
中国水稻科学 ›› 2020, Vol. 34 ›› Issue (5): 406-412.DOI: 10.16819/j.1001-7216.2020.0104
徐善斌1, 郑洪亮1, 刘利锋3, 卜庆云2, 李秀峰2,*( ), 邹德堂1,*(
), 邹德堂1,*( )
)
收稿日期:2020-01-10
修回日期:2020-04-16
出版日期:2020-09-10
发布日期:2020-09-10
通讯作者:
李秀峰,邹德堂
基金资助:
Shanbin XU1, Hongliang ZHENG1, Lifeng LIU3, Qingyun BU2, Xiufeng Li2,*( ), Detang ZOU1,*(
), Detang ZOU1,*( )
)
Received:2020-01-10
Revised:2020-04-16
Online:2020-09-10
Published:2020-09-10
Contact:
Xiufeng Li, Detang ZOU
摘要:
【目的】CRISPR/Cas9基因编辑技术已成为水稻分子育种的重要手段。为了促进水稻育种的发展,本研究以非香型粳稻品种龙粳11为试验材料,对GS3、GS9和Badh2基因进行编辑,以期获得能稳定遗传的长粒香水稻材料。【方法】利用CRISPR/Cas9技术,以GS3、GS9和Badh2为靶基因,构建敲除载体pYLCRISPR/Cas9-GS3/ GS9/Badh2-gRNA,通过农杆菌介导法,在龙粳11的GS3、GS9和Badh2基因中引入了特定的突变。【结果】T2代无转基因的gs3/gs9/badh2纯合突变体与野生型龙粳11相比,粒长增加26.43%~27.01%,单株产量增加10.82%~12.11%,千粒重增加18.34%~41.36%,稻米变香,高效地将圆粒水稻变成长粒香型水稻。【结论】利用CRISPR/Cas9技术获得能够稳定遗传并具有长粒香品质的纯合突变株系,为组合多个品质性状提供了一种方便有效的方法,从育种角度加快了新品系创制过程。
中图分类号:
徐善斌, 郑洪亮, 刘利锋, 卜庆云, 李秀峰, 邹德堂. 利用CRISPR/Cas9技术高效创制长粒香型水稻[J]. 中国水稻科学, 2020, 34(5): 406-412.
Shanbin XU, Hongliang ZHENG, Lifeng LIU, Qingyun BU, Xiufeng Li, Detang ZOU. Improvement of Grain Shape and Fragrance by Using CRISPR/Cas9 System[J]. Chinese Journal OF Rice Science, 2020, 34(5): 406-412.
| 引物名称 Primer name | 引物序列(5'-3') Primer sequence(5'-3') |
|---|---|
| B1-F | GCCGAGTGACATGGCAATGGCGG |
| B1-R | AAACCCGCCATTGCCATGTCACT |
| B2-F | GCCGCGATTGCTTCCTGCTCGGTT |
| B2-R | AAACAACCGAGCAGGAAGCAATCG |
| B3-F | GCCGCAAGTACCTCCGCGCAATCG |
| B3-R | AAACCGATTGCGCGGAGGTACTTG |
| U-F | CTCCGTTTTACCTGTGGAATCG |
| gRNA-R | CGGAGGAAAATTCCATCCAC |
| B1' | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| B2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| B2' | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| B3 | AGCGTGGGTCTCGTCTTGGTCCATCCACTCCAAGCTC |
| B3' | TTCAGAGGTCTCTAAGACACTGGAATCGGCAGCAAAGG |
| BL | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| Seq1-F | TTCAAAGCAAAGCACCAAGC |
| Seq1-R | GCTGGGGAAAACTTACAATG |
| Seq2-F | TCCACTCGACTCCTCACTCA |
| Seq2-R | GATTAGCGTAGGCCCTCACG |
| Seq3-F | ATCCATCTCCGTATCTCT |
| Seq3-R | GGTAGTCACCACCCTA |
| Hyg-F | ACGGTGTCGTCCATCACAGTTTGCC |
| Hyg-R | TTCCGGAAGTGCTTGACATTGGGGA |
| NOFS | GCGGTGTCATCTATGTTACTAG |
| M13F-47 | CGCCAGGGTTTTCCCAGTCACGAC |
表1 本研究中所用的引物
Table 1 Primers used in this research.
| 引物名称 Primer name | 引物序列(5'-3') Primer sequence(5'-3') |
|---|---|
| B1-F | GCCGAGTGACATGGCAATGGCGG |
| B1-R | AAACCCGCCATTGCCATGTCACT |
| B2-F | GCCGCGATTGCTTCCTGCTCGGTT |
| B2-R | AAACAACCGAGCAGGAAGCAATCG |
| B3-F | GCCGCAAGTACCTCCGCGCAATCG |
| B3-R | AAACCGATTGCGCGGAGGTACTTG |
| U-F | CTCCGTTTTACCTGTGGAATCG |
| gRNA-R | CGGAGGAAAATTCCATCCAC |
| B1' | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| B2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| B2' | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| B3 | AGCGTGGGTCTCGTCTTGGTCCATCCACTCCAAGCTC |
| B3' | TTCAGAGGTCTCTAAGACACTGGAATCGGCAGCAAAGG |
| BL | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| Seq1-F | TTCAAAGCAAAGCACCAAGC |
| Seq1-R | GCTGGGGAAAACTTACAATG |
| Seq2-F | TCCACTCGACTCCTCACTCA |
| Seq2-R | GATTAGCGTAGGCCCTCACG |
| Seq3-F | ATCCATCTCCGTATCTCT |
| Seq3-R | GGTAGTCACCACCCTA |
| Hyg-F | ACGGTGTCGTCCATCACAGTTTGCC |
| Hyg-R | TTCCGGAAGTGCTTGACATTGGGGA |
| NOFS | GCGGTGTCATCTATGTTACTAG |
| M13F-47 | CGCCAGGGTTTTCCCAGTCACGAC |
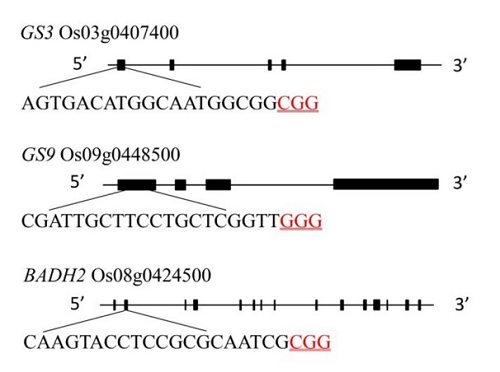
图1 GS3、GS9和Badh2基因结构和靶点位置黑色序列为靶点序列,红色下划线序列为PAM序列。
Fig. 1. Gene structure and target site of GS3, GS9 and Badh2. The black sequence is the target sequence and the red underlined sequence is the PAM sequence.
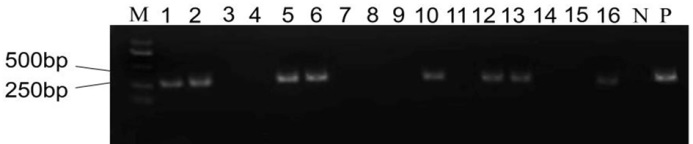
图3 T0代植株转基因检测 M-DM2000 DNA标记; N-阴性对照; P-阳性对照; 1~16代表T0植株。
Fig. 3. Transgenic detection of T0 generation plants. M, DM2000 DNA marker; N, Negative control; P, Positive control; Lanes 1-16, T0 generation plants.
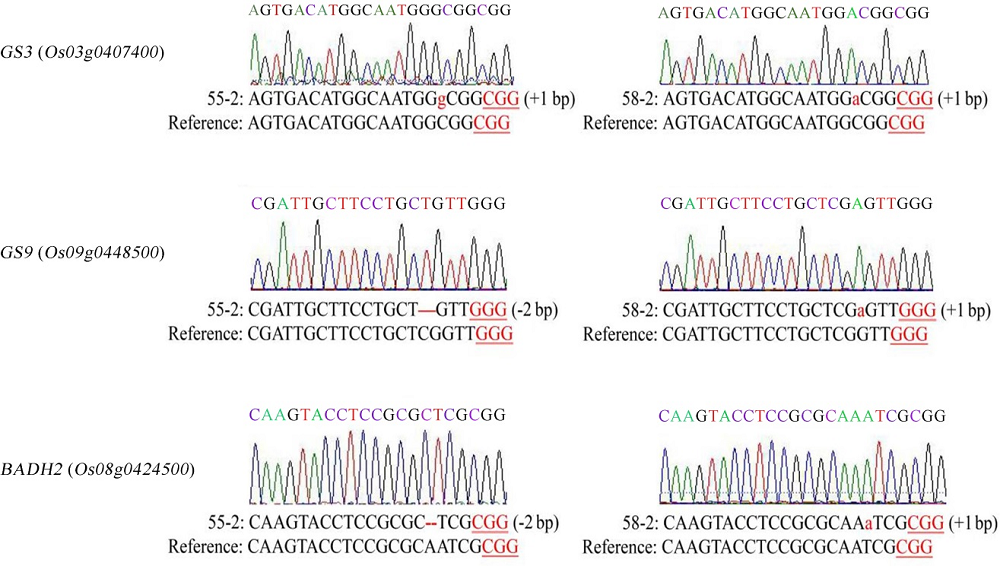
图4 三基因纯合突变植株测序结果红色小写字母表示插入1 bp的突变,红色连字符表示缺失的序列。
Fig. 4. Sequencing result of homozygous plants with triple gene mutation. Red lowercase letters represent 1 bp insertions and the deleted sequences are shown by red hyphens.
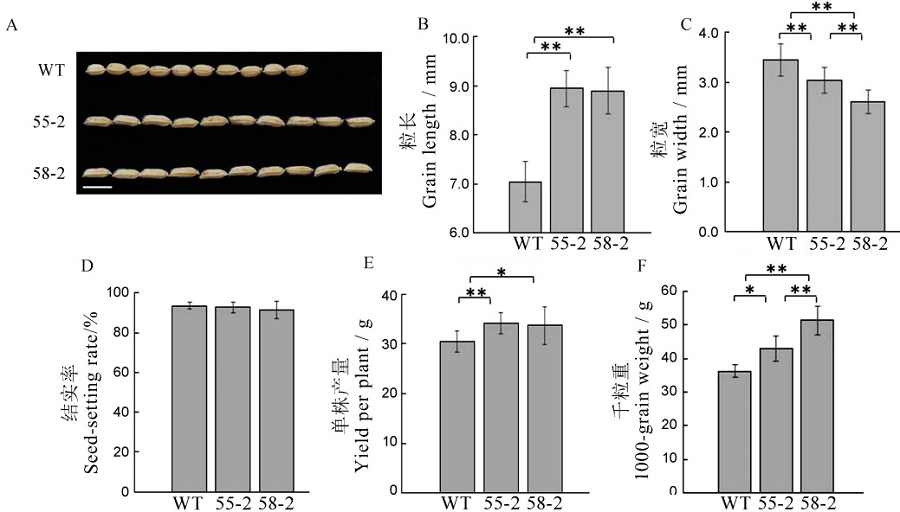
图5 T2代植株农艺性状考查数值用平均数±标准差表示,*,**分别表示在0.05和0.01水平上显著差异(t检验)。
Fig. 5. Agronomic traits of T2 generation plants. Values are shown as mean ± SD.*,**Significantly different at 0.05 and 0.01 levels by t-test, respectively.
| [1] | 郭韬, 余泓, 邱杰, 李家洋, 韩斌, 林鸿宣. 中国水稻遗传学研究进展与分子设计育种[J]. 中国科学: 生命科学, 2019, 49(10): 1185-1212. |
| Guo T, Yu H, Qiu J, Li J Y, Han B, Lin H X.Advances in rice genetics and breeding by molecular design in China[J]. Science in China: Life Sciences, 2019, 49(10): 1185-1212. (in Chinese with English abstract) | |
| [2] | Wang M G, Mao Y F, Lu Y M, Wang Z D, Tao X P, Zhu J K.Multiplex gene editing in rice with simplified CRISPR-Cpf1 and CRISPR-Cas9 systems[J]. Journal of Integrative Plant Biology, 2018, 60(8): 626-631. |
| [3] | Cai Y P, Chen L, Liu X J, Guo C, Sun S, Wu C X, Jiang B J, Han T F, Hou W S.CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean[J]. Plant Biotechnology Journal, 2018, 16(1): 176-185. |
| [4] | Qi W W, Zhu T, Tian Z R, Li C B, Zhang W, Song R T.High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize[J/OL].BMC Biotechnology, 2016, 16(1): 58. |
| [5] | Zhang S J, Zhang R Z, Song G Q, Gao J, Li W, Han X D, Chen M L, Li Y L, Li G Y.Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat[J/OL].BMC Plant Biology, 2018, 18(1): 302. |
| [6] | Wang P C, Zhang J, Sun L, Ma Y Z, Xu J, Liang S J, Deng J W, Tan J F, Zhang Q H, Tu L L, Daniell H, Jin S X, Zhang X L.High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system[J]. Plant Biotechnology Journal, 2018, 16(1): 137-150. |
| [7] | Liu G Q, Li J Q, Godwin I D.Genome editing by CRISPR/Cas9 in sorghum through biolistic bombardment[J]. Methods in Molecular Biology, 2019, 1931: 169-183. |
| [8] | Samanta M K, Dey A, Gayen S.CRISPR/Cas9: An advanced tool for editing plant genomes[J]. Transgenic Research, 2016, 25(5): 561-573. |
| [9] | Ji X, Zhang H W, Zhang Y, Wang Y P, Gao C X.Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants[J/OL].Nature Plants, 2015, 1: 15144. |
| [10] | Hsu P D, Lander E S, Zhang F.Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6): 1262-1278. |
| [11] | Smih F, Rouet P, Romanienko P J, Jasin M.Double strand breaks at the target locus stimulate gene targeting in embryonic stem cells[J]. Nucleic Acids Research, 1995, 23(24): 5012-5019. |
| [12] | Song X J, Huang W, Shi M, Zhu M Z, Lin H X.A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase[J]. Nature Genetics, 2007, 39(5): 623-630. |
| [13] | Fan C C, Xing Y Z, Mao H L, Lu T T, Han B, Xu C G, Li X H, Zhang Q F.GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein[J]. Theoretical and Applied Genetics, 2006, 112(6): 1164-1171. |
| [14] | Zhao D S, Li Q F, Zhang C Q, Zhang C, Yang Q Q, Pan L X, Ren X Y, Lu J, Gu M H, Liu Q Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality[J/OL]. Nature Communications, 2018, 9(1): 1240. |
| [15] | Liu J F, Chen J, Zheng X M, Wu F Q, Lin Q B, Heng Y Q, Tian P, Cheng Z J, Yu X W, Zhou K N, Zhang X, Guo X P, Wang J L, Wang H Y, Wan J M.GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice[J/OL].Nature Plants, 2017, 3: 17043. |
| [16] | Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B I, Onishi A, Miyagawa H, Katoh E.Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield[J]. Nature Genetics, 2013, 45(6): 707-711. |
| [17] | Che R H, Tong H N, Shi B H, Liu Y Q, Fang S R, Liu D P, Xiao Y H, Hu B, Liu L C, Wang H R, Zhao M F, Chu C C. Control of grain size and rice yield by GL2-mediated brassinosteroid responses[J/OL].Nature Plants, 2015, 2: 15195. |
| [18] | Qi P, Lin Y S, Song X J, Shen J B, Huang W, Shan J X, Zhu M Z, Jiang L, Gao J P, Lin H X.The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3[J]. Cell Research, 2012, 22(12): 1666-1680. |
| [19] | Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu C J, Li X H, Xiao J H, He Y Q, Zhang Q F.Natural variation in GS5 plays an important role in regulating grain size and yield in rice[J]. Nature Genetics, 2011, 43(12): 1266-1269. |
| [20] | Wang Y X, Xiong G S, Hu J, Jiang L, Yu H, Xu J, Fang Y X, Zeng L J, Xu E B, Xu J, Ye W J, Meng X B, Liu R F, Chen H Q, Jing Y H, Wang Y H, Zhu X D, Li J Y, Qian Q.Copy number variation at the GL7 locus contributes to grain size diversity in rice[J]. Nature Genetics, 2015, 47(8): 944-948. |
| [21] | Si L Z, Chen J Y, Huang X H, Gong H, Luo J H, Hou Q Q, Zhou T Y, Lu T T, Zhu J J, Shang G Y, Chen E W, Gong C X, Zhao Q, Jing Y F, Zhao Y, Li Y, Cui L L, Fan D L, Lu Y Q, Weng Q J, Wang Y C, Zhan Q L, Liu K Y, Wei X H, An K, An G, Han B.OsSPL13 controls grain size in cultivated rice[J]. Nature Genetics, 2016, 48(4): 447-456. |
| [22] | Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D.Control of grain size, shape and quality by OsSPL16 in rice[J]. Nature Genetics, 2012, 44(8): 950-954. |
| [23] | Li M R, Li X X, Zhou Z J, Wu P Z, Fang M C, Pan X P, Lin Q P, Luo W B, Wu G J, Li H Q.Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system[J/OL].Frontiers in Plant Science, 2016, 7: 377. |
| [24] | Sakthivel K, Sundaram R M, Rani N S, Balachandran S M, Neeraja C N.Genetic and molecular basis of fragrance in rice[J]. Biotechnology Advances, 2009, 27(4): 468-473. |
| [25] | Chen S H, Yang Y, Shi W W, Ji Q, He F, Zhang Z D, Cheng Z K, Liu X N, Xu M L.Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance[J]. Plant Cell, 2008, 20(7): 1850-1861. |
| [26] | Shan Q W, Zhang Y, Chen K L, Zhang K, Gao C X.Creation of fragrant rice by targeted knockout of the OsBadh2 gene using TALEN technology[J]. Plant Biotechnology Journal, 2015, 13(6): 791-800. |
| [27] | 孙慧宇, 宋佳, 王敬国, 刘化龙, 孙健, 莫天宇, 徐善斌, 郑洪亮, 邹德堂. 利用CRISPR/Cas9技术编辑Badh2基因改良粳稻香味[J]. 华北农学报, 2019, 34(4): 1-8. |
| Sun H Y, Song J, Wang J G, Liu H L, Sun J, Mo T Y, Xu S B, Zheng H L, Zou D T.Editing Badh2 gene to improve the fragrance of japonica rice by CRISPR/Cas9 technology[J]. Acta Agricultuae Boreali-Sinica, 2019, 34(4): 1-8. (in Chinese with English abstract) | |
| [28] | 曾栋昌, 马兴亮, 谢先荣, 祝钦泷, 刘耀光. 植物CRISPR/Cas9多基因编辑载体构建和突变分析的操作方法[J]. 中国科学: 生命科学, 2018, 48(7): 783-794. |
| Zeng D C, Ma X L, Xie X R, Zhu Q L, Liu Y G.A protocol for CRISPR/Cas9-based multi-gene editing and sequence decoding of mutant sites in plants[J]. Science in China: Life Sciences, 2018, 48(7): 783-794. (in Chinese with English abstract) | |
| [29] | Xie X R, Ma X, Zhu Q L, Zeng D C, Li G S, Liu Y G, CRISPR-GE: A convenient software toolkit for CRISPR- based genome editing[J]. Molecular Plant, 2017, 10(9): 1246-1249. |
| [30] | Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G.A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants[J]. Molecular Plant, 2015, 8(8): 1274-1284. |
| [31] | Liu W Z, Xie X R, Ma X L, Li J, Chen J H, Liu Y G.DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations[J]. Molecular Plant, 2015, 8(9): 1431-1433. |
| [32] | Zhang Y C, Li S M, Xue S J, Yang S H, Huang J, Wang L.Phylogenetic and CRISPR/Cas9 studies in deciphering the evolutionary trajectory and phenotypic impacts of rice ERECTA genes[J/OL]. Frontiers in Plant Science, 2018, 9: 473. |
| [33] | Wang M, Mao Y F, Lu Y M, Wang Z D, Tao X P, Zhu J K.Multiplex gene editing in rice with simplified CRISPR-Cpf1 and CRISPR-Cas9 systems[J]. Journal of Integrative Plant Biology, 2018, 60(8): 626-631. |
| [34] | Zhang J S, Zhang H, Botella J R, Zhu J K.Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties[J]. Journal of Integrative Plant Biology, 2018, 60(5): 369-375. |
| [35] | Wang Y, Geng L Z, Yuan M L, Wei J, Jin C, Li M, Yu K, Zhang Y, Jin H B, Wang E, Chai Z J, Fu X D, Li X G.Deletion of a target gene in indica rice via CRISPR/Cas9[J]. Plant Cell Reports, 2017, 36(8): 1333-1343. |
| [36] | Nan J Z, Feng X M, Wang C, Zhang X H, Wang R S, Liu J X, Yuan Q B, Jiang G Q, Lin S Y.Improving rice grain length through updating the GS3 locus of an elite variety Kongyu 131[J]. Rice, 2018, 11(1): 21. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 丁正权, 潘月云, 施扬, 黄海祥. 基于基因芯片的嘉禾系列长粒优质食味粳稻综合评价与比较[J]. 中国水稻科学, 2024, 38(4): 397-408. |
| [5] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [6] | 吕宙, 易秉怀, 陈平平, 周文新, 唐文帮, 易镇邪. 施氮量与移栽密度对小粒型杂交水稻产量形成的影响[J]. 中国水稻科学, 2024, 38(4): 422-436. |
| [7] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [8] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [9] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [10] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [11] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [12] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [13] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [14] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [15] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||