
中国水稻科学 ›› 2019, Vol. 33 ›› Issue (1): 75-84.DOI: 10.16819/j.1001-7216.2019.8035
徐红星1, 王国荣2, 鲁艳辉1,*( ), 杨亚军1, 郑许松1, 田俊策1, 吕仲贤1,*(
), 杨亚军1, 郑许松1, 田俊策1, 吕仲贤1,*( )
)
收稿日期:2018-03-27
修回日期:2018-05-15
出版日期:2019-01-10
发布日期:2019-01-10
通讯作者:
鲁艳辉,吕仲贤
基金资助:
Hongxing XU1, Guorong WANG2, Yanhui LU1,*( ), Yajun YANG1, Xusong ZHENG1, Junce TIAN1, Zhongxian LÜ1,*(
), Yajun YANG1, Xusong ZHENG1, Junce TIAN1, Zhongxian LÜ1,*( )
)
Received:2018-03-27
Revised:2018-05-15
Online:2019-01-10
Published:2019-01-10
Contact:
Yanhui LU, Zhongxian LÜ
摘要:
【目的】筛选特定试验条件下二化螟(Chilo suppressalis)稳定表达的内参基因,为二化螟基因表达研究奠定基础。【方法】根据二化螟转录组测序结果挑选出11个候选基因,利用实时荧光定量PCR测定其在二化螟不同发育历期、不同组织、温度处理、杀虫剂处理、取食不同饲料、取食不同水稻、dsRNA处理和混合样品的表达量,利用RefFinder、BestKeeper、GeNorm和NormFinder等软件和ΔCt值对11个候选基因的稳定性进行评估。【结果】5种分析方法表明,在二化螟不同发育历期稳定性较高的内参基因是AK、RPL10和EF1,不同组织中稳定性较高的内参基因是EF1、TUB和ACTB,不同温度下较稳定的内参基因为TUB、RPL10和EF1,杀虫剂处理样品中较稳定的是TF4和ACTA,取食不同饲料较稳定的是TUB、TF4、EF1和RPL10,取食不同品种水稻较稳定的是TUB和EF1,dsRNA处理样品中较稳定的是TUB、AK、ACTB和EF1,混合样品中较稳定的是EF1、TUB和ACTB。【结论】为不同试验条件下选择合适的内参基因提供了参考,也有利于在二化螟基因表达研究中获得更加可靠准确的数据。
中图分类号:
徐红星, 王国荣, 鲁艳辉, 杨亚军, 郑许松, 田俊策, 吕仲贤. 二化螟实时荧光定量PCR内参基因筛选和表达稳定性评价[J]. 中国水稻科学, 2019, 33(1): 75-84.
Hongxing XU, Guorong WANG, Yanhui LU, Yajun YANG, Xusong ZHENG, Junce TIAN, Zhongxian LÜ. Screening Reference Genes and Evaluating of Their Expression Stability for qRT-PCR Normalization in Chilo suppressalis (Lepidoptera: Pyralididae)[J]. Chinese Journal OF Rice Science, 2019, 33(1): 75-84.
| 基因名(缩写) Gene name (Gene symbol) | 引物序列 Sequence (5′-3′) | 基因功能 Putative function | PCR扩增效率E/% | 决定系数Coefficient of determination | ||
|---|---|---|---|---|---|---|
| Beta微管蛋白 | F: CTCCGACTTACAGTTAGAGC | 细胞骨架结构蛋白 Cytoskeleton structural protein | 104.2 | 0.994 | ||
| Beta-tubulin(TUB) | R: AGTACTGAATCGACAAGCTC | |||||
| 延长因子-1α | F: CTGGGTATTGGACAAACTGA | 核糖体的组成结构 Structural constituent of ribosome | 107.0 | 0.997 | ||
| Elongation factor-1α(EF1) | R: GAGGTTCCTGTGATCATGTT | |||||
| 转录因子4 | F: ATTGCTGTGATAAAGAAGAAC | 催化GTP结合到核糖体的受体 Catalyze GTP binding to the acceptor of ribosome | 98.7 | 0.993 | ||
| Transcription factor 4(TF4) | R: AGAAGGTGGTGGACTCAAC | |||||
| 甘油醛-3-磷酸脱氢酶Glyceraldehyde-3-phosphate dehydrogenase(GAPDH) | F: GGGTATTCTTGACTACAC | 糖酵解酶Glycolytic enzyme | 105.9 | 0.995 | ||
| R: CTGGATGTACTTGATGAG | ||||||
| 肌动蛋白A1 | F: GTCGCTTCCCAAATTACATC | 参与细胞的运动、结构和完整性Involved in cell motility, structure and integrity | 103.5 | 0.987 | ||
| Actin A1(ACTA) | R: CTCCATATCGTTCCAGTCG | |||||
| Beta肌动蛋白 | F: GATCATGTTTGAGACCTT | 参与细胞的运动、结构和完整性Involved in cell motility, structure and integrity | 97.6 | 0.992 | ||
| Beta-actin(ACTB) | R: GATCTTCATGAGGTAGTC | |||||
| 精氨酸激酶 | F: CTGAAGAAGTACCTTACC | 细胞能量代谢关键酶Key enzyme F cellular energy metabolism | 99.4 | 0.996 | ||
| Argininase kinase(AK) | R: CAATCCAGCAGAGTTGAG | |||||
| 核糖体蛋白S2 | F: CAACGATGAGGTCTTGAAGA | 核糖体的组成结构 Structural constituent of ribosome | 102.1 | 0.990 | ||
| Ribosomal protein S2(RPS2) | R: CGATCTTGTTACCCCAGTAG | |||||
| 核糖体蛋白S3 | F: CGGAGATCATCATTATGG | 核糖体的组成结构 Structural constituent of ribosome | 103.5 | 0.989 | ||
| Ribosomal protein S3(RPS3) | R: GAGTTTGTATCTGAGAGAC | |||||
| 核糖体蛋白S5 | F: TACTGCCATAATCAACTCCG | 核糖体的组成结构 Structural constituent of ribosome | 105.4 | 0.992 | ||
| Ribosomal protein S5(RPS5) | R: TTAGATGAACCCTTAGCAGC | |||||
| 核糖体蛋白L10 | F: GACTTGGGTAAGAAGAAG | 核糖体的组成结构 Structural constituent of ribosome | 101.4 | 0.989 | ||
| Ribosomal protein L10(RPL10) | R: GATGACATGGAATGGATG | |||||
表1 备选基因的功能、引物序列和产物特性
Table 1 Function, primer sequence and amplicon characteristics of the candidate reference genes used in this study.
| 基因名(缩写) Gene name (Gene symbol) | 引物序列 Sequence (5′-3′) | 基因功能 Putative function | PCR扩增效率E/% | 决定系数Coefficient of determination | ||
|---|---|---|---|---|---|---|
| Beta微管蛋白 | F: CTCCGACTTACAGTTAGAGC | 细胞骨架结构蛋白 Cytoskeleton structural protein | 104.2 | 0.994 | ||
| Beta-tubulin(TUB) | R: AGTACTGAATCGACAAGCTC | |||||
| 延长因子-1α | F: CTGGGTATTGGACAAACTGA | 核糖体的组成结构 Structural constituent of ribosome | 107.0 | 0.997 | ||
| Elongation factor-1α(EF1) | R: GAGGTTCCTGTGATCATGTT | |||||
| 转录因子4 | F: ATTGCTGTGATAAAGAAGAAC | 催化GTP结合到核糖体的受体 Catalyze GTP binding to the acceptor of ribosome | 98.7 | 0.993 | ||
| Transcription factor 4(TF4) | R: AGAAGGTGGTGGACTCAAC | |||||
| 甘油醛-3-磷酸脱氢酶Glyceraldehyde-3-phosphate dehydrogenase(GAPDH) | F: GGGTATTCTTGACTACAC | 糖酵解酶Glycolytic enzyme | 105.9 | 0.995 | ||
| R: CTGGATGTACTTGATGAG | ||||||
| 肌动蛋白A1 | F: GTCGCTTCCCAAATTACATC | 参与细胞的运动、结构和完整性Involved in cell motility, structure and integrity | 103.5 | 0.987 | ||
| Actin A1(ACTA) | R: CTCCATATCGTTCCAGTCG | |||||
| Beta肌动蛋白 | F: GATCATGTTTGAGACCTT | 参与细胞的运动、结构和完整性Involved in cell motility, structure and integrity | 97.6 | 0.992 | ||
| Beta-actin(ACTB) | R: GATCTTCATGAGGTAGTC | |||||
| 精氨酸激酶 | F: CTGAAGAAGTACCTTACC | 细胞能量代谢关键酶Key enzyme F cellular energy metabolism | 99.4 | 0.996 | ||
| Argininase kinase(AK) | R: CAATCCAGCAGAGTTGAG | |||||
| 核糖体蛋白S2 | F: CAACGATGAGGTCTTGAAGA | 核糖体的组成结构 Structural constituent of ribosome | 102.1 | 0.990 | ||
| Ribosomal protein S2(RPS2) | R: CGATCTTGTTACCCCAGTAG | |||||
| 核糖体蛋白S3 | F: CGGAGATCATCATTATGG | 核糖体的组成结构 Structural constituent of ribosome | 103.5 | 0.989 | ||
| Ribosomal protein S3(RPS3) | R: GAGTTTGTATCTGAGAGAC | |||||
| 核糖体蛋白S5 | F: TACTGCCATAATCAACTCCG | 核糖体的组成结构 Structural constituent of ribosome | 105.4 | 0.992 | ||
| Ribosomal protein S5(RPS5) | R: TTAGATGAACCCTTAGCAGC | |||||
| 核糖体蛋白L10 | F: GACTTGGGTAAGAAGAAG | 核糖体的组成结构 Structural constituent of ribosome | 101.4 | 0.989 | ||
| Ribosomal protein L10(RPL10) | R: GATGACATGGAATGGATG | |||||
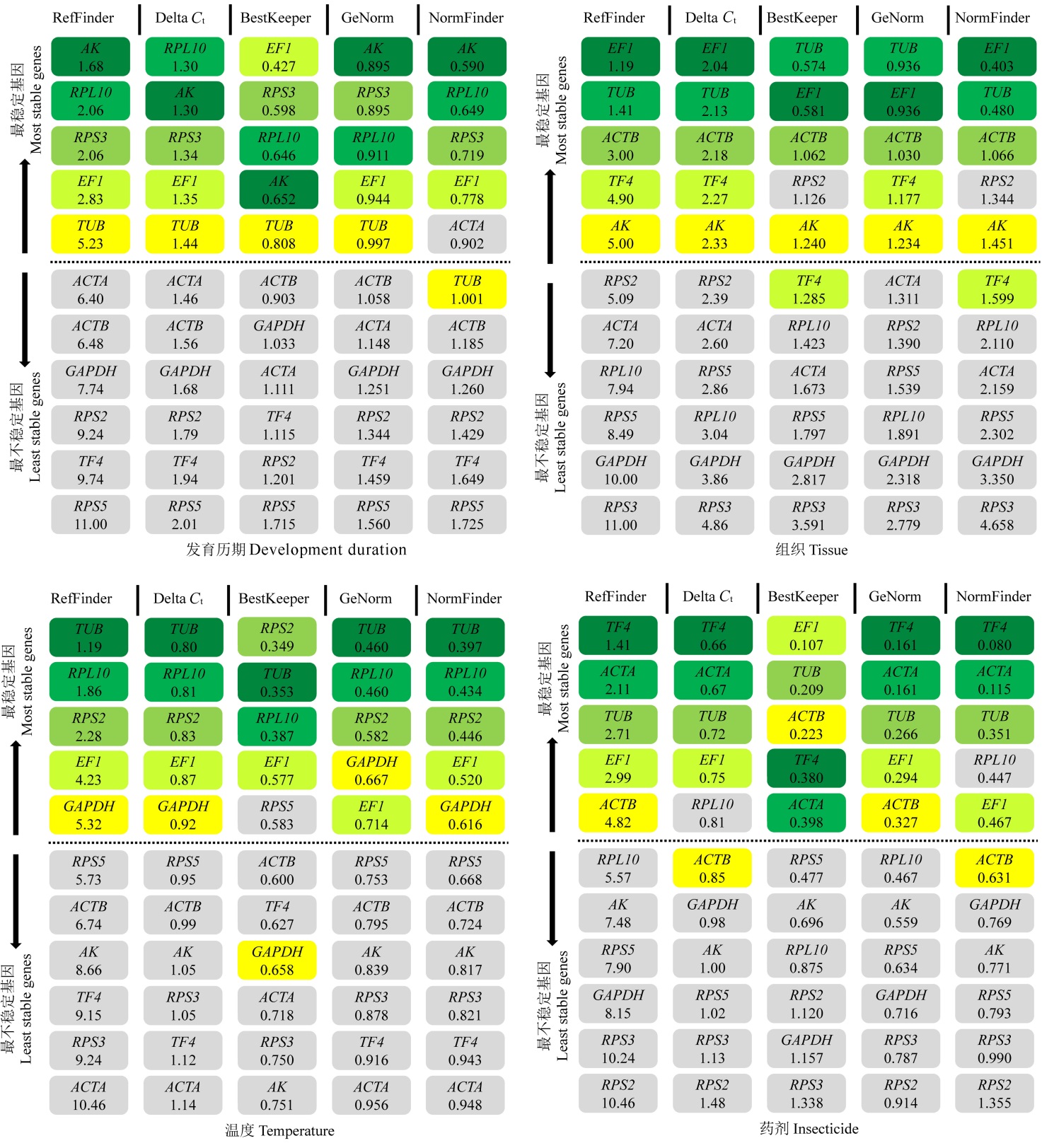
图2 候选内参基因在不同发育历期、不同组织、不同温度处理、不同杀虫剂处理下的表达稳定性分析
Fig. 2. Comparison of the rankings in candidate reference genes expression stability in development durations of C. suppressalis, across different tissues of C. suppressalis, exposed to different temperatures or insecticides.
| 发育历期 Development duration | 不同组织Tissue | 温度处理 Temperature | 杀虫剂处理Insecticide | 喂食处理Diet | 转基因处理 Transgenic rice | dsRNA | 综合分析 Total |
|---|---|---|---|---|---|---|---|
| AK | EF1 | TUB | TF4 | TUB | TUB | TUB | EF1 |
| RPL10 | TUB | RPL10 | ACTA | TF4 | EF1 | AK | TUB |
| EF1 | ACTB | EF1 | EF1 | ACTB | ACTB | ||
| RPL10 | EF1 |
表2 不同实验条件下的理想内参基因
Table 2 Ideal reference genes across different experimental conditions according to software analysis.
| 发育历期 Development duration | 不同组织Tissue | 温度处理 Temperature | 杀虫剂处理Insecticide | 喂食处理Diet | 转基因处理 Transgenic rice | dsRNA | 综合分析 Total |
|---|---|---|---|---|---|---|---|
| AK | EF1 | TUB | TF4 | TUB | TUB | TUB | EF1 |
| RPL10 | TUB | RPL10 | ACTA | TF4 | EF1 | AK | TUB |
| EF1 | ACTB | EF1 | EF1 | ACTB | ACTB | ||
| RPL10 | EF1 |
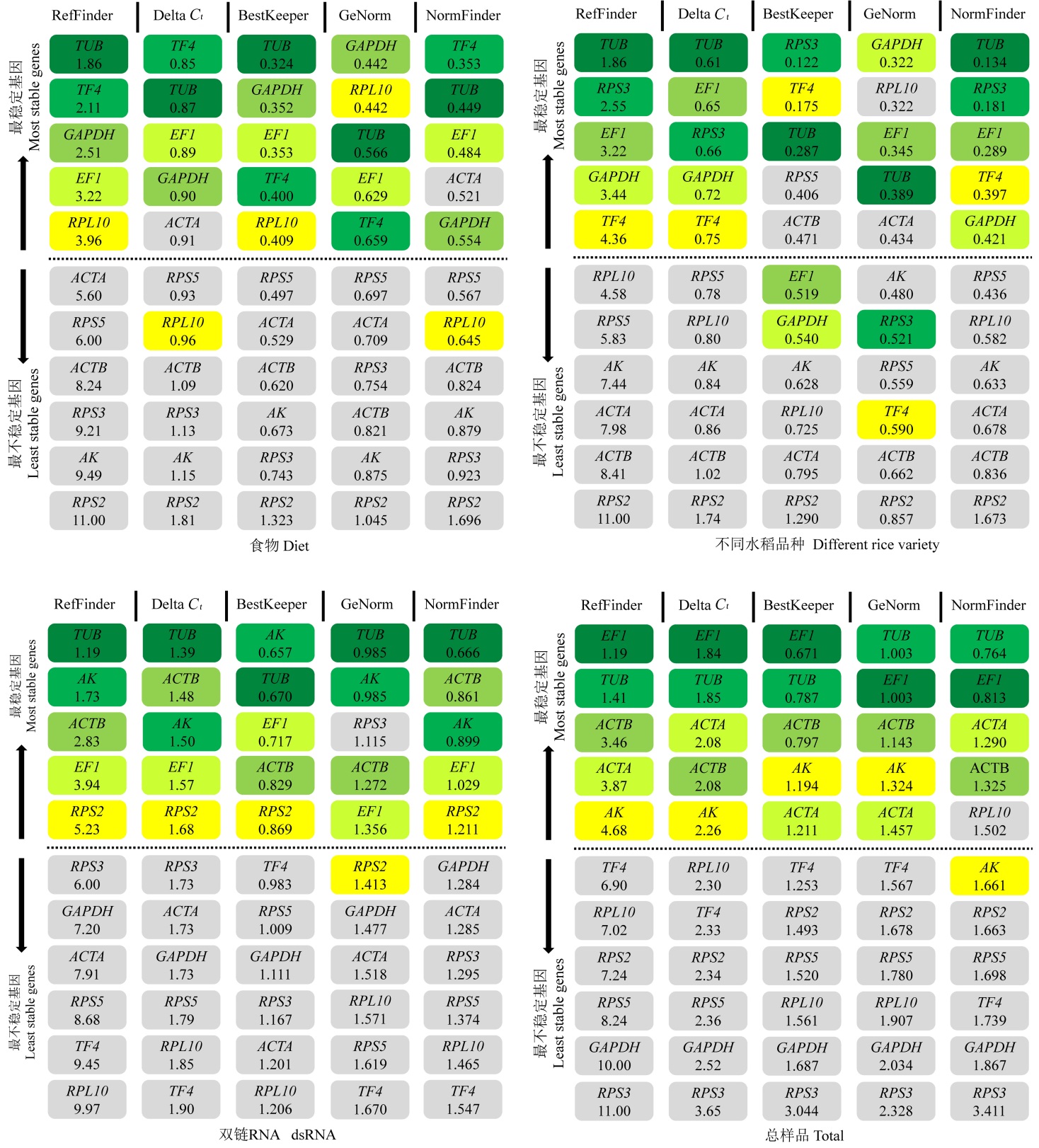
图3 候选内参基因在不同喂食处理、取食不同水稻品种、经dsRNA处理、在所有供试样品中的表达稳定性分析
Fig. 3. Comparison of the rankings in candidate reference gene expression stability of C. suppressalis fed on different diets, reared on different rice varieties, under dsRNA treatments or in all the tested samples of C. suppressalis.
| [1] | Bustin S A.Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays.J Mol Endocrinol, 2000, 25: 169-193. |
| [2] | Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E.Housekeeping genes as internal standards: Use and limits.J Biotechnol, 1999, 75: 291-295. |
| [3] | de Jonge H J, Fehrmann R S, de Bont E S, Hofstra R M, Gerbens F, Kamps W A, de Vries E G, van der Zee A G, te Meerman G J, ter Elst A. Evidence based selection of housekeeping genes.PLoS One, 2007, 2: e898. |
| [4] | Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O.Towards a systematic validation of references in real-time RT-PCR.Plant Cell, 2008, 20: 1734-1735. |
| [5] | Huggett J, Dheda K, Bustin S, Zumla A.Real-time RT-PCR normalisation; strategies and considerations.Genes Immun, 2005, 6: 279-284. |
| [6] | Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol, 2002, 3: Research0034. |
| [7] | 符伟, 谢文, 张卓, 吴青君, 王少丽, 张友军. Bt毒素诱导下小菜蛾实时定量PCR 内参基因的筛选. 昆虫学报, 2012, 55(12): 1406-1412. |
| Fu W, Xie W, Zhang Z, Wu Q J, Wang S L, Zhang Y J.Selection of valid reference genes for gene expression studies by quantitative real-time PCR in Plutella xylostella(Lepidoptera: Plutellidae) after exposure to Bt toxin. Acta Entomol Sin, 2012, 55(12): 1406-1412. (in Chinese with English abstract) | |
| [8] | Lu Y H, Yuan M, Gao X W, Kang T H, Zhan S, Wan H, Li J H.Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura(Lepidoptera: Noctuidae). PLoS One, 2013, 8: e68059. |
| [9] | Chandra GS, Asokan R, Manamohan M, Kumar N K, Sita T.Evaluation of reference genes for quantitative real-time PCR normalization in cotton bollworm,Helicoverna armigera. Mol Biol, 2014, 48: 927-938. |
| [10] | Shakeel M, Zhu X, Kang T, Wan H, Li J.Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm,Helicoverpa armigera(Lepidoptera: Noctuidae). J Asia-Pac Entomol, 2015, 18: 123-130. |
| [11] | Zhang S D, An S H, Li Z, Wu F M, Yang Q P, Liu Y C, Cao J J, Zhang H J, Zhang Q W, Liu X X.Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera(Lepidoptera: Noctuidae). Gene, 2015, 555: 393-402. |
| [12] | Zhu X, Yuan M, Shakeel M, Zhang Y J, Wang S L, Wang X, Zhan S, Kang T H, Li J H.Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua(Hubner),(Lepidoptera: Noctuidae). PLoS One, 2014, 9: e84730. |
| [13] | Arun A, Baumle V, Amelot G, Nieberding C M.Selection and validation of reference genes for qRT-PCR expression analysis of candidate genes involved in olfactory communication in the butterfly Bicyclus anynana. PLoS One, 2015, 10: e0120401. |
| [14] | Lu Y H, Zheng X S, Liang Q, Xu H X, Yang Y J, Tian J C, He X C, Lu Z X.Evaluation and validation of reference genes for SYBR Green qRT-PCR normalization in Sesamia inferens(Lepidoptera: Noctuidae). J Asia-Pac Entomol, 2015, 18: 669-675. |
| [15] | Sun M, Lu M X, Tang X T, Du Y Z.Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens(Lepidoptera: Noctuidae). PLoS One, 2015, 10: e0115979. |
| [16] | Pan H P, Yang X W, Bidne K, Hellmich R L, Siegfried B D, Zhou X G.Selection of reference genes for RT-qPCR analysis in the monarch butterfly, Danaus plexippus(L.), a migrating bio-Indicator. PLoS One, 2015, 10: e0129482. |
| [17] | Liu G Q, Qiu X H, Cao L, Zhang Y, Zhan Z B, Han R C.Evaluation of reference genes for reverse transcription quantitative PCR studies of physiological responses in the ghost moth,Thitarodes armoricanus(Lepidoptera, Hepialidae). PLoS One, 2016, 11: e0159060. |
| [18] | Zhang L, Zhang Q L, Wang X T, Yang X Z, Li X P, Yuan M L.Selection of reference genes for qRT-PCR and expression analysis of high-altitude-related genes in grassland caterpillars (Lepidoptera: Erebidae: Gynaephora) along an altitude gradient.Ecol Evol, 2017, 7: 9054-9065. |
| [19] | 杨苓, 胡晓静, 徐志峰, 何林, 肖伟. 桃蛀螟实时荧光定量PCR内参基因的筛选. 昆虫学报, 2017, 60(11): 1266-1277. |
| Yang L, Hu X J, Xu Z F, He L, Xiao W.Screening of reference genes for qRT-PCR in Conogethes punctiferails(Lepidoptera: Crambidae). Acta Entomol Sin, 2017, 60(11): 1266-1277. (in Chinese with English abstract) | |
| [20] | Teng X L, Zhang Z, He G L, Yang L W, Li F.Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects.J Insect Sci, 2011, 12: 60. |
| [21] | Xu J, Lu M X, Cui Y D, Du Y Z.Selection and evaluation of reference genes for expression analysis using qRT-PCR in Chilo suppressalis(Lepidoptera: Pyralidae). J Econ Entomol, 2017, 110: 683-691. |
| [22] | 胡阳, 郑永利, 曹国连, 傅强. 利用半人工饲料大规模简便化饲养二化螟中国水稻科学, 2013, 27(5): 535-538. |
| Hu Y, Zheng Y L, Cao G L, Fu Q. A technique for rearing Chilo suppressalis in the large scale with an oligidic diet in laboratory. Chin J Rice Sci, 2013, 27(5): 535-538. (in Chinese with English abstract) | |
| [23] | 吴敏, 张真真, 高聪芬. 水稻二化螟抗药性监测方法. 应用昆虫学报, 2013, 50(2): 548-552. |
| Wu M, Zhang Z Z, Gao C F.Methods of insecticides resistance monitoring of the striped stem borer, Chilo suppressalis.Chin J Appl Entomol, 2013, 50(2):548-552. (in Chinese with English abstract) | |
| [24] | Hui X M, Yang W, He G L, Yang Q P, Han Z, Li F.RNA interference of ace1 and ace2 in Chilo suppressalis reveals their different contributions to motor ability and larval growth. Insect Mol Biol, 2011, 4: 507-518. |
| [25] | Pfaffl M W, Tichopad A, Prgomet C, Neuvians T P.Determination of stable housekeeping genes, differentially regulated target genes and sample integrity. BestKeeper: Excel-based tool using pair-wise correlations.Biotechnol Lett, 2004, 26: 509-515. |
| [26] | Andersen C L, Jensen J L, Orntoft T F.Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets.Cancer Res, 2004, 64: 5245-5250. |
| [27] | Schmittgen T D, Livak K J.Analyzing real-time PCR data by the comparative CT method.Nat Protoc, 2008, 3: 1101-1108. |
| [28] | Radonic A, Thulke S, Mackay I M, Landt O, Siegert W, Nitsche A.Guideline to reference gene selection for quantitative real-time PCR.Biochem Biophys Res Commun, 2004, 313: 856-862. |
| [29] | Huis R, Hawkins S, Neutelings G.Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol, 2010, 10: 71. |
| [30] | Kozera B, Rapacz M.Reference genes in real-time PCR. J Appl Genet, 2013, 54: 391-406. |
| [31] | 冯波, 郭前爽, 毛必鹏, 杜永均. 松墨天牛化学感受组织荧光定量PCR内参基因的鉴定与筛选. 昆虫学报, 2016, 59(4): 427-437. |
| Feng B, Guo Q, Mao B P, Du Y J.Identification and selection of valid reference genes for assaying gene expression in the chemosensory tissues of Monochamus alternatus(Coleoptera: Cerambycidae) by RT-PCR. Acta Entomol Sin, 2016, 59(4): 427-437. (in Chinese with English abstract) | |
| [32] | Nielsen M G, Gadagkar S R, Gutzwiller L.Tubulin evolution in insects: Gene duplication and subfunction- alization provide specialized isoforms in a functionally constrained gene family.BMC Evol Biol, 2010, 10: 113. |
| [33] | Yuan M, Lu Y H, Zhu X, Wan H, Shakeel M, Zhan S, Jin B R, Li J H.Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper,Nilaparvata lugens(Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One, 2014, 9: e86503. |
| [34] | An X K, Hou M L, Liu Y D.Reference gene selection and evaluation for gene expression studies using qRT-PCR in the white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae).J Econ Entomol, 2016, 109: 879-886. |
| [35] | Dai T M, Lu Z C, Liu W X, Wan F H.Selection and validation of reference genes for qRT-PCR analysis during biological invasions: The thermal adaptability of Bemisia tabaci MED. PLoS One, 2017, 12: e0173821. |
| [36] | Ponton F, Chapuis M P, Pernice M, Sword GA, Simpson S J.Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster.J Insect Physiol, 2011, 57: 840-850. |
| [37] | Yang Q P, Li Z, Cao J J, Zhang S D, Zhang H J, Wu X Y, Zhang Q W, Liu X X.Selection and assessment of reference genes for quantitative PCR normalization in migratory locust Locusta migratoria (Orthoptera: Acrididae).PLoS One, 2014, 9: e98164. |
| [38] | Zheng Y T, Li H B, Lu M X, Du Y Z.Evaluation and validation of reference genes for qRT-PCR normalization in Frankliniella occidentalis(Thysanoptera: Thripidae). PLoS One, 2014, 9: e111369. |
| [39] | Pan H P, Yang X W, Siegfried B D, Zhou X G.A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle,Hippodamia convergens(Coleoptera: Coccinellidae). PLoS One, 2015, 10: e0125868. |
| [40] | Qiu L, Wang P, Wu T, Li B, Wang X, Lei C, Lin Y, Zhao J, Ma W.Downregulation of Chilo suppressalis alkaline phosphatase genes associated with resistance to three transgenic Bacillus thuringiensis rice lines. Insect Mol Biol, 2018, 27: 83-89. |
| [41] | Xu G, Gu G X, Teng Z W, Wu S F, Huang J, Song Q S, Ye G Y, Fang Q.Identification and expression profiles of neuropeptides and their G protein-coupled receptors in the rice stem borer Chilo suppressalis. Sci Rep, 2016, 6: 28976. |
| [42] | Xu G, Wu S F, Teng Z W, Yao H W, Fang Q, Huang J, Ye G Y.Molecular characterization and expression profiles of nicotinic acetylcholine receptors in the rice striped stem borer,Chilo suppressalis(Lepidoptera: Crambidae). Insect Sci, 2017, 24: 371-384. |
| [43] | Xu G, Wu S F, Wu Y S, Gu G X, Fang Q, Ye G Y.De novo assembly and characterization of central nervous system transcriptome reveals neurotransmitter signaling systems in the rice striped stem borer,Chilo suppressalis. BMC Genomics, 2015, 16: 525. |
| [1] | 黄奇娜, 徐有祥, 林光号, 党洪阳, 郑振权, 张燕, 王晗, 邵国胜, 尹献远. 硅对镉胁迫下水稻苗期抗氧化酶系统及镉离子吸收和转运相关基因表达水平的影响[J]. 中国水稻科学, 2023, 37(5): 486-496. |
| [2] | 刘艳, 何林凤, 汪书超, 杨凤霞, 高聪芬, 吴顺凡. 二化螟对甲氧虫酰肼的抗性风险、交互抗性及亚致死效应研究[J]. 中国水稻科学, 2023, 37(4): 427-435. |
| [3] | 刘琴, 夏杨, 韩光杰, 李传明, 陆玉荣, 黄立鑫, 祁建杭, 徐健. 水稻二化螟病原线虫N-Yz1的分离鉴定及其感染特性[J]. 中国水稻科学, 2022, 36(6): 639-646. |
| [4] | 黄奇娜, 江苏, 汪利民, 张燕, 俞林飞, 李春福, 丁利群, 邵国胜. 低温胁迫后水分对水稻幼苗根系活力和水孔蛋白相关基因表达的影响[J]. 中国水稻科学, 2022, 36(4): 367-376. |
| [5] | 朱春权, 徐青山, 曹小闯, 朱练峰, 孔亚丽, 金千瑜, 张均华. 不同属性特征基质对早稻秧苗耐低温的影响[J]. 中国水稻科学, 2021, 35(5): 503-512. |
| [6] | 张珏锋, 张琴, 李芳, 钟海英, 陈建明. 氯虫苯甲酰胺胁迫下二化螟中肠细菌类微生物的多样性[J]. 中国水稻科学, 2020, 34(6): 586-594. |
| [7] | 宋瑞雪, 鲁涵, 鲁艳辉, 郑许松, 吕仲贤. 取食香根草后水稻螟虫对杀虫剂敏感度变化[J]. 中国水稻科学, 2019, 33(3): 282-286. |
| [8] | 邓卉, 鄂志国, 牛百晓, 王磊, 陈忱. DNA甲基化抑制剂5-氮脱氧胞苷对水稻基因组甲基化及幼苗生长发育的影响[J]. 中国水稻科学, 2019, 33(2): 108-117. |
| [9] | 窦玲玲, 胡海超, 马龙, 柯笑楠, 刘明月, 练旺民, 金珂, 谢玲娟, 刘庆坡. 一个水稻铜锌SOD酶基因在应答亚砷酸盐胁迫中的作用[J]. 中国水稻科学, 2018, 32(5): 437-444. |
| [10] | 彭喜旭, 白宁宁, 王海华. 响应镉胁迫的水稻WRKY15转录因子基因的分离与表达特征[J]. 中国水稻科学, 2018, 32(2): 103-110. |
| [11] | 李路, 徐以华, 梁梦琦, 王玲, 刘连盟, 侯雨萱, 黎起秦, 黄世文. 水稻对穗枯病的抗病机理初步研究[J]. 中国水稻科学, 2017, 31(5): 551-558. |
| [12] | 赵丹丹, 周丽琪, 张帅, 姚蓉, 邱运霞, 高聪芬. 二化螟对双酰胺类杀虫剂的抗药性监测和交互抗性研究[J]. 中国水稻科学, 2017, 31(3): 307-314. |
| [13] | 钟海英, 张珏锋, 李芳, 陈建明. 二化螟水稻、茭白种群幼虫口器和触角及其感器扫描电镜观察[J]. 中国水稻科学, 2017, 31(2): 195-206. |
| [14] | 胡训霞, 史春阳, 丁艳, 张萍, 葛永胜, 刘玉金, 王泽港, 葛才林. 水稻根系中磷高效吸收和利用相关基因表达对低磷胁迫的应答[J]. 中国水稻科学, 2016, 30(6): 567-576. |
| [15] | 潘鹏屹, 朱建平, 王云龙, 郝媛媛, 蔡跃, 张文伟, 江玲, 王益华, 万建民. 水稻粉质胚乳突变体ws的表型分析及基因克隆[J]. 中国水稻科学, 2016, 30(5): 447-457. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||