
Chinese Journal OF Rice Science ›› 2021, Vol. 35 ›› Issue (2): 103-111.DOI: 10.16819/j.1001-7216.2021.0905
• Research Papers • Next Articles
Kai LU, Tao CHEN, Shu YAO, Wenhua LIANG, Xiaodong WEI, Yadong ZHANG*( ), Cailin WANG*(
), Cailin WANG*( )
)
Received:2020-09-05
Revised:2020-09-18
Online:2021-03-10
Published:2021-03-10
Contact:
Yadong ZHANG, Cailin WANG
路凯, 陈涛, 姚姝, 梁文化, 魏晓东, 张亚东*( ), 王才林*(
), 王才林*( )
)
通讯作者:
张亚东,王才林
基金资助:Kai LU, Tao CHEN, Shu YAO, Wenhua LIANG, Xiaodong WEI, Yadong ZHANG, Cailin WANG. Functional Analysis on Four Receptor-like Protein Kinases Under Salt Stress in Rice[J]. Chinese Journal OF Rice Science, 2021, 35(2): 103-111.
路凯, 陈涛, 姚姝, 梁文化, 魏晓东, 张亚东, 王才林. 盐胁迫下四个水稻类受体蛋白激酶的功能分析[J]. 中国水稻科学, 2021, 35(2): 103-111.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2021.0905
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| Os04g0275100-Target-1F Os04g0275100-Target-1R Os04g0275100-Target-2F Os04g0275100-Target-2R Os09g0353200-Target-1F Os09g0353200-Target-1R Os09g0353200-Target-2F Os09g0353200-Target-2R Os07g0541900-Target-1F Os07g0541900-Target-1R Os07g0541900-Target-2F Os07g0541900-Target-2R Os01g0852100-Target-1F Os01g0852100-Target-1R Os01g0852100-Target-2F Os01g0852100-Target-2R | GGCACGGCTCTGCTATGCCTCCT AAACAGGAGGCATAGCAGAGCCG GCCGCTACATCGGCGGCGACAACG AAACCGTTGTCGCCGCCGATGTAG GGCAGCCTCACTATCCGAAGCAG AAACCTGCTTCGGATAGTGAGGC GCCGCTGCGGTTCGATCTGCATCT AAACAGATGCAGATCGAACCGCAG GGCACCTCATCAGCGCCCTCCAG AAACCTGGAGGGCGCTGATGAGG GCCGCCTCCTGAACGCCACCGTG AAACCACGGTGGCGTTCAGGAGG GCCGTATAAGGGGAAGCTCAGGGA AAACTCCCTGAGCTTCCCCTTATA GCCGTGGCCGATTTCGGCTTCGCA AAACTGCGAAGCCGAAATCGGCCA |
| Os04g0275100-T1T2-F Os04g0275100-T1T2-R Os09g0353200-T1T2-F Os09g0353200-T1T2-R Os07g0541900-T1T2-F Os07g0541900-T1T2-R Os01g0852100-T1-F Os01g0852100-T1-R Os01g0852100-T2-F Os01g0852100-T2-R U-F | GTGGGACGGATGAAGTAGTATATCTTCG AATCAAGCAGCAAGGGGACCT ACGTGCAAGGATCTATTCAAAACT GGAGAGAACACATACCGGGTAAG ACATCCAAAATGATACGCATTCCA AGCTCCGACCTCAGGTTACACC GGTACTCGCACCAGGAATTTAGAG CATCATGCTTAGGGGTTCAACA TTGTCCTGAATAAACCAACAGTAGCACT CAACAACTTGGGAAACGTTAAACG CTCCGTTTTACCTGTGGAATCG |
| U-R | CGGAGGAAAATTCCATCCAC |
| Uctcg-B1’ | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| gRctga-B2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| Uctga-B2’ | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| gRcggt-BL | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| Cas9-F | GATCCTTACTTTCCGTATTCCTTACTACG |
| Cas9-R OsActin1-qPCR-F OsActin1-qPCR-R | ATACCCTCCTCAATCCTCTTCATG GATGACCCAGATCATGTTTG GGGCGATGTAGGAAAGC |
Table 1 Primers used in this study.
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| Os04g0275100-Target-1F Os04g0275100-Target-1R Os04g0275100-Target-2F Os04g0275100-Target-2R Os09g0353200-Target-1F Os09g0353200-Target-1R Os09g0353200-Target-2F Os09g0353200-Target-2R Os07g0541900-Target-1F Os07g0541900-Target-1R Os07g0541900-Target-2F Os07g0541900-Target-2R Os01g0852100-Target-1F Os01g0852100-Target-1R Os01g0852100-Target-2F Os01g0852100-Target-2R | GGCACGGCTCTGCTATGCCTCCT AAACAGGAGGCATAGCAGAGCCG GCCGCTACATCGGCGGCGACAACG AAACCGTTGTCGCCGCCGATGTAG GGCAGCCTCACTATCCGAAGCAG AAACCTGCTTCGGATAGTGAGGC GCCGCTGCGGTTCGATCTGCATCT AAACAGATGCAGATCGAACCGCAG GGCACCTCATCAGCGCCCTCCAG AAACCTGGAGGGCGCTGATGAGG GCCGCCTCCTGAACGCCACCGTG AAACCACGGTGGCGTTCAGGAGG GCCGTATAAGGGGAAGCTCAGGGA AAACTCCCTGAGCTTCCCCTTATA GCCGTGGCCGATTTCGGCTTCGCA AAACTGCGAAGCCGAAATCGGCCA |
| Os04g0275100-T1T2-F Os04g0275100-T1T2-R Os09g0353200-T1T2-F Os09g0353200-T1T2-R Os07g0541900-T1T2-F Os07g0541900-T1T2-R Os01g0852100-T1-F Os01g0852100-T1-R Os01g0852100-T2-F Os01g0852100-T2-R U-F | GTGGGACGGATGAAGTAGTATATCTTCG AATCAAGCAGCAAGGGGACCT ACGTGCAAGGATCTATTCAAAACT GGAGAGAACACATACCGGGTAAG ACATCCAAAATGATACGCATTCCA AGCTCCGACCTCAGGTTACACC GGTACTCGCACCAGGAATTTAGAG CATCATGCTTAGGGGTTCAACA TTGTCCTGAATAAACCAACAGTAGCACT CAACAACTTGGGAAACGTTAAACG CTCCGTTTTACCTGTGGAATCG |
| U-R | CGGAGGAAAATTCCATCCAC |
| Uctcg-B1’ | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| gRctga-B2 | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| Uctga-B2’ | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| gRcggt-BL | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| Cas9-F | GATCCTTACTTTCCGTATTCCTTACTACG |
| Cas9-R OsActin1-qPCR-F OsActin1-qPCR-R | ATACCCTCCTCAATCCTCTTCATG GATGACCCAGATCATGTTTG GGGCGATGTAGGAAAGC |

Fig. 1. Relative expression level of RLKs under NaCl, ABA and mannitol treatment. qPCR was conducted to detect the relative expression of RLK. ** represent significant difference at P<0.01.
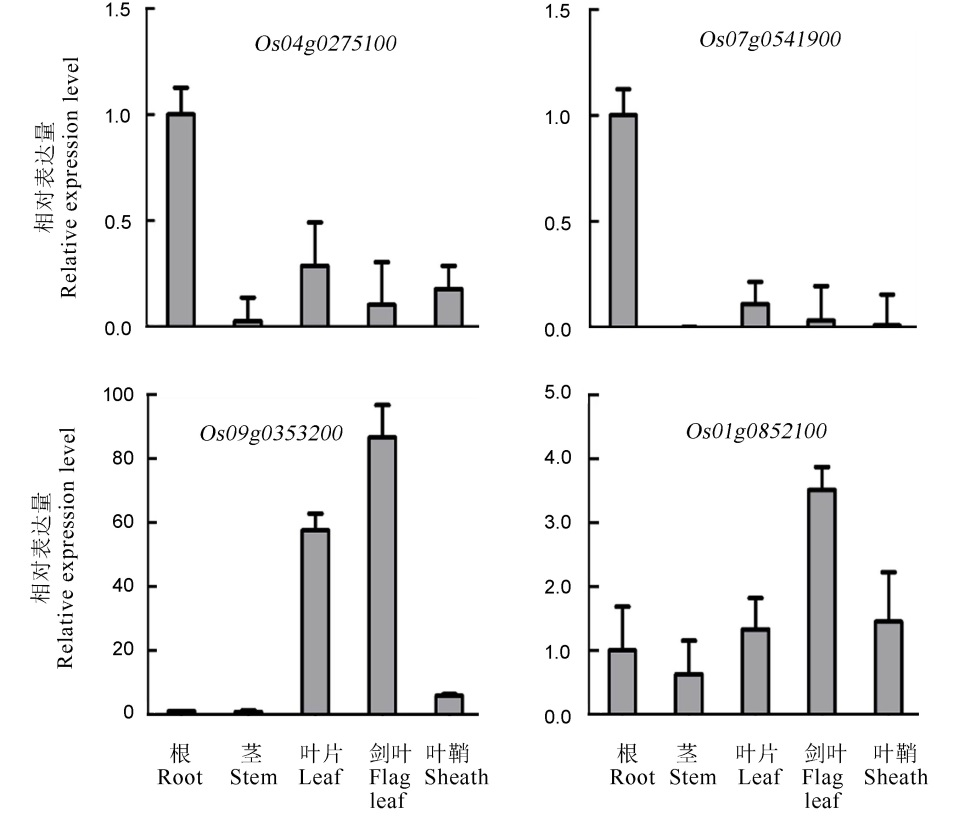
Fig. 2. Relative expression level of RLKs in different tissues and organs. qPCR was conducted to detect the relative RLK expression in root, shoot, leaf, flag leaf and sheath at the filling stage.
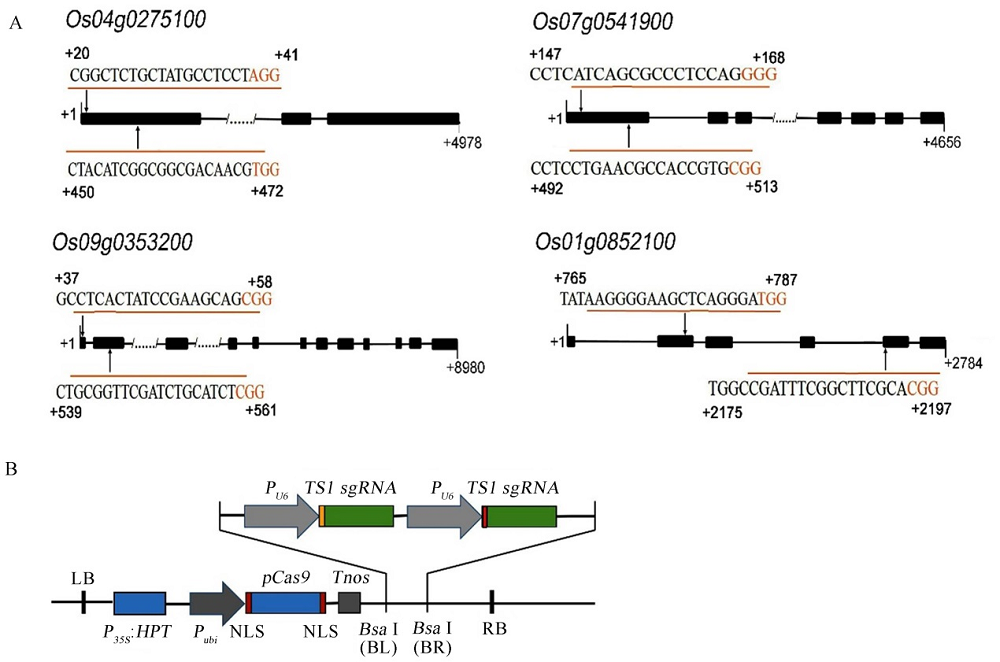
Fig. 3. Target site of RLKs and diagram of pYLCRISPR/Cas9PUbi-RLKs-sgRNA vector. A, Target site of RLKs; B, Diagram of pYLCRISPR/Cas9PUbi-RLKs-sgRNA vector.
Fig. 4. Identification of the transgenic plants by amplification of Cas9. A, Identification of the Os04g0275100-edited plant; B, Identification of the Os07g0541900-edited plant; C, Identification of the Os09g0353200-edited plant; D, Identification of the Os01g0852100- edited plant.
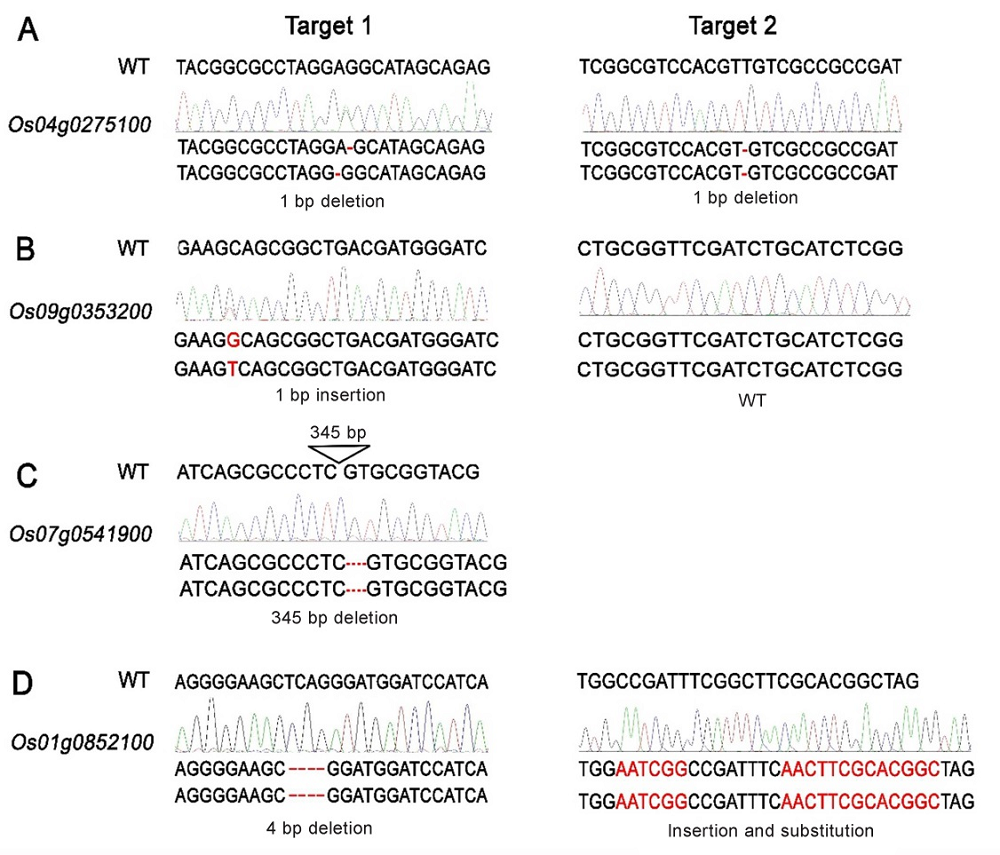
Fig. 5. Identification of the loss-of-function mutants of RLKs. A, Identification of the mutation type of Os04g0275100; B, Identification of the mutation type of Os09g0353200; C, Identification of the mutation type of Os07g0541900; D, Identification of the mutation type of Os01g0852100.
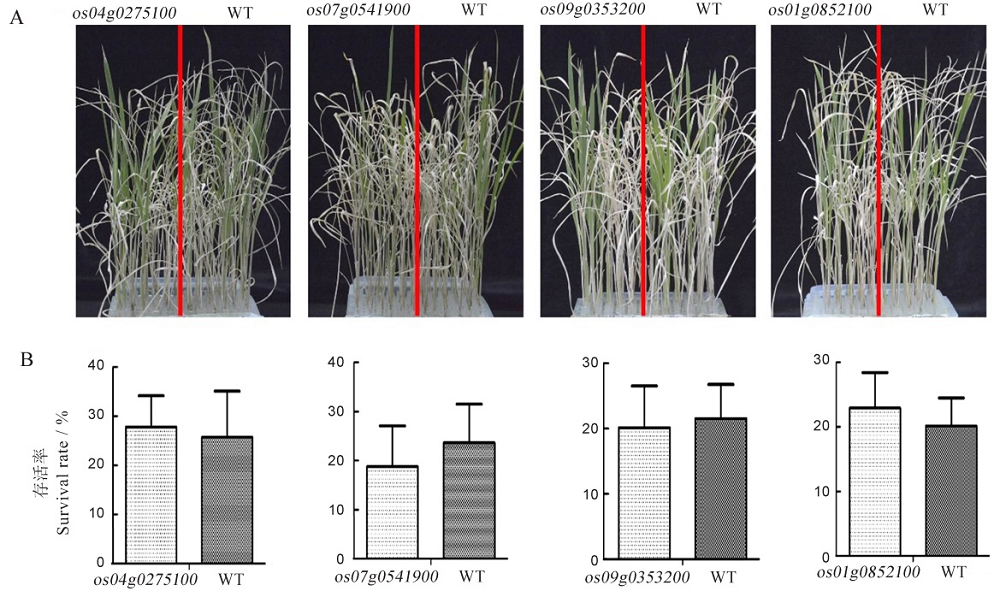
Fig. 6. Salt tolerance analysis of the loss-of-function mutants of RLKs. A, Phenotypic analysis on salt tolerance of mutants and WT; B, Comparison of survival rate between mutants and WT.
| [1] | Gong Z, Xiong L M, Shi H Z, Yang S H, Herrera-Estrella L R, Xu G H, Chao D Y, Li J R, Wang P Y, Qin F, Li J, Ding Y L, Shi Y T, Wang Y, Yang Y Q, Guo Y, Zhu J K. Plant abiotic stress response and nutrient use efficiency.Science China: Life Sciences, 2020, 63(5): 635-674. |
| [2] | Zhu J K.Plant salt tolerance.Trends in Plant Science, 2001, 6(2): 66-71. |
| [3] | Flowers T J.Improving crop salt tolerance.Journal of Experimental Botany, 2004, 55(396): 307-319. |
| [4] | Sahi C, Singh A, Kumar K, Blumwald E, Grover A.Salt stress response in rice: Genetics, molecular biology, and comparative genomics.Functional & Integrative Genomics, 2006, 6(4): 263-284. |
| [5] | Zelm E V, Zhang Y X, Testerink C.Salt tolerance mechanisms of plants.Annual Review of Plant Biology, 2020, 71(1): 403-433. |
| [6] | Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K.New insights on plant salt tolerance mechanisms and their potential use for breeding.Frontiers in Plant Science, 2016, 7(11): 1787. |
| [7] | Sivakumar P, Sharmila P, Saradhi P P.Proline suppresses rubisco activity in higher plants.Biochemical and Biophysical Research Communications, 1998, 252(2): 428-432. |
| [8] | Lemmon M A, Schlessinger J.Cell signaling by receptor tyrosine kinases.Cell, 2010, 141(7): 1117-1134. |
| [9] | Morillo S A, Tax F E.Functional analysis of receptor-like kinases in monocots and dicots.Current Opinion in Plant Biology, 2006, 9(5): 460-469. |
| [10] | Ouyang S Q, Liu Y F, Liu P, Lei G, He S J, Ma B, Zhang W K, Zhang J S, Chen S Y.Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant Journal, 2010, 62(2): 316-329. |
| [11] | Li C H, Wang G, Zhao J L, Zhang L Q, Ai L F, Han Y F, Sun D Y, Zhang S W, Sun Y.The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice.Plant Cell, 2014, 26(6): 2538-2553. |
| [12] | Shi C C, Feng C C, Yang M M, Li J L, Li X X, Zhao B C, Huang Z J, Ge R C. Overexpression of the receptor-like protein kinase genes AtRPK1 and OsRPK1 reduces the salt tolerance of Arabidopsis thaliana. Plant Science, 2014, 217-218: 63-70. |
| [13] | Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran L S.Sensing the environment: Key roles of membrane-localized kinases in plant perception and response to abiotic stress.Journal of Experimental Botany, 2013, 64(2): 445-458. |
| [14] | Shiu S H, Bleecker A B.Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences of the United States, 2001, 98(19): 10763-10768. |
| [15] | Tanaka H, Osakabe Y, Katsura S, Mizuno S, Maruyama K, Kusakabe K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K.Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant Journal, 2012, 70(4): 599-613. |
| [16] | Wang X F, Kota U, He K, Blackburn K, Li J, Goshe M B, Huber S C, Clouse S D.Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling.Developmental Cell, 2008, 15(2): 220-235. |
| [17] | Shiu S H, Karlowski W M, Pan R, Tzeng Y H, Mayer K F, Li W H.Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell, 2004, 16(5): 1220-1234. |
| [18] | Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G.A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants.Molecular Plant, 2015, 8(8): 1274-1284. |
| [19] | 王才林, 张亚东, 赵凌, 路凯, 朱镇, 陈涛, 赵庆勇, 姚姝, 周丽慧, 赵春芳, 梁文化, 孙明法, 严国红. 耐盐碱水稻研究现状、问题与建议. 中国稻米, 2019, 25(1): 1-6. |
| Wang C L, Zhang Y D, Zhao L, Lu K, Zhu Z, Chen T, Zhao Q Y, Yao S, Zhou L H, Zhao C F, Liang W H, Shun M F, Yan G H.Research status, questions and suggestions on salt tolerance rice varieties.China Rice, 2019, 25(1): 1-6. (in Chinese) | |
| [20] | 黄忠明, 周延彪, 唐晓丹, 赵新辉, 周在为, 符星学, 王凯, 史江伟, 李艳锋, 符辰建, 杨远柱. 基于CRISPR/Cas9 技术的水稻温敏不育基因tms5突变体的构建. 作物学报, 2018, 44(6): 844-851. |
| Huang Z M, Zhou Y B, Tang X D, Zhao X H, Zhou Z W, Fu X X, Wang K, Shi J W, Li Y F, Fu C J, Yang Y Z.Construction of tms5 mutants in rice based on CRISPR/Cas9 technology. Acta Agronomica Sinica, 2018, 44(6): 844-851. (in Chinese with English abstract) | |
| [21] | Zhao D S, Li Q F, Zhang C Q, Zhang C, Yang Q Q, Pan L X, Ren X Y, Lu J, Gu M H, Liu Q Q.GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nature Communications, 2018, 9(1): 1240. |
| [22] | 王才林, 张亚东, 朱镇, 姚姝, 赵庆勇, 陈涛, 周丽慧, 赵凌. 优良食味粳稻新品种南粳9108的选育与利用. 江苏农业科学, 2013, 41(9): 86-88. |
| Wang C L, Zhang Y D, Zhu Z, Yao S, Zhao Q Y, Chen T, Zhou L H, Zhao L.Breeding of the japonica rice varieties with good eating quality.Jiangsu Agricultural Sciences, 2013, 41(9): 86-88. (in Chinese) | |
| [23] | Zhou Y, Liu C, Tang D, Yan L, Wang D, Yang Y Z, Liu X M.The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice.The Plant Cell, 2018, 30(5): 1100-1118. |
| [24] | Gao L L, Xue H W.Global analysis of expression profiles of rice receptor-like kinase genes.Molecular Plant, 2012, 5(1): 143-153. |
| [1] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [2] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [3] | JING Xiu, ZHOU Miao, WANG Jing, WANG Yan, WANG Wang, WANG Kai, GUO Baowei, HU Yajie, XING Zhipeng, XU Ke, ZHANG Hongcheng. Effect of Drought Stress on Root Morphology and Leaf Photosynthetic Characteristics of Good Taste japonica Rice from Late Stage of Panicle Differentiation to Early Stage of Grain Filling [J]. Chinese Journal OF Rice Science, 2024, 38(1): 33-47. |
| [4] | LI Jingfang, WEN Shuyue, ZHAO Lijun, CHEN Tingmu, ZHOU Zhenling, SUN Zhiguang, LIU Yan, CHEN Haiyuan, ZHANG Yunhui, CHI Ming, XING Yungao, XU Bo, XU Dayong, WANG Baoxiang. Development of Aromatic Salt-tolerant Rice Based on CRISPR/Cas9 Technology [J]. Chinese Journal OF Rice Science, 2023, 37(5): 478-485. |
| [5] | XIA Yang, LI Chuanming, LIU Qin, HAN Guangjie, XU Bin, HUANG Lixin, QI Jianhang, LU Yurong, XU Jian. Effects of Piriformospora indica on the Growth and Antioxidant System of Rice Seedlings Under Salt Stress [J]. Chinese Journal OF Rice Science, 2023, 37(5): 543-552. |
| [6] | LI Gang, GAO Qingsong, LI Wei, ZHANG Wenxia, WANG Jian, CHEN Baoshan, WANG Di, GAO Hao, XU Weijun, CHEN Hongqi, JI Jianhui. Directed Knockout of SD1 Gene Improves Lodging Resistance and Blast Resistance of Rice [J]. Chinese Journal OF Rice Science, 2023, 37(4): 359-367. |
| [7] | HUANG Yaru, XU Peng, WANG Lele, HE Yizhe, WANG Hui, KE Jian, HE Haibing, WU Liquan, YOU Cuicui. Effects of Exogenous Trehalose on Grain Filling Characteristics and Yield Formation of japonica Rice Cultivar W1844 [J]. Chinese Journal OF Rice Science, 2023, 37(4): 379-391. |
| [8] | DUAN Min, XIE Liujie, GAO Xiuying, TANG Haijuan, HUANG Shanjun, PAN Xiaobiao. Creation of Thermo-sensitive Genic Male Sterile Rice Lines with Wide Compatibility Based on CRISPR/Cas9 Technology [J]. Chinese Journal OF Rice Science, 2023, 37(3): 233-243. |
| [9] | WANG Yu, SUN Quanyi, DU Haibo, XU Zhiwen, WU Keting, YIN Li, FENG Zhiming, HU Keming, CHEN Zongxiang, ZUO Shimin. Improvement of the Resistance of Nanjing 9108 to Blast and Sheath Blight by Pyramiding Resistance Gene Pigm and Quantitative Trait Genes qSB-9TQ and qSB-11HJX [J]. Chinese Journal OF Rice Science, 2023, 37(2): 125-132. |
| [10] | ZHOU Zhenling, LIN Bing, ZHOU Qun, YANG Bo, LIU Yan, ZHOU Tianyang, WANG Baoxiang, GU Junfei, XU Dayong, YANG Jianchang. Responses of Rice Varieties Differing in Salt Tolerance to Salt Stress and Their Physiological Mechanisms [J]. Chinese Journal OF Rice Science, 2023, 37(2): 153-165. |
| [11] | LIU Shuli, ZHANG Rui, Shahid HUSSAIN, WANG Yang, CHEN Yinglong, WEI Huanhe, HOU Hongyan, DAI Qigen. Research Progress in Alleviating Effects of Exogenous Substances on Salt Stress in Rice [J]. Chinese Journal OF Rice Science, 2023, 37(1): 1-15. |
| [12] | PEI Feng, WANG Guangda, GAO Peng, FENG Zhiming, HU Keming, CHEN Zongxiang, CHEN Hongqi, CUI Ao, ZUO Shimin. Evaluation of New japonica Rice Lines with Low Cadmium Accumulation and Good Quality Generated by Knocking Out OsNramp5 [J]. Chinese Journal OF Rice Science, 2023, 37(1): 16-28. |
| [13] | WANG Shiguang, LU Zhanhua, LIU Wei, LU Dongbai, WANG Xiaofei, FANG Zhiqiang, WU Haoxiang, HE Xiuying. Generating Guangdong Simiao Rice Germplasms by Applying CRISPR/Cas9 Gene Editing and Marker-assisted Selection Technology [J]. Chinese Journal OF Rice Science, 2023, 37(1): 29-36. |
| [14] | CHEN Tao, ZHAO Qingyong, ZHU Zhen, ZHAO Ling, YAO Shu, ZHOU Lihui, ZHAO Chunfang, ZHANG Yadong, WANG Cailin. Development of New Low Glutelin Content japonica Rice Lines with Good Eating Quality and Fragrance by Molecular Marker-Assisted Selection [J]. Chinese Journal OF Rice Science, 2023, 37(1): 55-65. |
| [15] | WU Mingming, ZENG Wei, ZHAI Rongrong, YE Jing, ZHU Guofu, YU Faming, ZHANG Xiaoming, YE Shenghai. Research Progress in Molecular Mechanism and Breeding Status of Salt Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(6): 551-561. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||