
中国水稻科学 ›› 2022, Vol. 36 ›› Issue (1): 13-26.DOI: 10.16819/j.1001-7216.2022.210312
张涛荟1,#, 王海宇1,#, 万华1, 张莉萍1, 谢振威1, 陈可毅1, 何晓栋1, 赵志刚1,*( ), 万建民1,2
), 万建民1,2
收稿日期:2021-03-23
修回日期:2021-04-22
出版日期:2022-01-10
发布日期:2022-01-10
通讯作者:
赵志刚
作者简介:第一联系人:#共同第一作者;
基金资助:
ZHANG Taohui1,#, WANG Haiyu1,#, WAN Hua1, ZHANG Liping1, XIE Zhenwei1, CHEN Keyi1, HE Xiaodong1, ZHAO Zhigang1,*( ), WAN Jianmin1,2
), WAN Jianmin1,2
Received:2021-03-23
Revised:2021-04-22
Online:2022-01-10
Published:2022-01-10
Contact:
ZHAO Zhigang
About author:First author contact:#These authors contributed equally to this work;
摘要:
【目的】通过构建分离群体定位并克隆水稻雌雄不育基因OsFMA2,并对其在水稻育性调控中的功能进行探究。【方法】通过EMS诱变水稻宁粳4号得到稳定的不育突变体,对突变体进行表型及细胞学观察,利用精细定位和Mut-map相结合的方法克隆了水稻雌雄不育基因OsFMA2,通过实时荧光定量PCR检测其在各组织表达水平差异,利用水稻原生质体表达系统进行OsFMA2蛋白的亚细胞定位。【结果】细胞学观察发现突变体为雌雄不育。利用极端个体将其精细定位于水稻第6染色体长臂448 kb的区间内。结合全基因组重测序结果,最终确定OsFMA2为目标基因。该基因编码一个复制蛋白,在单子叶植物中高度保守。OsFMA2具有组织特异性,在幼穗中高表达。亚细胞定位结果显示OsFMA2蛋白定位于细胞核中。【结论】OsFMA2在幼穗中高表达,可能参与了水稻雄配子第一次减数分裂前期同源重组过程,同时,OsFMA2的表达对雌配子发育也产生了不利影响。
张涛荟, 王海宇, 万华, 张莉萍, 谢振威, 陈可毅, 何晓栋, 赵志刚, 万建民. 水稻雌雄不育突变体Osfma2的细胞学观察及基因图位克隆[J]. 中国水稻科学, 2022, 36(1): 13-26.
ZHANG Taohui, WANG Haiyu, WAN Hua, ZHANG Liping, XIE Zhenwei, CHEN Keyi, HE Xiaodong, ZHAO Zhigang, WAN Jianmin. Cytological Observation of a Female and Male Sterile Osfma2 Mutant in Rice and Its Map-based Cloning[J]. Chinese Journal OF Rice Science, 2022, 36(1): 13-26.
| 引物名称 | 正向引物序列 | 反向引物序列 | 实验目的 |
|---|---|---|---|
| Name | Forward sequence(5’-3’) | Reverse sequence(5’-3’) | Purpose |
| RM162 | GCCAGCAAAACCAGGGATCCGG | CCGCAAGCCGCACAAGACCTTG | 精细定位 Fine mapping |
| H971-5 | CCGCCCGCGTTTTATTTACT | GGCAACATTCTTCGGCCTC | 精细定位 Fine mapping |
| HF-2 | CGCCGCCAAGAGCTAATTAA | TGAGAGATCTCGATCGACTTCTC | 精细定位 Fine mapping |
| HF-3 | GCCTATGTTTAGCCGCGAAA | GGAAGGAGAAGTTGAGGGGG | 精细定位 Fine mapping |
| Hi-1 | CCGGACCGTGATTTCGTTAG | TCAATACTAAAATCTTCGCCCCT | 精细定位 Fine mapping |
| Hh-4 | CTTTCTCCCCGTCGATCCTT | GCCCCACGGAGCATATCTAG | 精细定位 Fine mapping |
| Hh-9 | GGCAGGAAGTCCAAAAAGCT | TCCATAAAGCAAGCTGATGCA | 精细定位 Fine mapping |
| He-6 | CGTAGGCGAGGTGGTACAAT | GAGAGGTCACTGGTCAGCTT | 精细定位 Fine mapping |
| RM5463 | ACCCTTGCAGACAACGTACC | GCATGCAGCTGCTGGTATAT | 精细定位 Fine mapping |
| QHG | AGCTCTGCAGCAACCAGATA | TGCTTTGGGCATTCCAGTTC | qRT-PCR |
表1 本研究用于精细定位和定量的引物序列
Table 1 Fine-mapping and quantitative primer sequences used in the study.
| 引物名称 | 正向引物序列 | 反向引物序列 | 实验目的 |
|---|---|---|---|
| Name | Forward sequence(5’-3’) | Reverse sequence(5’-3’) | Purpose |
| RM162 | GCCAGCAAAACCAGGGATCCGG | CCGCAAGCCGCACAAGACCTTG | 精细定位 Fine mapping |
| H971-5 | CCGCCCGCGTTTTATTTACT | GGCAACATTCTTCGGCCTC | 精细定位 Fine mapping |
| HF-2 | CGCCGCCAAGAGCTAATTAA | TGAGAGATCTCGATCGACTTCTC | 精细定位 Fine mapping |
| HF-3 | GCCTATGTTTAGCCGCGAAA | GGAAGGAGAAGTTGAGGGGG | 精细定位 Fine mapping |
| Hi-1 | CCGGACCGTGATTTCGTTAG | TCAATACTAAAATCTTCGCCCCT | 精细定位 Fine mapping |
| Hh-4 | CTTTCTCCCCGTCGATCCTT | GCCCCACGGAGCATATCTAG | 精细定位 Fine mapping |
| Hh-9 | GGCAGGAAGTCCAAAAAGCT | TCCATAAAGCAAGCTGATGCA | 精细定位 Fine mapping |
| He-6 | CGTAGGCGAGGTGGTACAAT | GAGAGGTCACTGGTCAGCTT | 精细定位 Fine mapping |
| RM5463 | ACCCTTGCAGACAACGTACC | GCATGCAGCTGCTGGTATAT | 精细定位 Fine mapping |
| QHG | AGCTCTGCAGCAACCAGATA | TGCTTTGGGCATTCCAGTTC | qRT-PCR |
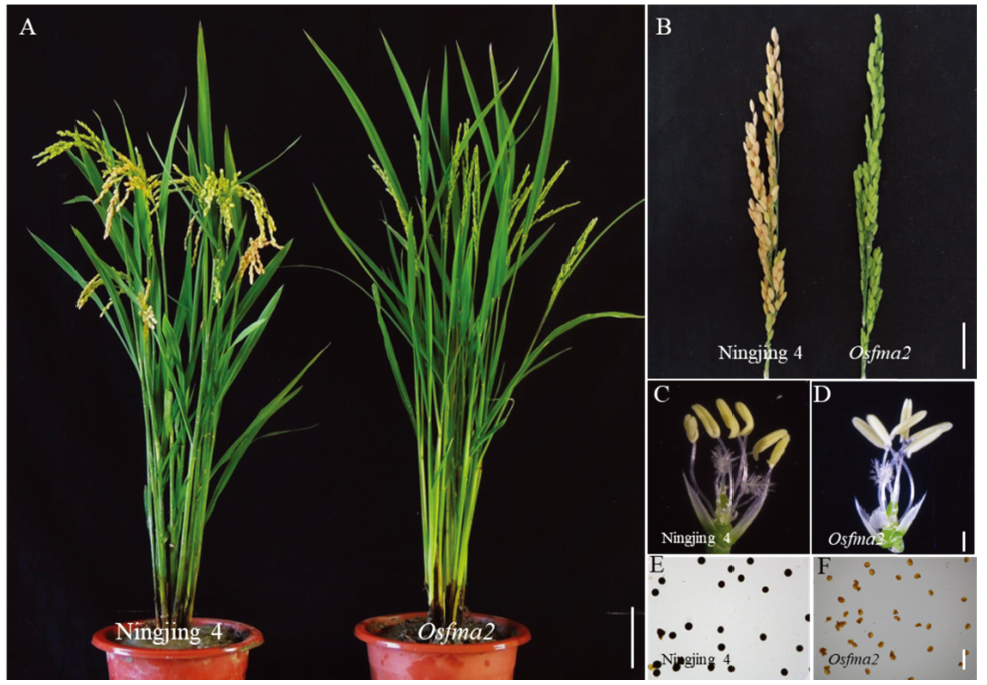
图1 野生型和突变体Osfma2表型和育性观察 A—野生型和突变体Osfma2的植株形态,比例尺为15 cm。B—野生型和突变体的小穗,比例尺为4 cm。C—野生型的花药,比例尺为600 μm。D—突变体的花药表型,比例尺为600 μm。E—野生型的花粉镜检,比例尺为50 μm。F—突变体的花粉镜检,比例尺为50 μm。
Fig. 1. Phenotypes and sterility of the wild type and the mutant Osfma2. A, Plant morphology of the wild type (Ningjing 4) and the Osfma2 mutant, bar=15 cm. B, Panicle of the wild type and the Osfma2 mutant, bar=4 cm. C, Anther of wild type, bar=600 μm. D, Anther of the mutant Osfma2, bar=600 μm. E, Pollen of Ningjing 4 stained with I2-KI, bar= 50 μm. F, Pollen of Osfma2 stained with I2-KI, bar= 50 μm.
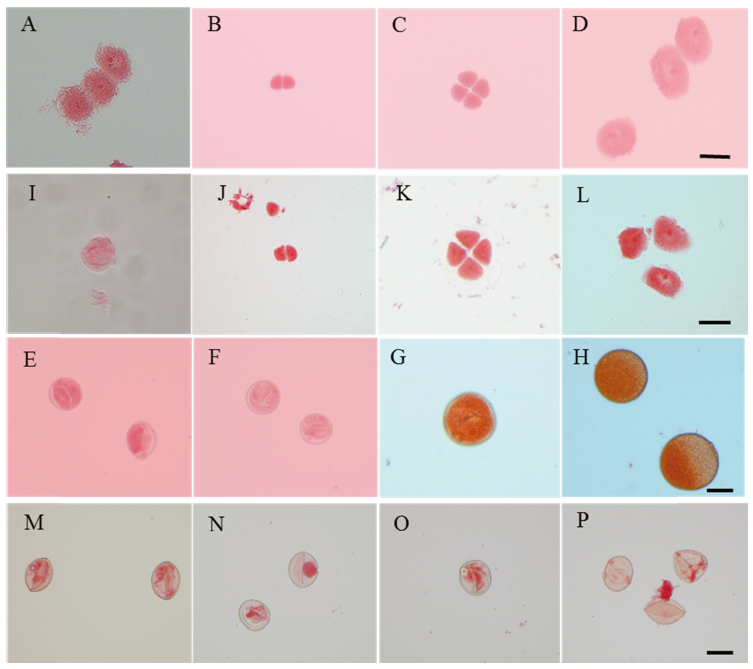
图2 野生型和突变体Osfma2的花粉发育过程 A~H为野生型花粉发育过程;I~P为突变体花粉发育过程。A和I—孢原细胞期;B和J—二分体期;C和K—四分体期;D和L—小孢子早期;E和M—小孢子晚期;F和N—二核早期;G和O—二核晚期;H和P—成熟花粉期。比例尺为10 μm。
Fig. 2. Developmental process of pollen in the wild type and the mutant Osfma2. A-H, Wild type; I-P, Mutant Osfma2. A and I, Archesporial cell; B and J, Dyads; C and K, Tetrads; D and L, Early microspore; E and M, Late microspore; F and N, Early bicellular pollen; G and O, Late bicellular pollen; H and P, Mature pollen. Bar=10 μm.

图3 野生型和突变体Osfma2的花药半薄切片观察 A~F为野生型花药;G~L为突变体花药。A和G—花药发育减数分裂之前;B和H—花药发育的8b阶段;C和I—花药发育的第9阶段;D和J—花药发育的第10阶段; E和K—花药发育的第11阶段;F和L—花药发育的成熟阶段。箭头指向: E—上皮; En—内层; ML—中间层; T—绒毡层; MMC—小孢子母细胞; Tds—四分体; Msp—小孢子壁细胞; BP—二核花粉; Mp—成熟花粉粒。比例尺为10 μm。
Fig. 3. Transverse section of anthers of the wild type and the mutant Osfma2. A-F, Wild type; G-L, Mutant. A and G, Before the stage of meiosis; B and H, 8b stage of anther development; C and I, The 9th stage of anther development; D and J, The 10th stage of anther development; E and K, The 11th stage of anther development; F and L, Mature stages of anther development. E, Epidermis; En, Endothecium; ML, Middle layer; T, Tapetum; MMC, Microspore mother cell; Tds, Tetrads; Msp, Microspore parietal cell; BP, Bicellular pollen; Mp, Mature pollen. Bar=10 μm.
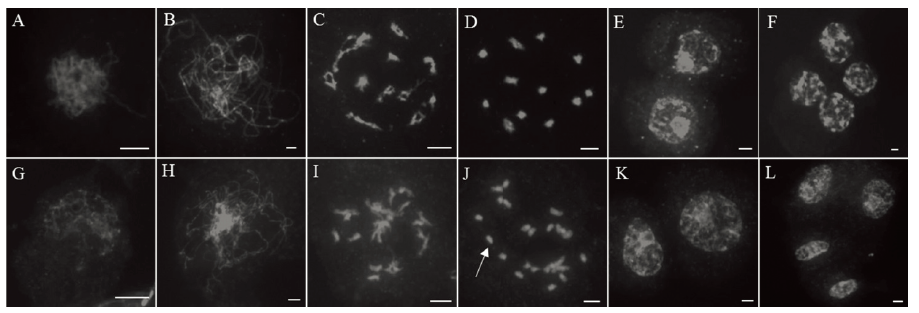
图4 野生型和突变体Osfma2孢原细胞染色体观察 A~F为野生型孢母细胞;G~L为突变体的孢母细胞。A和G—孢母细胞的偶线期;B和H—粗线期;C和I—双线期;D和J—终变期;E和K—二分体;F和L—四分体。白色箭头指向单价体。比例尺为5μm。
Fig. 4. Chromosome observation of archesporial cell of the wild type and the mutant Osfma2. A~F, Wild type; G~L, Mutant Osfma2. A and G, Zygotene; B and H, Pachytene; C and I, Diplotene; D and J, Diakinesis; E and K, Telophase I; F and L, Telophase II. The white arrows point to univalents. Bar=5μm.
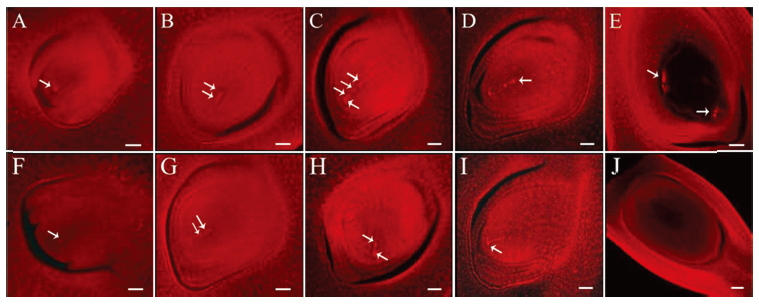
图5 野生型和突变体Osfma2胚囊发育的观察 A~E为野生型胚囊发育过程;F~J为突变体的胚囊发育过程。A和F—孢原细胞期;B和G—二分体期;C和H—四分体时期;D和I—功能大孢子期;E和J—成熟胚囊期。A~D、F~I的比例尺为30 μm;E和J的比例尺为60μm。
Fig. 5. Observation of embryo sac development of the wild type and the mutant Osfma2. A~E, Embryo sac development of wild type; F~J, Embryo sac development of mutant Osfma2. A and F, Archesporial cell stage; B and G, Dyad stage; C and H, Tetrad stage; D and I, Functional megaspore stage; E and J, Mature embryo stage. Bar=60 μm in E and J. Bar=30 μm in others.
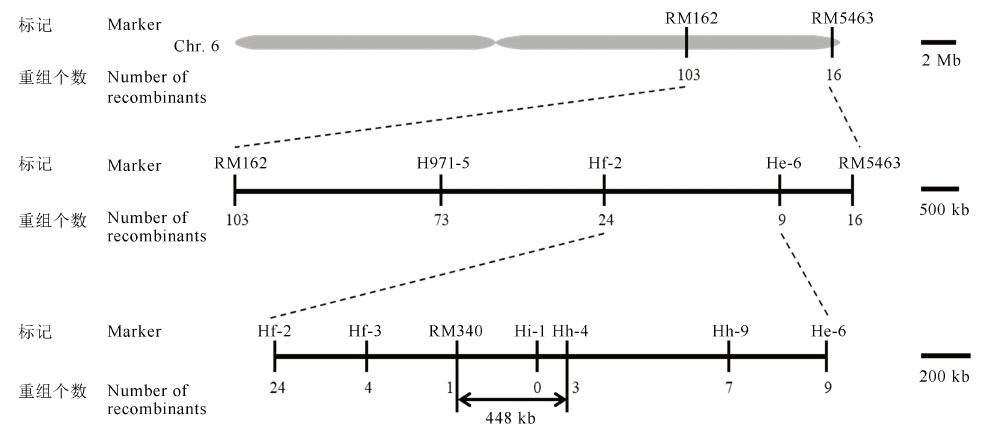
图6 OsFMA2基因在第6染色体上的分子定位 OsFMA2基因的初连锁和精细定位。OsFMA2基因最终定位在分子标记RM340和Hh-4之间448 kb的区间内。
Fig. 6. OsFMA2 gene mapping on chromosome 6. The preliminary and fine mapping of OsFMA2. The OsFMA2 gene was ultimately located in the 448-kb interval between the molecular markers RM340 and Hh-4.
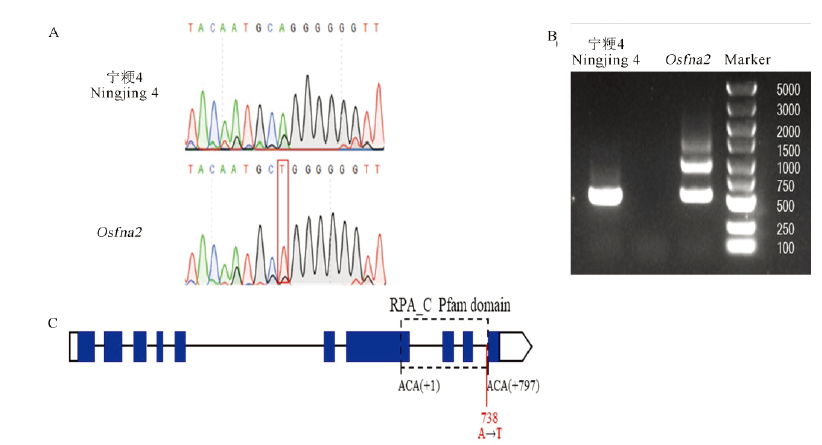
图8 不育基因OsFMA2的克隆和序列分析 A—野生型与突变体的测序差异;B—野生型和突变体中OsFMA2基因的差异剪切;C—OsFMA2的基因结构,蓝色框代表外显子,线条代表内含子,虚线框代表的是RPA_C保守结构域,从首位氨基酸的第一个碱基开始计算到末位氨基酸的最后一个碱基为止。红色实线代表突变位置。白色框分别代表5′ UTR和3′ UTR。
Fig. 8. Cloning and sequence analysis of the sterility gene OsFMA2. A, Sequencing differences between the wild type and the mutant; B, Differential splicing of the OsFMA2 gene in the wild type and the mutant; C, Gene structure of OsFMA2. The blue box represents the exon, the line represents the intron, and the dotted box represents the RPA_C conserved domain, which is calculated from the first base of the first amino acid to the last base of the last amino acid. The solid red line represents the mutation. The white boxes represent 5′ UTR and 3′ UTR.
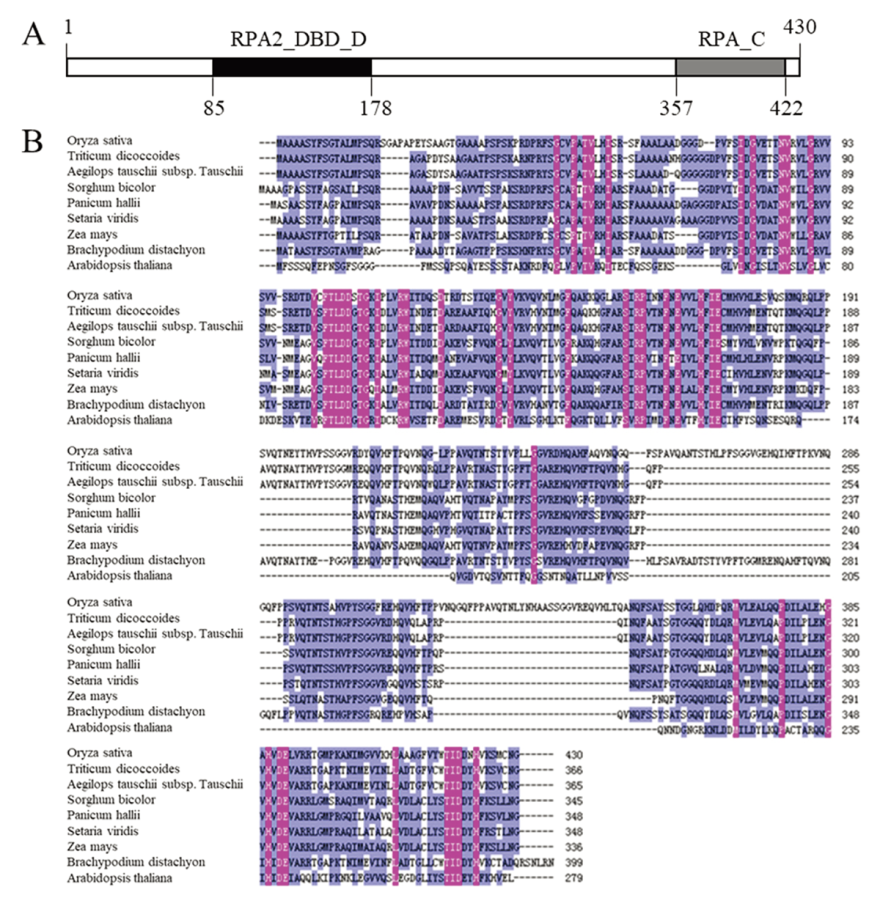
图9 OsFMA2生物信息学分析 A—OsFMA2蛋白结构预测,数字代表氨基酸残基位置;B—OsFMA2蛋白与其他物种中RPA蛋白序列同源比对。
Fig. 9. Bio-information analysis of OsFMA2. A, Protein-structure prediction of OsFMA2. The numbers indicate the position of amino acid residue at the OsFMA2; B, Multiple sequence alignment between OsFMA2 and RPA protein of other species.
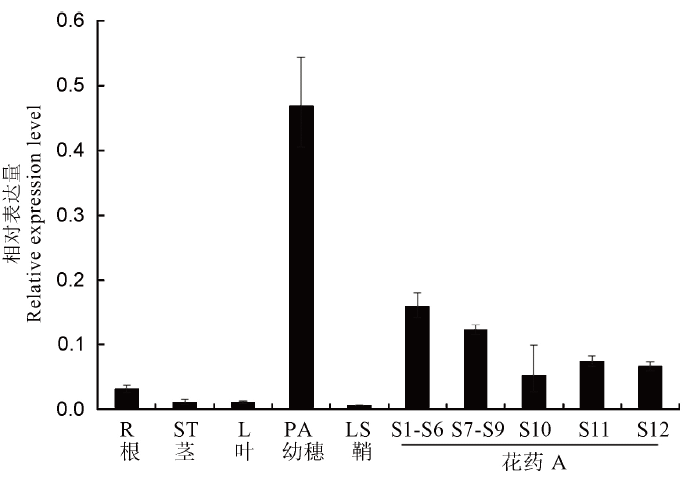
图11 OsFMA2基因在水稻各组织中的相对表达量 R—根;ST—茎秆;L—叶;PA—3mm幼穗;LS—叶鞘;A—花药;S1-S6表示小孢子母细胞形成期;S7-S9表示花药减数分裂期;S10—单核花粉期;S11—双核花粉期;S12—三核花粉期。内参为OsUbiquitin,误差线为n=3的标准差。
Fig. 11. Relative expression levels of OsFMA2 in different rice tissues. R, Root; ST, Stem; L, Leaf; PA, Panicle; LS, Leaf sheath; S1-S6, Microspore mother cell stage; S7-S9, Meiotic division of male gamete; S10, Mononucleate stage; S11, Binuclear stage; S12, Tri-nuclear stage. Osubiquitin was used as an internal control. Error bars show the SD (n=3).
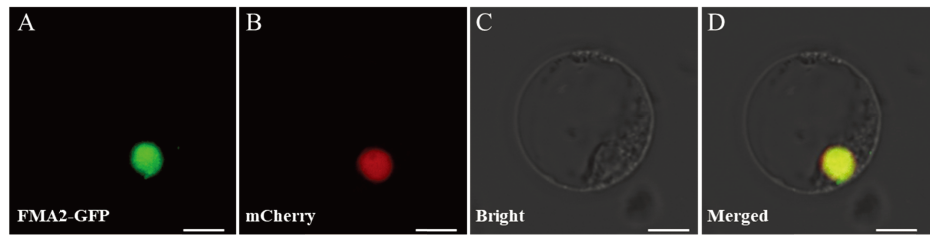
图12 OsFMA2蛋白在水稻原生质体中的亚细胞定位 A—OsFMA2-GFP融合蛋白在水稻原生质体中的亚细胞定位;B—核标签在水稻原生质体中的定位;C—明场视野中原生质体状态;D—GFP,核标签,明场的融合。比例尺为10 μm。
Fig. 12. Subcellular localization of OsFMA2 in rice protoplast. A, Subcellular localization of OsFMA2-GFP in rice protoplast; B, Localization of nuclear marker; C, Bright field of rice protoplast status; D, Merged image of GFP, nuclear marker and bright field. Bar=10 μm.
| [1] | 谭何新, 文铁桥, 张大兵. 水稻花粉发育的分子机理[J]. 植物学通报, 2007,24(3):330-339. |
| Tan H X, Wen T Q, Zhang D B. Molecular mechanisms of pollen development in Oryza sativa[J]. Chinese Bulletin of Botany, 2007,24(3):330-339. (in Chinese with English abstract) | |
| [2] | 马西青, 方才臣, 邓联武, 万向元. 水稻隐性核雄性不育基因研究进展及育种应用探讨[J]. 中国水稻科学, 2012,26(5):511-520. |
| Wan X Q, Fang C C, Deng L W, Wan X Y. Research progress and breeding application of recessive genic male sterility genes in rice[J]. Chinese Journal of Rice Science, 2012,26(5):511-520. (in Chinese with English abstract) | |
| [3] | 官文祥, 邓赟, 李小旭, 吴为人, 郑燕. 水稻雌性不育分子机理研究进展[J]. 分子植物育种, 2017,15(2):672-684. |
| Guan W X, Deng Y, Li X X, Wu W R, Zheng Y. Advances in research on molecular mechanism of female sterility in rice (Oryza sativa L.)[J]. Molecular Plant Breeding, 2017,15(2):672-684. (in Chinese with English abstract) | |
| [4] | 刘春宏, 方珊茹, 刘玉芹, 沈伟锋. 水稻雄性核不育基因的研究进展[J]. 台湾农业探索, 2012,19(1):71-75. |
| Liu C H, Fang S R, Liu Y Q, Shen W F. Research progress on genic male sterile genes in rice (Oryza sativa L)[J]. Taiwan Agricultural Research, 2012,19(1):71-75. (in Chinese with English abstract) | |
| [5] | Wang C, Liu Q, Shen Y, Hua Y, Wang J J, Lin J R, Wu M G, Sun T T, Cheng Z K, Mercier R. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes[J]. Nature Biotechnology, 2019,37(3):283-286. |
| [6] | Nonomur K, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N. The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis[J]. The Plant Cell, 2004,16(4):1008-1020. |
| [7] | Chang L, Ma H, Xue H W. Functional conservation of the meiotic genes SDS and RCK in male meiosis in the monocot rice[J]. Cell Research, 2009,19(6):768-782. |
| [8] | Yu H X, Wang M, Tang D, Wang K J, Chen F L, Gong Z Y, Gu M H, Cheng Z K. OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice[J]. Chromosoma, 2010,119(6):625-636. |
| [9] | Wang Y X, Copenhaver G. Meiotic recombination: mixing it up in plants[J]. Annual Review of Plant Biology, 2018,69(1):577-609. |
| [10] | Shingu Y, Tokai T, Agawa Y, Toyota K, Ahmed S, Kobayashi M, Komatsu A, Mikawa T, Yamamoto M, Wakasa K, Shibata T, Kusano K. The double-stranded break-forming activity of plant SPO11s and a novel rice SPO11 revealed by a Drosophila bioassay[J]. BMC Molecular Biology, 2012,13:1-16. |
| [11] | Zhang B W, Wang M, Tang D, Li Y F, Xu M, Gu M H, Cheng Z K, Yu H X. XRCC3 is essential for proper double-strand break repair and homologous recombination in rice meiosis[J]. Journal of Experimental Botany, 2015,66(19):5713-5725. |
| [12] | Deng Z Y, Wang T. OsDMC1 is required for homologous pairing in Oryza sativa[J]. Plant Molecular Biology, 2007,65(1-2):31-42. |
| [13] | Sheridan S, Yu X, Roth R, Heuser J, Sehorn M, Sung P, Egelman E, Bishop D. A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments[J]. Nucleic Acids Research, 2008,36(12):4057-4066. |
| [14] | Sakane I, Kamataki C, Takizawa Y, Nakashima M, Toki S, Ichikawa H, Ikawa S, Shibata T, Kurumizaka H. Filament formation and robust strand exchange activities of the rice DMC1A and DMC1B proteins[J]. Nucleic Acids Research, 2008,36(13):4266-4276. |
| [15] | Morozumi Y, Ino R, Ikawa S, Mimida N, Shimizu T, Toki S, Ichikawa H, Shibata T, Kurumizaka H. Homologous pairing activities of two rice RAD51 proteins, RAD51A1 and RAD51A2[J]. PloS ONE, 2013,8(10):e75451. |
| [16] | Vries S, Baart E, Dekker M, Siezen A, Rooij D, Boer P, Riele H. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis[J]. Gene & Development, 1999,13(5):523-531. |
| [17] | Mimitou E, Symington L. DNA end resection: Many nucleases make light work[J]. DNA Repair, 2009,8(9):983-995. |
| [18] | Youds J, Boulton S. The choice in meiosis defining the factors that influence crossover or non-crossover formation[J]. Journal of Cell Science, 2011,124(4):501-513. |
| [19] | Fairman M, Stillman B. Cellular factors required for multiple stages of SV40 DNA-replication in vitro[J]. The EMBO Journal, 1988,7(4):1211-1218. |
| [20] | Iftode C, Daniely Y, Borowiec J. Replication Protein A (RPA): The Eukaryotic SSB[J]. Critical Reviews in Biochemistry and Molecular Biology, 1999,34(3):141-180. |
| [21] | Osman K, Sanchez-Moran E, Mann S, Jones G, Franklin F. Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair[J]. The EMBO Journal, 2009,28(4):394-404. |
| [22] | Takashi Y, Kobayashi Y, Tanaka K, Tamura K. Arabidopsis replication protein A 70a is required for DNA damage response and telomere length homeostasis[J]. Plant Cell Physiology, 2009,50(11):1965-1976. |
| [23] | Aklilu B, Soderquist R, Culligan K. Genetic analysis of the replication protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication[J]. Nucleic Acids Research, 2014,42(5):3104-3118. |
| [24] | Chang Y X, Gong L, Yuan W Y, Li X W, Chen G X, Li X H, Zhang Q F, Wu C Y. Replication Protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice[J]. Plant Physiology, 2009,151(4):2162-2173. |
| [25] | Li X W, Chang Y X, Xin X D, Zhu C M, Li X H, Higgins J, Wu C Y. Replication protein A2c coupled with replication protein A1c regulates crossover formation during meiosis in rice[J]. The Plant Cell, 2013,25(10):3885-3899. |
| [26] | 冯九焕, 卢永根, 刘向东, 徐雪宾. 水稻花粉发育过程及其分期[J]. 中国水稻科学, 2001,15(1):22-29. |
| Feng J H, Lu Y G, Liu X D, Xu X B. Pollen development and its stages in rice (Oryza sativa L.)[J]. Chinese Journal of Rice Science, 2001,15(1):22-29. (in Chinese with English abstract) | |
| [27] | Zhang D B, Luo X, Zhu L. Cytological analysis and genetic control of rice anther development[J]. Journal of Genetics and Genomics, 2011,38(9):379-390. |
| [28] | Xia R, Wang J G, Liu C Y, Wang Y Q, Zhai J X, Liu J, Hong X H, Cao X F, Zhu J K, Gong Z Z. ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis[J]. The Plant Cell, 2006,18(1):85-103. |
| [29] | Belanger K, Griffith A, Baker H, Hansen J, Kovacs L, Seconi J, Strine A. The karyopherin Kap95 and the C-termini of Rfa1, Rfa2, and Rfa3 are necessary for efficient nuclear import of functional RPA complex proteins in Saccharomyces cerevisiae[J]. DNA and Cell Biology, 2011,30(9):641-651. |
| [30] | Keshav K F, Chen C, Dutta A. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex[J]. Molecular and Cellular Biology, 1995,15(6):3119-3128. |
| [31] | Zhang J, Han F P. Centromere pairing precedes meiotic chromosome pairing in plants[J]. Science China Life Sciences, 2017,60(11):1197-1202. |
| [32] | Simonet J, Zick D. Genes involved in caryogamy and meiosis in Podospora anserine[J]. Molecular Genetics Genomics, 1978,162(3):237-242. |
| [33] | Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M. Genome sequencing reveals agronomically important loci in rice using MutMap[J]. Nature Biotechnology, 30(2):174-178. |
| [34] | Ishibashi T, Kimura S, Sakaguchi K. A higher plant has three different types of RPA heterotrimeric complex[J]. Journal of Biochemistry, 2006,139(1):99-104. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||