏荣桃, 王剑, 卢代华, 张鸿, 龚学书, 陈雪娟, 任鸿志, 毛建辉. 水稻稻曲病抗性鉴定技术及影响因子研究[J]. 中国农学通报, 2015, 31(18): 266-272. |
| Fu R T, Wang J, Lu D H, Zhang H, Gong X S, Chen X J, Ren H Z, Mao J H. Resistance identification and influence factor of rice false smut[J]. Chinese Agricultural Science Bulletin, 2015, 31(18): 266-272. (in Chinese with English abstract) | |
| [4] | Hu M L, Luo L X, Wang S, Liu Y F, Li J Q. Infection processes of Ustilaginoidea virens during artificial inoculation of rice panicles[J]. European Journal of Plant Pathology, 2014, 139: 67-77. |
| [5] | Meng J J, Sun W B, Mao Z L, Dan X, Wang X, Lu S, Yang L, Zhou L, Zhang G. Main ustilaginoidins and their distribution in rice false smut balls[J]. Toxins, 2015, 7(10): 4023-4034. |
| [6] | Lai D W, Meng J J, Zhang X P, Xu D, Dai J G, Zhou L G. Ustilobisorbicillinol A, a cytotoxic sorbyl-containing aromatic polyketide from Ustilaginoidea virens[J]. Organic Letters, 2019, 21(5): 1311-1314. |
| [7] | Fu X X, Xie R S, Wang J. Development of colloidal gold-based lateral flow immunoassay for rapid qualitative and semi-quantitative analysis of ustiloxins A and B in rice samples[J]. Toxins, 2017, 9(3): 79. |
| [8] | Meng J J, Gu G, Dang P Q, Zhang X P, Wang W X, Dai J G, Liu Y, Lai D W, Zhou L G. Sorbicillinoids from the fungus Ustilaginoidea virens and their phytotoxic, cytotoxic, and antimicrobial activities[J]. Frontiers in Chemistry, 2019, 7: 435. |
| [9] | Li Y, Koiso Y, Kobayashi H, Hashimoto Y, Iwasaki S. Ustiloxins, new antimitotic cyclic peptides: Interaction with porcine brain tubulin[J]. Biochemical Pharmacology, 1995, 49(10): 1367-1372. |
| [10] | Hu Z, Dang Y, Liu C S, Zhou L, Liu H. Acute exposure to ustiloxin A affects growth and development of early life zebrafish, Danio rerio[J]. Chemosphere, 2019, 226: 851-857. |
| [11] | 陈美军, 胡东维, 徐颖. 稻曲病菌毒素的活性测定、抗体制备与细胞定位[J]. 实验生物学报, 2004, 37(4): 310-314. |
| Chen M J, Hu D W, Xu Y. Activity assay, antiserum preparation and cellular localization of ustiloxins[J]. Acta Biologiae Experimentalis Sinica, 2004, 37(4): 310-314. (in Chinese with English abstract) | |
| [12] | Hamed K A, Wayne T S, Cartwright R D, Sciumbato G L. Ustilaginoidea virens infection of rice in Arkansas: Toxicity of false smut galls, their extracts and the ustiloxin fraction[J]. American Journal of Plant Sciences, 2014, 5(21): 3166-3176. |
| [13] | Wang X H, Wang J, Lai D W, Wang W X, Liu Y. Ustiloxin G, a new cyclopeptide mycotoxin from rice false smut balls[J]. Toxins, 2017, 9(2): 54-63. |
| [14] | Fu R T, Wang J, Chen C, Gong X S, Lu D H. Effect of crude toxins of Ustilaginoidea virens on rice seed germination[J]. African Journal of Microbiology Research, 2017, 11(32): 1267-1273. |
| [15] | Luduena R F, Roach M C, Prasad V. Interaction of ustiloxin A with bovine brain tubulin[J]. Biochemical Pharmacology, 1994, 47(9): 1593-1599. |
| [16] | 武斌, 温雪玮, 李衫衫, 胡东维, 梁五生. 稻曲病菌毒素对水稻幼根转录组的影响[J]. 农业生物技术报, 2018, 26(7):1093-1106. |
| Wu B, Wen X W, Li S S, Hu D W, Lang W S. Influences of mycotoxins of Villosiclava virens on the transcriptome of rice (Oryza sativa) seedling roots[J]. Journal of Agricultural Biotechnology, 2018, 26(7): 1093-1106. (in Chinese with English abstract) | |
| [17] | Yuan Z, Zhang Y, Xu G, Bi D, Qu H, Zou X, Gao X, Yang H, He H, Wang X, Bao J, Zuo S, Pan X, Zhou B, Wang G, Qu S. Comparative transcriptome analysis of Rhizoctonia solani-resistant and -susceptible rice cultivars reveals the importance of pathogen recognition and active immune responses in host resistance[J]. Journal of Plant Biology, 2018, 61(3):143-158. |
| [18] | 楚乐乐, 罗成科, 李芳兰, 路旭平, 马天利, 李培富. 盐胁迫下OsDSR2 RNAi转基因水稻的生理特性及转录组学分析[J]. 植物遗传资源学报, 2020, 21(4): 954-965. |
| Chu L L, Luo C K, Li F L, Lu X P, Ma T L, Li P F. Analysis of the physiological characteristics and transcriptome profiles of OsDSR2 RNAi transgenic rice under salt stress[J]. Journal of Plant Genetic Resources, 2020, 21(4): 954-965. (in Chinese with English abstract) | |
| [19] | Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency[J]. The Annals of Statistics, 2001, 29(4): 1165-1188. |
| [20] | Chao J, Jin J, Wang D, Han R, Zhu R S, Zhu Y G, Li S Q, Sun M X. Cytological and transcriptional dynamics analysis of host plant revealed stages specific biological processes related to compatible rice Ustilaginoidea virens interaction[J]. PLoS ONE, 2014, 9(3): e91391. |
| [21] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method[J]. Methods, 2001, 25(4): 402-408. |
| [22] | Ismaiel A, Papenbrock J. Mycotoxins: Producing fungi and mechanisms of phytotoxicity[J]. Agriculture, 2015, 5: 492-537. |
| [23] | Janardhanan K K, Husain A. Phytotoxic activity of tenuazonic acid isolated from Alternaria alternata (Fr.) Keissler causing leaf blight of Datura innoxia Mill. and its effect on host metabolism[J]. Journal of Phytopathology, 1984, 111(3-4): 305-311. |
| [24] | McLean M. The phytotoxicity of Fusarium metabolites: An update since 1989. Mycopathologia, 1996, 133: 163-179. |
| [25] | Ismaiel A A, Tharwat N A. Antifungal activity of silver ion on ultrastructure and production aflatoxin B1 and patulin by two mycotoxigenic strains, Aspergillus flavus OC1 and Penicillium vulpinum CM1[J]. Journal de Mycologie Medical, 2014, 24(3): 193-204. |
| [26] | Fujita K, Arase S, Hiratsuka H, Honda Y, Nozu M. The role of toxin(s) produced by germinating spores of Pyricularia oryzae in pathogenesis[J]. Journal of Phytopathology, 1994, 142(3-4): 245-252. |
| [27] | Vidhyasekaran P, Ruby Ponmalar T, Samiyappan R, Velazhahan R, Muthukrishnan S. Host-specific toxin production by Rhizoctonia solani, the rice sheath blight pathogen[J]. Phytopathology, 1997, 87(12): 1258-1263. |
| [28] | Samuel A T, Valentine I T. Effect of total aflatoxin on the growth characteristics and chlorophyll level of sesame (Sesamum indicum L.)[J]. New York Science Journal, 2014, 7: 8-13. |
| [29] | Aver’yanov A A, Lapikova V P, Lebrun M H. Tenuazonic acid, toxin of rice blast fungus, induces disease resistance and reactive oxygen production in plants[J]. Russian Journal of Plant Physiology, 2007, 54: 749-754. |
| [30] | 齐俊生, 李怀方. 一种检测棉花黄萎菌毒素致萎性的新方法: 叶片针刺涂抹法[J]. 棉花学报, 2006, 18(4): 228-232. |
| Qi J S, Li H F. A new detection method of wilting induction by phytotoxin from V. dahliae on cotton through leaf pricking and spreading[J]. Cotton Science, 2006, 18(4): 228-232. (in Chinese with English abstract) | |
| [31] | 杨艳丽, 肖浪涛, 胡先奇. 马铃薯晚疫病菌与寄主品种抗性关系研究. 中国农业科学, 2009, 42(6): 2202-2210. |
| Yang Y L, Xiao L T, Hu X Q. Study on the relationship between the toxin of Phytophthora infestans and resistance of potato[J]. Scientia Agricultura Sinica, 2009, 42(6): 2202-2210. (in Chinese with English abstract) | |
| [32] | Ambawat S, Sharma P, Yadav N R. MYB transcription factor genes as regulators for plant responses: An overview[J]. Physiology and Molecular Biology of Plants, 2013, 19(3): 307-321. |
| [33] | Wang Y M, Kwon S J, Wu J N, Choi J Y, Lee Y H, Agrawai G K, Tamogami S, Rakwal R, Park S R, Kim B G, Jung K H, Kang K Y, Kim S G, Kim S T. Transcriptome analysis of early responsive genes in rice during Magnaporthe oryzae infection[J]. The Plant Pathology Journal, 2014, 30(4): 343-354. |
| [34] | Han Y Q, Zhang K, Yang J, Zhang N, Zhang Y, Liu Y F, Chen Z Y, Hsiang T, Sun W X. Differential expression profiling of the early response to Ustilaginoidea virens between false smut resistant and susceptible rice varieties[J]. BMC Genomics, 2015, 16: 955. |
| [35] | 张艺丹, 曾英, 卢山. 水稻二萜合成途径中代谢流调控机制研究进展[J]. 植物生理学报, 2019, 55(12): 1762-1768. |
| Zhang Y D, Zeng Y, Lu S. Recent progress in the study of metabolic flux regulation in rice diterpene biosynthesis[J]. Plant Physiology Journal, 2019, 55(12): 1762-1768. (in Chinese with English abstract) | |
| [36] | Peters R J. Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants[J]. Phytochemistry, 2006, 67: 2307-2317 |
| [37] | Zi J, Mafu S, Peters R J. To gibberellins and beyond Surveying the evolution of (di)terpenoid metabolism[J]. Annual Review of Plant Biology, 2014, 65: 259-286. |
| [38] | 韩彦卿, 韩渊怀, 张春来, 孙文献. 水稻幼穗与Ustilaginoidea virens互作早期的转录组分析[J]. 植物病理学报, 2019, 49(3): 296-305. |
| Han Y Q, Han Y H, Zhang C L, Sun W X. Transcriptomic analysis of early interaction between rice young spikelets and Ustilaginoidea virens[J]. Acta Phytopathologica Sinica, 2019, 49(3): 296-305. (in Chinese with English abstract) | |
| [39] | 彭波, 彭宇, 彭娟, 孔冬艳, 何璐璐, 孙艳芳, 黄雅琴, 宋世枝. 水稻种子主要营养物质合成及调控研究与展望. 热带作物学报, 2018, 39(6): 1241-1251. |
| Peng B, Peng Y, Peng J, Kong D M, He L L, Sun Y F, Huang Y Q, Song S Z. Research advancement and prospects of main nutritious substances synthesis and regulation in rice seeds[J]. Chinese Journal of Tropical Crops, 2018, 39(6): 1241-1251. (in Chinese with English abstract) | |
| [40] | Thitisaksakul M, Jiménez R C, Arias M C, Beckles D M. Effects of environmental factors on cereal starch biosynthesis and composition[J]. Journal of Cereal Science, 2012, 56(1): 67-80. |
| [41] | Fujita N. Starch biosynthesis in rice endosperm[J]. Agri-bioscience Monographs, 2014, 4(1): 1-18. |
| [42] | Fan J, Yang L, Zheng A P, Wang W M, Guo X Y, Li L, Huang F, Sun W X, Yan L, Huang Y Y. Infection of Ustilaginoidea virens intercepts rice seed formation but activates grain-filling-related genes[J]. Journal of Integrative Plant Biology, 2015, 57(6): 577-590. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 伏荣桃, 陈诚, 王剑, 赵黎宇, 陈雪娟, 卢代华. 转录组和代谢组联合分析揭示稻曲病菌的致病因子[J]. 中国水稻科学, 2024, 38(4): 375-385. |
| [5] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [6] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [7] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [8] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [9] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [10] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [11] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [12] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [13] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [14] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [15] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||