
中国水稻科学 ›› 2022, Vol. 36 ›› Issue (2): 139-149.DOI: 10.16819/j.1001-7216.2022.210308
王永祥, 燕海刚, 徐含聪, 傅玉双, 单壮壮, 胡晓晴, 张文伟*( ), 江玲
), 江玲
收稿日期:2021-03-19
修回日期:2021-05-12
出版日期:2022-03-10
发布日期:2022-03-11
通讯作者:
张文伟
基金资助:
WANG Yongxiang, YAN Hangang, XU Hancong, FU Yushuang, SHAN Zhuangzhuang, HU Xiaoqing, ZHANG Wenwei*( ), JIANG Ling
), JIANG Ling
Received:2021-03-19
Revised:2021-05-12
Online:2022-03-10
Published:2022-03-11
Contact:
ZHANG Wenwei
摘要:
【目的】 研究水稻EARLY STARVATION1 (OsESV1)基因对水稻淀粉代谢的影响。【方法】 通过CRISPR-Cas9技术获得osesv1突变体,考查osesv1的表型及胚乳淀粉的理化特性,分析OsESV1的表达特性及相关功能。【结果】 OsESV1蛋白在植物界中十分保守。osesv1突变体株高、穗长、每穗粒数低于野生型,分蘖数显著多于野生型,叶片中淀粉含量显著下降,籽粒中的直链淀粉含量上升,而总淀粉含量无明显变化。OsESV1呈组成型表达,并且具有昼夜节律表达的特征。OsESV1蛋白定位在叶绿体内且呈点状分布。酵母双杂交和双分子荧光互补实验结果表明OsESV1蛋白可以与ADP-葡萄糖焦磷酸化酶小亚基(OsAGPS) 2a和OsAGPS1互作。【结论】 OsESV1基因影响水稻叶片的淀粉合成途径,而对胚乳淀粉合成的影响不明显。
王永祥, 燕海刚, 徐含聪, 傅玉双, 单壮壮, 胡晓晴, 张文伟, 江玲. OsESV1基因对水稻淀粉合成的影响[J]. 中国水稻科学, 2022, 36(2): 139-149.
WANG Yongxiang, YAN Hangang, XU Hancong, FU Yushuang, SHAN Zhuangzhuang, HU Xiaoqing, ZHANG Wenwei, JIANG Ling. Effects of OsESV1 on Starch Synthesis in Rice[J]. Chinese Journal OF Rice Science, 2022, 36(2): 139-149.
| 引物 Primer | 正向引物序列 Forward primer sequence (5′-3′) | 反向引物序列 Reverse primer sequence (5′-3′) |
|---|---|---|
| Crispr-OsESV1 | GGCAAGCACCTGGTACAGGGAGAG | AAACCTCTCCCTGTACCAGGTGCT |
| OsESV1-TI | AGCTCCTCAATCCCGTGATA | CTCATACGAAAATGTGGT |
| PAN580-OsESV1-GFP | TCCGGAGCTAGCTCTAGAATGGCCGCGTGCTCCAGG | CGCCCTTGCTCACCATGGATCCTTCCAGAGGCGAAGGAGGCA |
| 1305-OsESV1-GFP | TCCGGAGCTAGCTCTAGAATGGCCGCGTGCTCCAGG | CGCCCTTGCTCACCATGGATCCTTCCAGAGGCGAAGGAGGCA |
| QRT-OsESV1 | GTGATACTCCGCCGAAGAGA | TCGTCTCCACTCTCCCTGTA |
| Actin | TGCTATGTACGTCGCCATCCAG | AATGAGTAACCACGCTCCGTCA |
| AD-OsESV1 | CATGGAGGCCGAATTCATGGCCGCGTGCTCCAGG | CGAGCTCGATGGATCCTCATTCCAGAGGCGAAGGAGG |
| BD-OsESV1 | CATGGAGGCCGAATTCATGGCCGCGTGCTCCAG | GCAGGTCGACGGATCCTCATTCCAGAGGCGAAGGAGG |
| AD-OsAGPS1 | GGAGGCCAGTGAATTCATGGCGATGATGGCGATG | CGAGCTCGATGGATCCTTATATGACTGTTCCGCTAG |
| BD-OsAGPS1 | CATGGAGGCCGAATTCATGGCGATGATGGCGATG | GCAGGTCGACGGATCCTTATATGACTGTTCCGCTAG |
| AD-OsAGPS2a | GGAGGCCAGTGAATTCATGGCGATGGCGGCAGCCAT | CGAGCTCGATGGATCCTCATATAACTGTTCCGCTAG |
| AD-OsAGPS2b | CATGGAGGCCGAATTCATGAATGTATTGGCATCTAAG | CGAGCTCGATGGATCCTCATATAACTGTTCCGCTAGG |
| AD-OsAGPL1 | CATGGAGGCCGAATTCATGCAGTTCAGCAGTGTGTTT | CGAGCTCGATGGATCCCTATATGACCTTCCCGTCC |
| AD-SSⅠ | CATGGAGGCCGAATTCATGGCGACGGCGGCGGGGAT | CGAGCTCGATGGATCCTTACATGACATATGGTTGATC |
| AD-SSⅡa | CATGGAGGCCGAATTCATGTCGTCGGCCGTCGTCGCGTC | CGAGCTCGATGGATCCTCACCATTGGTACTTGGCCTT |
| AD-SSⅡb | CATGGAGGCCGAATTCTTCACCTCCTCTTCGCCGCG | CGAGCTCGATGGATCCTCACCACTGGTACTTGGCCTT |
| AD-BEI | GGAGGCCAGTGAATTCATGCTGTGTCTCACC | CGAGCTCGATGGATCCCTCATTTGCAGTCTTC |
| AD-BEⅡb | CATGGAGGCCGAATTCATGGCGGCGCCGGCGTCTG | CGAGCTCGATGGATCCTCATTCCGCTGGAGCATA |
| p2YC-OsESV1 | ATTTACGAACGATAGTTAATTAAATGGCGATGGCCGCGTGCTCCAGG | CACTGCCACCTCCTCCACTAGTTTCCAGAGGCGAAGGAGG |
| p2YN-OsAGPS2a | ATTTACGAACGATAGTTAATTAAATGGCGATGGCGGCAGCCAT | CACTGCCACCTCCTCCACTAGTTATAACTGTTCCGCTAGGG |
| p2YN-OsAGPS1 | ATTTACGAACGATAGTTAATTAAATGGCGATGGCGATGATGGCGATG | CACTGCCACCTCCTCCACTAGTTATGACTGTTCCGCTA |
表1 实验所用引物
Table 1 Primers used in this study.
| 引物 Primer | 正向引物序列 Forward primer sequence (5′-3′) | 反向引物序列 Reverse primer sequence (5′-3′) |
|---|---|---|
| Crispr-OsESV1 | GGCAAGCACCTGGTACAGGGAGAG | AAACCTCTCCCTGTACCAGGTGCT |
| OsESV1-TI | AGCTCCTCAATCCCGTGATA | CTCATACGAAAATGTGGT |
| PAN580-OsESV1-GFP | TCCGGAGCTAGCTCTAGAATGGCCGCGTGCTCCAGG | CGCCCTTGCTCACCATGGATCCTTCCAGAGGCGAAGGAGGCA |
| 1305-OsESV1-GFP | TCCGGAGCTAGCTCTAGAATGGCCGCGTGCTCCAGG | CGCCCTTGCTCACCATGGATCCTTCCAGAGGCGAAGGAGGCA |
| QRT-OsESV1 | GTGATACTCCGCCGAAGAGA | TCGTCTCCACTCTCCCTGTA |
| Actin | TGCTATGTACGTCGCCATCCAG | AATGAGTAACCACGCTCCGTCA |
| AD-OsESV1 | CATGGAGGCCGAATTCATGGCCGCGTGCTCCAGG | CGAGCTCGATGGATCCTCATTCCAGAGGCGAAGGAGG |
| BD-OsESV1 | CATGGAGGCCGAATTCATGGCCGCGTGCTCCAG | GCAGGTCGACGGATCCTCATTCCAGAGGCGAAGGAGG |
| AD-OsAGPS1 | GGAGGCCAGTGAATTCATGGCGATGATGGCGATG | CGAGCTCGATGGATCCTTATATGACTGTTCCGCTAG |
| BD-OsAGPS1 | CATGGAGGCCGAATTCATGGCGATGATGGCGATG | GCAGGTCGACGGATCCTTATATGACTGTTCCGCTAG |
| AD-OsAGPS2a | GGAGGCCAGTGAATTCATGGCGATGGCGGCAGCCAT | CGAGCTCGATGGATCCTCATATAACTGTTCCGCTAG |
| AD-OsAGPS2b | CATGGAGGCCGAATTCATGAATGTATTGGCATCTAAG | CGAGCTCGATGGATCCTCATATAACTGTTCCGCTAGG |
| AD-OsAGPL1 | CATGGAGGCCGAATTCATGCAGTTCAGCAGTGTGTTT | CGAGCTCGATGGATCCCTATATGACCTTCCCGTCC |
| AD-SSⅠ | CATGGAGGCCGAATTCATGGCGACGGCGGCGGGGAT | CGAGCTCGATGGATCCTTACATGACATATGGTTGATC |
| AD-SSⅡa | CATGGAGGCCGAATTCATGTCGTCGGCCGTCGTCGCGTC | CGAGCTCGATGGATCCTCACCATTGGTACTTGGCCTT |
| AD-SSⅡb | CATGGAGGCCGAATTCTTCACCTCCTCTTCGCCGCG | CGAGCTCGATGGATCCTCACCACTGGTACTTGGCCTT |
| AD-BEI | GGAGGCCAGTGAATTCATGCTGTGTCTCACC | CGAGCTCGATGGATCCCTCATTTGCAGTCTTC |
| AD-BEⅡb | CATGGAGGCCGAATTCATGGCGGCGCCGGCGTCTG | CGAGCTCGATGGATCCTCATTCCGCTGGAGCATA |
| p2YC-OsESV1 | ATTTACGAACGATAGTTAATTAAATGGCGATGGCCGCGTGCTCCAGG | CACTGCCACCTCCTCCACTAGTTTCCAGAGGCGAAGGAGG |
| p2YN-OsAGPS2a | ATTTACGAACGATAGTTAATTAAATGGCGATGGCGGCAGCCAT | CACTGCCACCTCCTCCACTAGTTATAACTGTTCCGCTAGGG |
| p2YN-OsAGPS1 | ATTTACGAACGATAGTTAATTAAATGGCGATGGCGATGATGGCGATG | CACTGCCACCTCCTCCACTAGTTATGACTGTTCCGCTA |
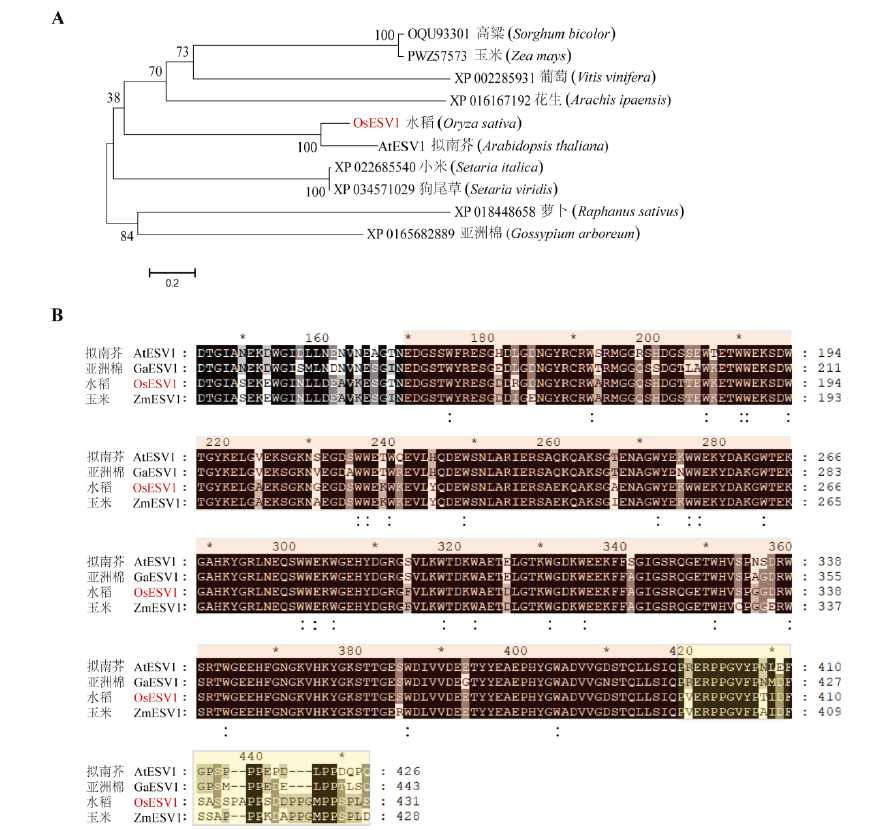
图1 OsESV1蛋白的保守性分析 A—OsESV1及其同源蛋白的进化树分析,蛋白名采用相应的GenBank 序列登录号;B—多氨基酸序列比对,星号表示20个氨基酸的间隔,浅红色区域为富含色氨酸(W)区,保守色氨酸残基下用“:”标注,黄色区域为富含脯氨酸(P)区。
Fig. 1. The conservation analysis of OsESV1. A, Neighbor-joining tree of OsESV1 and its homologs. The proteins are named by GenBank protein accession numbers; B, Multiple sequence alignment of OsESV1 and its homologs. Asterisks represent the interval of twenty amino acids. The tryptophan (W)-rich region is marked by light red and those conserved tryptophan residues are indicated by colons. The proline (P)-rich region is marked by yellow.
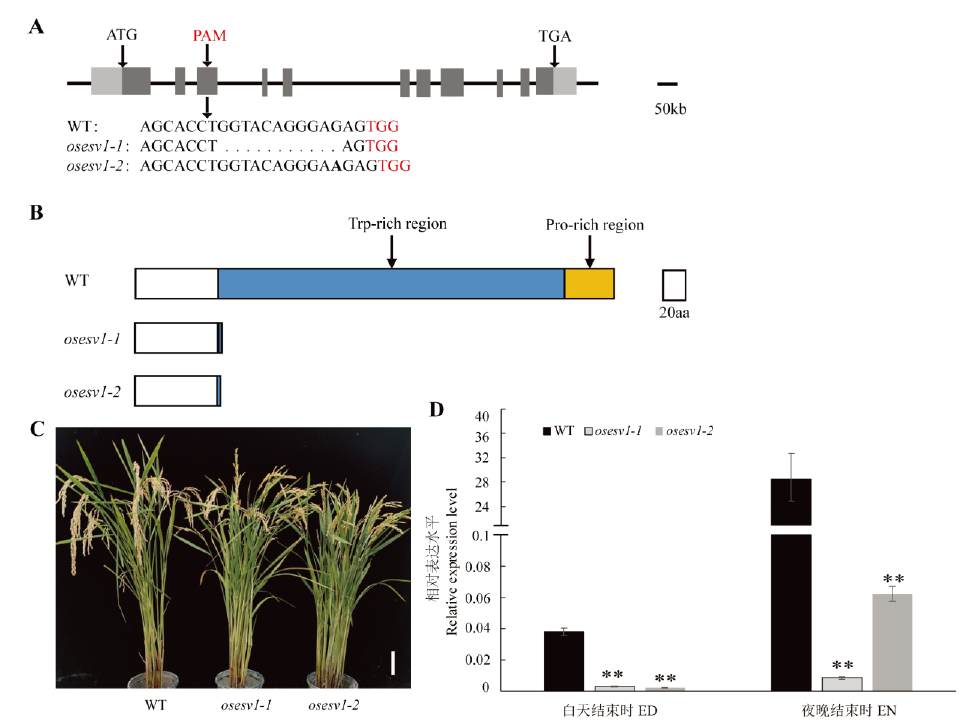
图2 osesv1突变体的表型 A—OsESV1的基因结构。深灰和浅灰方块分别表示外显子区域和UTR区域,方块之间的黑色连线表示内含子,下面为野生型和osesv1靶位点基因序列比较,连续的点表示缺失的序列,插入的碱基加粗标示,红色标注为原间隔基序(protospacer adjacent motif , PAM) TGG;B—野生型和突变体的OsESV1蛋白结构。箭头标注富含色氨酸区(Trp-rich region)和富含脯氨酸区(Pro-rich region);C—野生型和osesv1植株,标尺为10 cm;D—野生型和osesv1幼苗在白天和夜晚结束时OsESV1基因的表达水平。数据表示为平均值±标准差(n=3)。*和**分别表示差异达0.05和0.01显著水平(t 检验)。下同。
Fig. 2. Phenotype of the osesv1 mutants. A, Structure of the OsESV1 gene. The dark gray and light gray boxes represent the exon and UTR regions, respectively. The black lines connecting boxes are introns. The corresponding sequences of the wild type (WT) and osesv1 were compared below and a succession of points indicates the deleted sequence in osesv1. The inserted base is shown in bold. TGG (PAM) is marked by red; B, The protein structure of WT and osesv1; Arrows indicate the tryptophan-rich region and proline-rich region; C, WT and osesv1 plants. Bar=10cm; D, Expression level of OsESV1 in WT and osesv1 seedlings at end of day(ED) and end of night(EN). Values are mean ± SD (n=3). * and **indicate significant difference at 0.05 and 0.01 level, respectively (t-test). The same below.
| 材料 Materials | 株高 Plant height /cm | 穗长 Length of panicle /cm | 分蘖数 Tiller number | 每穗粒数 Grain number per panicle | 一次枝梗数 Primary rachis branch number | 千粒重 1000-grain weight /g |
|---|---|---|---|---|---|---|
| 野生型WT | 94.80±3.10 | 19.31±0.77 | 26.13±8.55 | 127.58±9.33 | 8.60±0.83 | 22.57±1.07 |
| osesv1-1 | 90.96±4.44** | 17.82±0.52** | 33.43±10.45* | 112.11±15.42* | 8.44±0.96 | 21.50±0.56 |
| osesv1-2 | 90.47±5.40** | 17.69±0.82** | 43.08±13.85** | 113.63±10.71** | 9.00±0.95 | 23.01±0.85 |
表2 野生型和osesv1的农艺性状
Table 2 Agronomic traits of the wild type (WT) and osesv1.
| 材料 Materials | 株高 Plant height /cm | 穗长 Length of panicle /cm | 分蘖数 Tiller number | 每穗粒数 Grain number per panicle | 一次枝梗数 Primary rachis branch number | 千粒重 1000-grain weight /g |
|---|---|---|---|---|---|---|
| 野生型WT | 94.80±3.10 | 19.31±0.77 | 26.13±8.55 | 127.58±9.33 | 8.60±0.83 | 22.57±1.07 |
| osesv1-1 | 90.96±4.44** | 17.82±0.52** | 33.43±10.45* | 112.11±15.42* | 8.44±0.96 | 21.50±0.56 |
| osesv1-2 | 90.47±5.40** | 17.69±0.82** | 43.08±13.85** | 113.63±10.71** | 9.00±0.95 | 23.01±0.85 |

图3 野生型和osesv1的叶片淀粉水平 A—白天和夜晚结束时野生型(WT)和osesv1叶片的碘染观察; B—白天和夜晚结束时野生型(WT)和osesv1叶片的淀粉含量测定。
Fig. 3. Starch contents in leaves of WT and osesv1. A and B show iodine staining and starch contents of WT and osesv1 leaves at end of day and end of night, respectively.
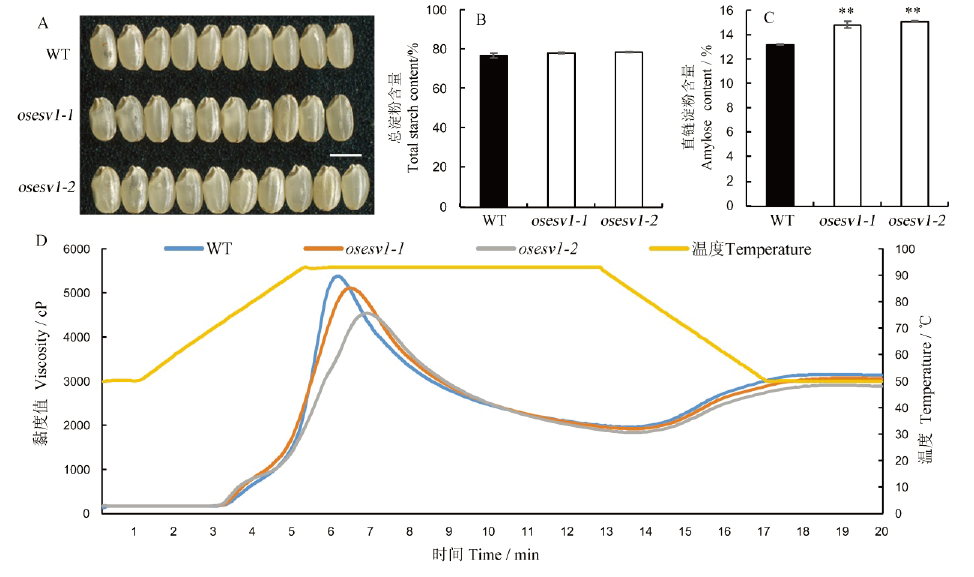
图4 osesv1种子表型及淀粉理化性质分析 A—野生型(WT)和osesv1的成熟种子表型,标尺为5 mm;B, C—野生型(WT)和osesv1种子总淀粉含量、直链淀粉含量;D—野生型和osesv1胚乳淀粉的黏度曲线。
Fig. 4. Phenotype of osesv1 grains and physicochemical properties of endosperm starch. A, Phenotype of WT and osesv1 mature seeds. Bar=5 mm; B, Total starch contents of WT and osesv1 grains; C, Amylose content of WT and osesv1 grains; D, Viscosity profiles of endosperm starch of WT and osesv1.
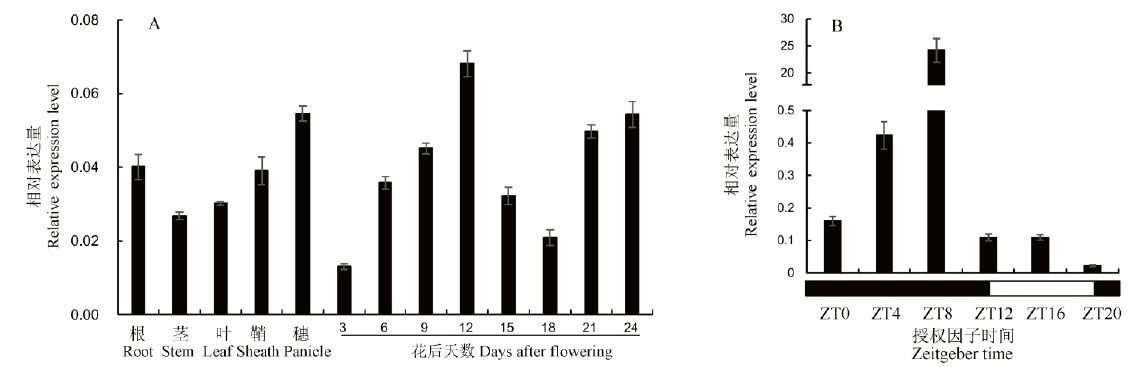
图5 OsESV1的表达模式分析 A—OsESV1在野生型不同组织及胚乳发育不同时期的表达水平;B—OsESV1的昼夜节律表达。黑框和白框分别表示黑夜和白天的时间轴。
Fig. 5. Expression pattern of OsESV1. A, Expression level of OsESV1 in various tissues and endosperm at different developmental stages of WT; B, Diurnal expression pattern of OsESV1. The black and white boxes represent the timelines of night and day, respectively.
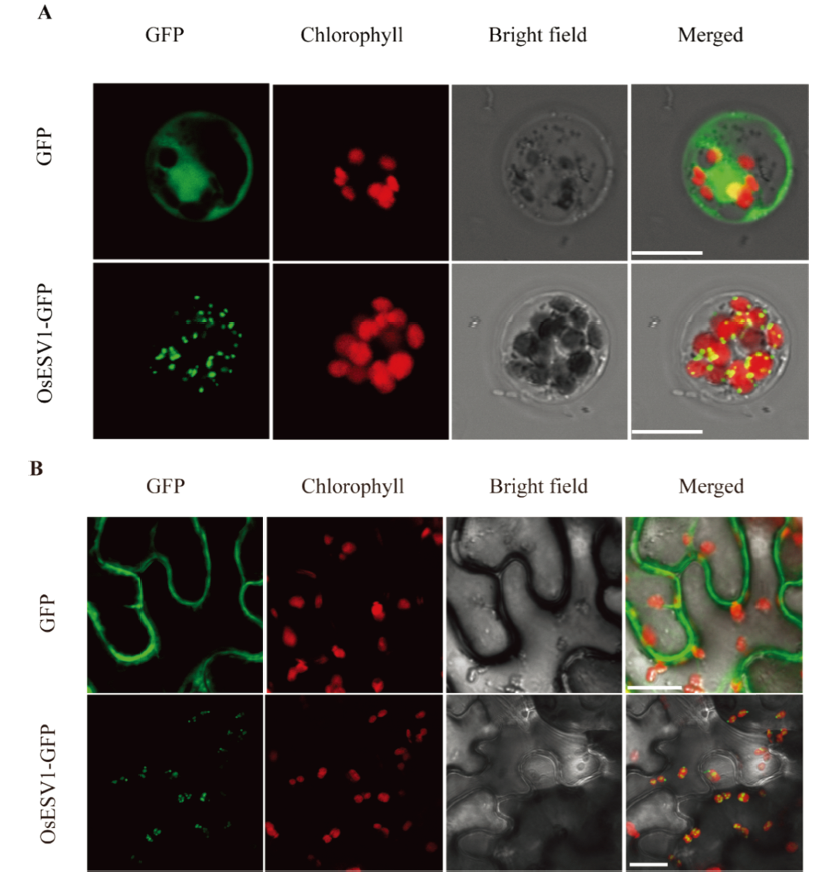
图6 OsESV1的亚细胞定位 A—OsESV1-GFP融合蛋白在水稻原生质体中的表达,pAN580-GFP空载体作为对照,标尺为20 μm;B—OsESV1-GFP融合蛋白在烟草表皮细胞中的表达,p1305-GFP空载体作为对照,标尺为20 μm。
Fig. 6. Subcellular localization of OsESV1. A, Transient expression of OsESV1-GFP fusion protein in rice protoplasts. The empty vector pAN580-GFP was used as control. Bars=20 μm; B, Transient expression of OsESV1-GFP fusion protein in tobacco epidermal cells. The empty vector p1305-GFP was used as control. Bars=20 μm.
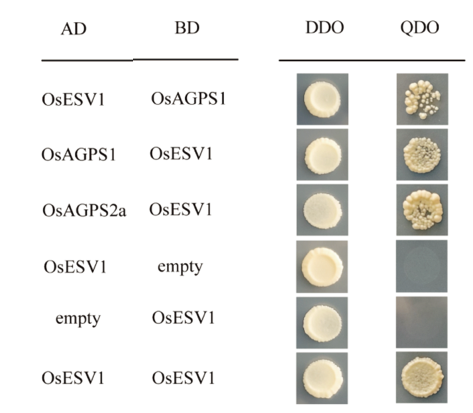
图7 酵母双杂实验表明OsESV1可以与OsAGPS2a和OsAGPS1互作 二缺:SD-Leu/-Trp;四缺:SD-Leu/-Trp/-His/-Ade。
Fig. 7. Yeast two-hybrid assays show that OsESV1 can interact with OsAGPS2a and OsAGPS1. DDO(double dropout supplements), SD-Leu/-Trp; QDO(quadruple dropout supplements), SD-Leu/-Trp/ -His/-Ade.
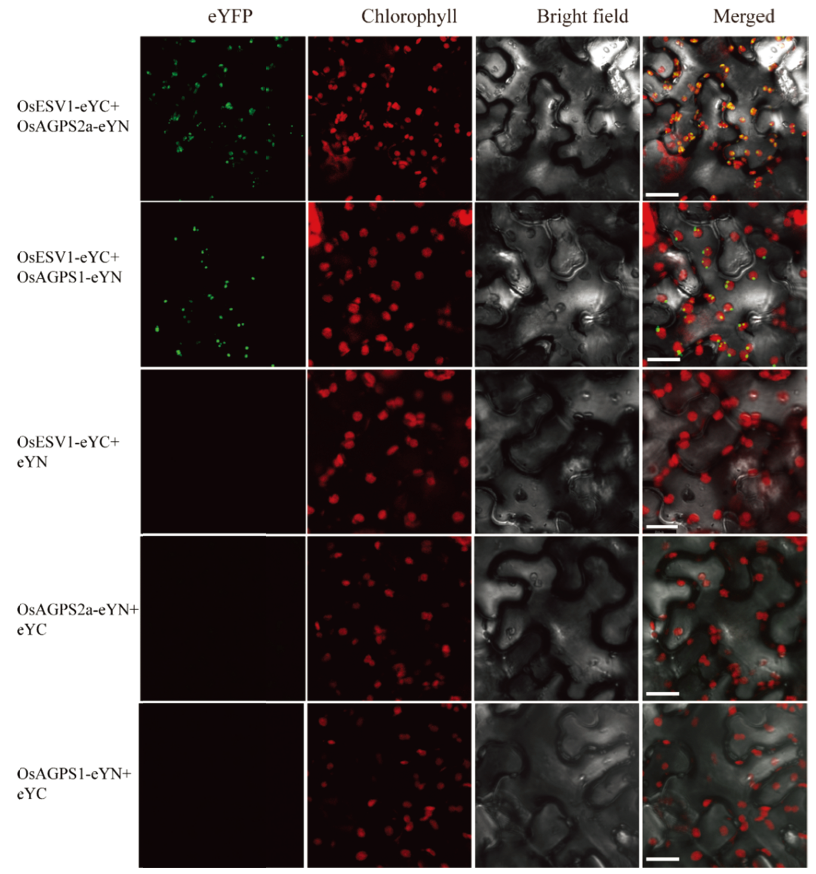
图8 双分子荧光互补实验证明OsESV1可以与OsAGPS2a和OsAGPS1在烟草表皮细胞中互作
Fig. 8. BiFC assays show that OsESV1 can interact with OsAGPS2a and OsAGPS1 in tobacco epidermal cells. Bars = 10 μm.
| [1] | 侯立刚, 周广春, 严永峰, 全成哲, 马巍. 吉林省水稻产业发展现状与未来发展对策[J]. 北方水稻, 2015,45(2):73-75. |
| Hou L G, Zhou G C, Yan Y F, Quan C Z, Ma W. Analysis on the development status and the counter- measures of rice industry in Jilin[J]. North Rice, 2015,45(2):73-75. (in Chinese with English abstract) | |
| [2] | 方鹏飞, 李三峰, 焦桂爱, 谢黎虹, 胡培松, 魏祥进, 唐绍清. 水稻粉质胚乳突变体flo7的理化性质及基因定位[J]. 中国水稻科学, 2014,28(5):447-457. |
| Fang P F, Li S F, Jiao G A, Xie L H, Hu P S, Wei X J, Tang S Q. Physicochemical property analysis and gene mapping of a floury endosperm mutant flo7 in rice[J]. Chinese Journal of Rice Science, 2014,28(5):447-457. (in Chinese with English abstract) | |
| [3] | 蔡跃. 水稻心白胚乳突变体w59的基因克隆及功能分析[D]. 南京: 南京农业大学, 2018. |
| Cai Y. Gene cloning and functional analysis of rice white-core endosperm mutant w59[D]. Nanjing: Nanjing Agricultural University, 2018. (in Chinese with English abstract) | |
| [4] | Tetlow I J, Emes M J. Starch biosynjournal in the developing endosperms of grasses and cereals[J]. Agronomy, 2017,7(4):81. |
| [5] | Okita T W, Nakata P A, Anderson J M, Sowokinos J, Morell M, Preiss J. The subunit structure of potato tuber ADPglucose pyrophosphorylase[J]. Plant Physiology, 1990,93(2):785-790. |
| [6] | Geigenberger P. Regulation of starch biosynjournal in response to a fluctuating environment[J]. Plant Physiology, 2011,155(4):1566-1577. |
| [7] | Kaushik R P, Khush G S. Genetic analysis of endosperm mutants in rice Oryza sativa L.[J] Theoretical and Applied Genetics, 1991,83(2):146-152. |
| [8] | Wu Y, Pu C, Lin H, Huang H, Huang Y, Hong C, Chang M, Lin Y. Three novel alleles of FLOURY ENDOSPERM2 (FLO2) confer dull grains with low amylose content in rice[J]. Plant Science, 2015,233:44-52. |
| [9] | Nishio T, Iida S. Mutants having a low content of 16-kDa allergenic protein in rice (Oryza sativa L)[J]. Theoretical & Applied Genetics, 1993,86(2-3):317-321. |
| [10] | Kang H G, Park S, Matsuoka M, An G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate ortho- phosphate dikinase gene (OsPPDKB)[J]. The Plant Journal, 2010,42(6):901-911. |
| [11] | Ryoo N, Yu C, Park C S, Baik M Y, Baik M Y, Park I M, Cho M H, Bhoo S H, An G, Hahn T R, Jeon J S. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.)[J]. Plant Cell Reports, 2007,26(7):1083-1095. |
| [12] | Peng C, Wang Y, Liu F, Ren Y, Zhou K, Lü J, Zheng M, Zhao S, Zhang L, Wang C, Jiang L, Zhang X, Guo X, Bao Y, Wan J. FLOURY ENDOSPERM 6 encodes a CBM 48 domain-containing protein involved in compound granule formation and starch synjournal in rice endosperm[J]. The Plant Journal, 2014,77(6):917-930. |
| [13] | Zhang L, Ren Y, Lu B, Yang C, Feng Z, Liu Z, Chen J, Ma W, Wang Y, Yu X, Wang Y, Zhang W, Wang Y, Liu S, Wu F, Zhang X, Guo X, Bao Y, Jiang L, Wan J. FLOURY ENDOSPERM7 encodes a regulator of starch synjournal and amyloplast development essential for peripheral endosperm development in rice[J]. Journal of Experimental Botany, 2016(3):633-647. |
| [14] | Long W, Dong B, Wang Y, Pan P, Wang Y, Liu L, Chen X, Liu X, Liu S, Tian Y, Chen L, Wan J. FLOURY ENDOSPERM8, encoding the UDP-glucose pyrophos- phorylase 1, affects the synjournal and structure of starch in rice endosperm[J]. Journal of Plant Biology, 2017,60(5):513-522. |
| [15] | 刘艺, 朱小品, 刘喜, 田云录, 刘世家, 王云龙, 张文伟, 江玲, 王益华, 万建民. 水稻胚乳粉质突变体 flo9 的表型分析和基因定位[J]. 南京农业大学学报, 2018,41(4):616-624. |
| Liu Y, Zhu X P, Liu X, Tian Y L, Liu S J, Wang Y L, Zhang W W, Jiang L, Wang Y H, Wan J M. Phenotyping and gene-mapping of a floury endosperm mutant flo9 in rice[J]. Journal of Nanjing Agricultural University, 2018,41(4):616-624. (in Chinese with English abstract) | |
| [16] | Wu M, Ren Y, Cai M, Wang Y, Zhu S, Zhu J, Hao Y, Teng X, Zhu X, Jing R, Zhang H, Zhong M, Wang Y, Lei C, Zhang X, Guo X, Cheng Z, Lin Q, Wang J, Jiang L, Bao Y, Wang Y, Wan J. Rice FLOURY ENDOSPERM 10 encodes a pentatricopeptide repeat protein that is essential for the trans-splicing of mitochondrial nad1 intron 1 and endosperm development[J]. New Phytologist, 2019,223(2):736-750. |
| [17] | Zhu X, Teng X, Wang Y, Hao Y, Jing R, Wang Y, Liu Y, Zhu J, Wu M, Zhong M, Chen X, Zhang Y, Zhang W, Wang C, Wang Y, Wan J. FLOURY ENDOSPERM11 encoding a plastid heat shock protein 70 is essential for amyloplast development in rice[J]. Plant Science, 2018,277:89-99. |
| [18] | Zhong M, Liu X, Liu F, Ren Y, Wang Y, Zhu J, Teng X, Duan E, Wang F, Zhang H, Wu M, Hao Y, Zhu X, Jing R, Guo X, Jiang L, Wang Y, Wan J. FLOURY ENDOSPERM12 encoding alanine aminotransferase 1 regulates carbon and nitrogen metabolism in rice[J]. Journal of Plant Biology, 2019,62(1):61-73. |
| [19] | Hu T, Tian Y, Zhu J, Wang Y, Jing R, Lei J, Sun Y, Yu Y, Li J, Chen X, Zhu X, Hao Y, Liu L, Wang Y, Wan J. OsNDUFA9 encoding a mitochondrial complex I subunit is essential for embryo development and starch synjournal in rice[J]. Plant Cell Reports, 2018,37(12):1667-1679. |
| [20] | Xue M, Liu L, Yu Y, Zhu J, Gao H, Wang Y, Wan J. Lose-of-function of a rice nucleolus-localized pentatrico- peptide repeat protein is responsible for the floury endosperm14 mutant phenotypes[J]. Rice, 2019,12(1):1-15. |
| [21] | You X, Zhang W, Hu J, Jing R, Cai Y, Feng Z, Kong F, Zhang J, Yan H, Chen W, Chen X, Ma J, Tang X, Wang P, Zhu S, Liu L, Jiang L, Wan J. FLOURY ENDOSPERM15 encodes a glyoxalase I involved in compound granule formation and starch synjournal in rice endosperm[J]. Plant Cell Reports, 2019,38(3):345-359. |
| [22] | Teng X, Zhong M, Zhu X, Wang C, Ren Y, Wang Y, Zhang H, Jiang L, Wang D, Hao Y, Wu M, Zhu J, Zhang X, Guo X, Wang Y, Wan J. FLOURY ENDOSPERM16 encoding a NAD-dependent cytosolic malate dehydrogenase plays an important role in starch synjournal and seed development in rice[J]. Plant Biotechnology Journal, 2019,17(10):1914-1927. |
| [23] | Feike D, Seung D, Graf A, Bischof S, Ellick T, Coiro M, Soyk S, Eicke S, Mettler-Altmann T, Lu K J, Trick M, Zeeman S C, Smith A M. The starch granule-associated protein EARLY STARVATION1 is required for the control of starch degradation in Arabidopsis thaliana leaves[J]. The Plant Cell, 2016,28(6):1472-1489. |
| [24] | Malinova I, Mahto H, Brandt F, AL-Rawi S, Qasim H, Brust H, Hejazi M, Fettk J. EARLY STARVATION 1 specifically affects the phosphorylation action of starch- related dikinases[J]. The Plant Journal, 2018,95(1):126-137. |
| [25] | 汪秉琨, 张慧, 洪汝科, 张锦, 杨睿, 罗琼, 曾千春. CRISPR/Cas9系统编辑水稻Wx基因[J]. 中国水稻科学, 2018,32(1):35-42. |
| Wang B K, Zhang H, Hong R K, Zhang J W, Yang R, Luo Q, Zeng Q C. Wx gene editing via CRISPR/Cas9 system in rice[J]. Chinese Journal of Rice Science, 2018,32(1):35-42. (in Chinese with English abstract) | |
| [26] | 曾千春, 李旭刚, 马炳田, 陈松彪, 徐鸿林, 孟昆, 魏晓丽, 朱祯. 有效去除农杆菌和籼稻转化系统优化[J]. 分子植物育种, 2003,1(5):783-790. |
| Zeng Q C, Li X G, Ma B T, Chen S B, Xu H L, Meng K, Wei X L, Zhu Z. Efficient elimination of A. tumefaciens and optimization of Agrobacterium-mediated transformation of indica rice[J]. Molecular Plant Breeding, 2003,1(5):783-790. (in Chinese with English abstract) | |
| [27] | 王慧娜, 初志战, 马兴亮, 李日清, 刘耀光. 高通量的PCR 模板植物基因组 DNA 制备方法[J]. 作物学报, 2013,39(7):1200-1205. |
| Wang H N, Chu Z Z, Ma X L, Li R Q, Liu Y G. A high through-put protocol of plant genomic DNA preparation for PCR[J]. Acta Agronomica Sinica, 2013,39(7):1200-1205. (in Chinese with English abstract) | |
| [28] | Wang W, Wei X, Jiao G, Chen W, Wu Y, Sheng Z, Hu S, Xie L, Wang J, Tang S, Hu P. GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield[J]. Journal of Integrative Plant Biology, 2020,62(7):948-966. |
| [29] | Tong C, Chen Y, Tang F, Xu F, Huang Y, Chen H, Bao J. Genetic diversity of amylose content and RVA pasting parameters in 20 rice accessions grown in Hainan, China[J]. Food Chemistry, 2014,161:239-245. |
| [30] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method[J]. Methods, 2001,25(4):402-408. |
| [31] | Chen S, Tao L, Zeng L, Vega-Sanchez M E, Umemura K, Wang G. A highly efficient transient protoplast system for analyzing defence gene expression and protein- protein interactions in rice[J]. Molecular Plant Pathology, 2010,7(5):417-427. |
| [32] | Waadt R, Kudla J. In planta visualization of protein interactions using bimolecular fluorescence comple- mentation (BiFC)[J]. Cold Spring Harbor Protocols, 2008(4): pdb, prot4995. |
| [33] | Ohdan T, Francisco P B, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. Expression profiling of genes involved in starch synjournal in sink and source organs of rice[J]. Journal of Experimental Botany, 2005,56(422):3229-3244. |
| [34] | Lee S K, Hwang S K, Han M, Eom J S, Kang H G, Han Y, Choi S B, Cho M H, Bhoo S H, An G, Hahn T R, Okita T W, Jeon J S. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synjournal in the leaf and seed endosperm of rice (Oryza sativa L.)[J]. Plant Molecular Biology, 2007,65(4):531-546. |
| [35] | Seung D, Soyk S, Coiro M, Maier B A, Eicke S, Zeeman S C. PROTEIN TARGETING TO STARCH is required for localizing GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synjournal in Arabidopsis[J]. PLOS Biology, 2015,13(2):e1002080. |
| [36] | Abt M R, Zeeman S C. Evolutionary innovations in starch metabolism[J]. Current Opinion in Plant Biology, 2020,55:109-117. |
| [37] | Sakulsingharoj C, Choi S B, Hwang S K, Edwards G E, Bork J, Meyer C R, Preiss J, Okita T W. Engineering starch biosynjournal for increasing rice seed weight: the role of the cytoplasmic ADP-glucose pyropho- sphorylase[J]. Plant Science, 2004,167(6):1323-1333. |
| [38] | Cook F R, Fahy B, Trafford K. A rice mutant lacking a large subunit of ADP-glucose pyrophosphorylase has drastically reduced starch content in the culm but normal plant morphology and yield[J]. Functional Plant Biology, 2012,39:1068-1078. |
| [39] | Comparot-Moss S, Kötting O, Stettler M, Edner C, Graf A, Weise S E, Streb S, Lue W L, MacLean D, Mahlow S, Ritte G, Steup M, Chen J, Zeeman S C, Smith A M. A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves[J]. Plant Physiology, 2010,152(2):685-697. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 吕宙, 易秉怀, 陈平平, 周文新, 唐文帮, 易镇邪. 施氮量与移栽密度对小粒型杂交水稻产量形成的影响[J]. 中国水稻科学, 2024, 38(4): 422-436. |
| [6] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [7] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [8] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [9] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [10] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [11] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [12] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [13] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [14] | 肖正午, 方升亮, 曹威, 胡丽琴, 黎星, 解嘉鑫, 廖成静, 康玉灵, 胡玉萍, 张珂骞, 曹放波, 陈佳娜, 黄敏. 米粉质构特性与稻米理化性状的关系[J]. 中国水稻科学, 2024, 38(3): 316-323. |
| [15] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||