
中国水稻科学 ›› 2017, Vol. 31 ›› Issue (4): 355-363.DOI: 10.16819/j.1001-7216.2017.6165 355
孙立亭1,3, 林添资1,3, 王云龙1, 牛梅1, 胡婷婷1, 刘世家1, 王益华1, 万建民1,2,*( )
)
收稿日期:2016-12-20
修回日期:2017-02-07
出版日期:2017-07-25
发布日期:2017-07-10
通讯作者:
万建民
基金资助:
Liting SUN1,3, Tianzi LIN1,3, Yunlong WANG1, Mei NIU1, Tingting HU1, Shijia LIU1, Yihua WANG1, Jianmin WAN1,2,*( )
)
Received:2016-12-20
Revised:2017-02-07
Online:2017-07-25
Published:2017-07-10
Contact:
Jianmin WAN
摘要:
目的叶色突变相关基因鉴定和克隆有助于研究光合作用,补充并完善叶绿体发育机理和色素合成代谢途径,为开展水稻的高光效育种提供理论依据。方法从粳稻品种Dongjin的组培后代中分离出一个白条纹突变体st13,成熟期测定野生型和st13的主要农艺性状,苗期测定色素含量并观察叶绿体的超微结构;将st13和Dongjin进行正反交,观察F1植株表型,并对F2表型分离进行卡方检验,对st13进行遗传分析;利用st13×南京11(籼稻品种)的F2和F2:3群体,对st13突变基因定位;采用qPCR分析叶绿体发育和叶绿素合成相关基因在st13与野生型相对表达量。结果与野生型Dongjin相比,该突变体的株高、单株有效穗数、穗长、结实率和千粒重等主要农艺性状显著下降。苗期的色素含量降低,分蘖期无差异。突变体的叶绿体中既有含丰富的类囊体膜结构的正常叶绿体,也存在无类囊体结构的叶绿体。遗传分析和基因定位结果表明,st13的突变表型受1对隐性核基因控制,突变基因位于第3条染色体长臂InDel(Insertion-Deletion)标记I3-21和I3-22之间。进一步在这两个标记之间设计了6对InDel标记,最终将基因定位在94 kb区间内,此区间共有8个候选基因。结论这8个候选基因中,有5个假定的蛋白,其他三个都是有功能注释的蛋白,而这三个蛋白在水稻中均未见报道,因此,st13突变是由一个新的叶色基因突变引起的;同时st13中叶绿体发育、叶绿素合成和光合系统相关基因的表达也发生了显著改变,推测ST13可能是调控叶绿体发育的关键基因。
孙立亭, 林添资, 王云龙, 牛梅, 胡婷婷, 刘世家, 王益华, 万建民. 水稻白条纹突变体st13的表型分析及基因定位[J]. 中国水稻科学, 2017, 31(4): 355-363.
Liting SUN, Tianzi LIN, Yunlong WANG, Mei NIU, Tingting HU, Shijia LIU, Yihua WANG, Jianmin WAN. Phenotypic Analysis and Gene Mapping of a White Stripe Mutant st13 in Rice[J]. Chinese Journal OF Rice Science, 2017, 31(4): 355-363.
| 材料 Material | 株高 Plant height/cm | 有效穗数 No. of effective panicles | 剑叶长 Flag leaf length /cm | 穗长 Panicle length /cm | 结实率 Seed setting rate/% | 千粒重 1000-grain weight /g | |
|---|---|---|---|---|---|---|---|
| WT | 112.2±2.9 | 9.6±0.6 | 31.5±2.4 | 23.8±0.2 | 95.5±1.5 | 27.9±1.6 | |
| st13 | 70.8±4.7** | 5.8±0.5** | 31.9±1.5 | 17.9±0.5** | 74.0±1.8** | 20.3±0.9** | |
表1 野生型Dongjin与突变体st13的主要农艺性状比较
Table 1 Comparison of major agronomic traits between wild-type Dongjin and the st13 mutant.
| 材料 Material | 株高 Plant height/cm | 有效穗数 No. of effective panicles | 剑叶长 Flag leaf length /cm | 穗长 Panicle length /cm | 结实率 Seed setting rate/% | 千粒重 1000-grain weight /g | |
|---|---|---|---|---|---|---|---|
| WT | 112.2±2.9 | 9.6±0.6 | 31.5±2.4 | 23.8±0.2 | 95.5±1.5 | 27.9±1.6 | |
| st13 | 70.8±4.7** | 5.8±0.5** | 31.9±1.5 | 17.9±0.5** | 74.0±1.8** | 20.3±0.9** | |
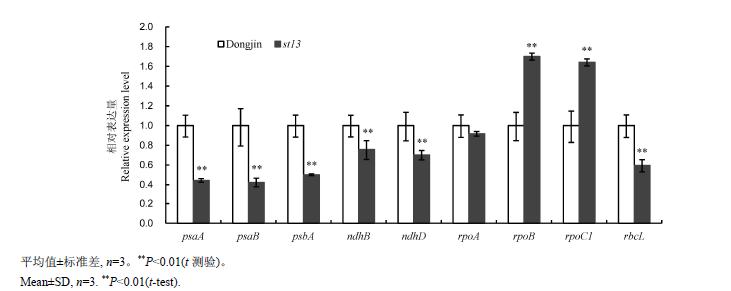
图4 苗期野生型Dongjin和st13突变体中叶绿体发育及光合系统相关基因的荧光定量PCR分析
Fig. 4. Quantitative RT-PCR analyses of genes associated with chloroplast development and photosynthetic system in wild-type Dongjin and st13 mutant at the seedling stage.
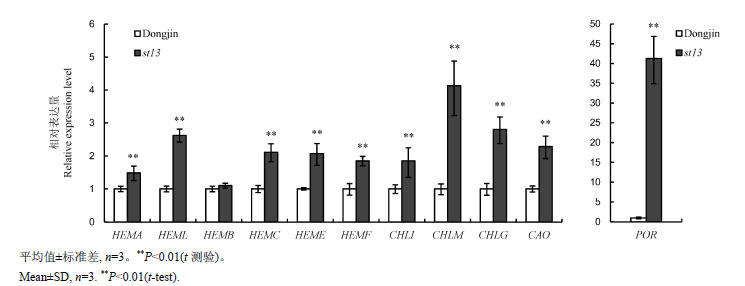
图5 苗期野生型Dongjin和st13突变体中叶绿体生物合成相关基因的荧光定量PCR分析
Fig. 5. Quantitative RT-PCR analyses of genes associated with chlorophyll biogenesis in wild-type Dongjin and st13 mutant at the seedling stage.
| 组合 Combination | 正常株数 No. of normal plants | 白条纹株数 No. of plants with white stripes | 实际分离比 Segration ratio | χ2(3:1) | χ20.05 |
|---|---|---|---|---|---|
| Dongjin×st13 | 286 | 98 | 2.92:1 | 0.03 | 3.84 |
| st13×Dongjin | 281 | 89 | 3.16:1 | 0.13 |
表2 野生型Dongjin与st13突变体正反交组合的F2分离比
Table 2 Segregation of F2 populations from wild-type Dongjin and st13 mutant.
| 组合 Combination | 正常株数 No. of normal plants | 白条纹株数 No. of plants with white stripes | 实际分离比 Segration ratio | χ2(3:1) | χ20.05 |
|---|---|---|---|---|---|
| Dongjin×st13 | 286 | 98 | 2.92:1 | 0.03 | 3.84 |
| st13×Dongjin | 281 | 89 | 3.16:1 | 0.13 |
| 引物名称 Primer name | 前引物 Forward primer | 后引物 Reverse primer | BAC克隆 BAC clone | |
|---|---|---|---|---|
| I3-21 | GCGAGATGGGCAGCTACTAC | ACACAATGTCCAGCTTGCAG | OSJNBa0010D22 | |
| P5 | CCTCCTCATAGCCCCAATCC | TCACTCTCCCATAGGTGGTC | OSJNBb0042N11 | |
| P3-13 | CCTCCGCACGAACCCT | GTGGGAAACTGGAGACGAG | OSJNBa0087M10 | |
| P3-14 | GGTTACATCTCCTTTTCGTTT | TCTTTGTTTGCTGCCATCT | OJ1519_A12 | |
| S-57 | AAAACCAAAACTAGAAGC | ATGTCCCTGGAAATGTA | OSJNBa0004G03 | |
| S-24 | CTCCGAGCGGCTGATTA | GGATAGTACTTCGTTTCGTA | OSJNBa0004G03 | |
| P3-29 | CAAACGCAATCTCACATCACA | AAGCGAGTGGGAGGGTG | OSJNBa0091E13 | |
| I3-22 | AGGTCTCGTGTCGTTCATCC | TGGAGGGAGCATGTCTATCA | OSJNBa0063J18 | |
表3 目标区域内有多态的InDel标记
Table 3 Polymorphic InDel markers used in this study.
| 引物名称 Primer name | 前引物 Forward primer | 后引物 Reverse primer | BAC克隆 BAC clone | |
|---|---|---|---|---|
| I3-21 | GCGAGATGGGCAGCTACTAC | ACACAATGTCCAGCTTGCAG | OSJNBa0010D22 | |
| P5 | CCTCCTCATAGCCCCAATCC | TCACTCTCCCATAGGTGGTC | OSJNBb0042N11 | |
| P3-13 | CCTCCGCACGAACCCT | GTGGGAAACTGGAGACGAG | OSJNBa0087M10 | |
| P3-14 | GGTTACATCTCCTTTTCGTTT | TCTTTGTTTGCTGCCATCT | OJ1519_A12 | |
| S-57 | AAAACCAAAACTAGAAGC | ATGTCCCTGGAAATGTA | OSJNBa0004G03 | |
| S-24 | CTCCGAGCGGCTGATTA | GGATAGTACTTCGTTTCGTA | OSJNBa0004G03 | |
| P3-29 | CAAACGCAATCTCACATCACA | AAGCGAGTGGGAGGGTG | OSJNBa0091E13 | |
| I3-22 | AGGTCTCGTGTCGTTCATCC | TGGAGGGAGCATGTCTATCA | OSJNBa0063J18 | |
| 基因 | 功能注释 |
|---|---|
| Gene | Annotation |
| Os03g0603300 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0603500 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0603600 | 甘油磷酸二酯酶 Glycerophosphoryl diester phosphodiesterase family protein |
| Os03g0603700 | A22A肽酶 Peptidase A22A, presenilin family protein |
| Os03g0604200 | 尿苷二磷酸葡萄糖脱氢酶 Similar to UDP-glucose dehydrogenase |
| Os03g0604500 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0604600 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0604700 | 假定蛋白 Hypothetical protein |
表4 94 kb区间内的预测基因及其可能的功能
Table 4 Predicted genes and their putative functions in the 94 kb region.
| 基因 | 功能注释 |
|---|---|
| Gene | Annotation |
| Os03g0603300 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0603500 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0603600 | 甘油磷酸二酯酶 Glycerophosphoryl diester phosphodiesterase family protein |
| Os03g0603700 | A22A肽酶 Peptidase A22A, presenilin family protein |
| Os03g0604200 | 尿苷二磷酸葡萄糖脱氢酶 Similar to UDP-glucose dehydrogenase |
| Os03g0604500 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0604600 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0604700 | 假定蛋白 Hypothetical protein |
| [1] | Fambrini M, Castagna A, Dalla V F, Degl’Innocenti E, Ranieri A, Vernieri P, Pardossi A, Guidi L, Rascio N, Pugliesi C. Characterization of a pigment-deficient mutant of sunflower (Helianthus annuus L.) with abnormal chloroplast biogenesis, reduced PSII activity and low endogenous level of abscisic acid. Plant Breeding, 2004, 6: 645-650. |
| [2] | Agrawal G K, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H.Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion: Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol, 2001, 125(3): 1248-1257. |
| [3] | Parks B M, Quail P H.Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell, 1991, 3(11): 1177-1186. |
| [4] | Kusumi K, Sakata C, Nakamura T, Kawasaki S, Yoshimura A, Iba K.A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions.Plant J, 2011, 68(6): 1039-1050. |
| [5] | Lee S, Kim J H, Yoo E S, Lee C H, Hirochika H, An G.Differential regulation of chlorophyll a oxygenase genes in rice.Plant Mol Biol, 2005 57: 805-818. |
| [6] | Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K.The rice nuclear gene,VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J, 2007, 52: 512-527. |
| [7] | Zhang H, Li J, Yoo J, Yoo S, Cho S, Koh H, Seo H S, Paek N.Rice chlorina-1 and chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol, 2006, 62: 325-337. |
| [8] | Jung K H, Hur J, Ryu C H, Choi Y, Chung Y Y, Miyao A, Hirochika H, An G.Characterization of a rice chlorophyll- deficient mutant using the T-DNA gene-trap system.Plant Cell Physiol, 2003, 44: 463-472. |
| [9] | Kong W, Yu X, Chen H, Liu L, Xiao Y, Wang Y, Wang C, Lin Y, Yu Y, Wang C, Jiang L, Zhai H, Zhao Z, Wan J.The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice.Plant Mol Biol, 2016, 92(1-2): 177-191. |
| [10] | Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Sun X, Deng X.Divinyl chlorophyll (ide) a can be converted to monovinyl chlorophyll (ide) a by a divinyl reductase in rice.Plant Physiol, 2010, 153: 994-1003. |
| [11] | Sakuraba Y, Rahman M L, Cho S H, Kim Y S, Koh H J, Yoo S C, Paek N C.The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J, 2013, 74: 122-133. |
| [12] | Wu Z M, Zhang X, He B, Diao L, Sheng S, Wang J, Guo X, Su N, Wang L, Jiang L, Wang C, Zhai H, Wan J.A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis.Plant Physiol, 2007, 145: 29-40. |
| [13] | Yang Y, Xu J, Huang L, Leng Y, Dai L, Rao Y, Chen L, Wang Y, Tu Z, Hu J, Ren D, Zhang G, Zhu L, Guo L, Qian Q, Zeng D.PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J Exp Bot, 2016, 67(5): 1297-1310. |
| [14] | Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M.Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice.Plant J, 2009, 57: 120-131. |
| [15] | Wang L, Wang C, Wang Y, Niu M, Ren Y, Zhou K, Zhang H, Lin Q, Wu F, Cheng Z, Wang J, Zhang X, Guo X, Jiang L, Lei C, Wang J, Zhu S, Zhao Z, Wan J.WSL3, a component of the plastid-encoded plastid RNA polymerase, is essential for early chloroplast development in rice.Plant Mol Biol, 2016, 92(4-5): 581-595. |
| [16] | Wang Y, Wang C, Zheng M, Lyu J, Xu Y, Li X, Niu M, Long W, Wang D, Wang H Y, William T, Wang Y, Wan J.WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development.Plant Physiol, 2016, 170(4): 2110-2123. |
| [17] | Lopez-Juez E.Plastid biogenesis between light and shadows.J Exp Bot, 2007, 58: 11-26. |
| [18] | Sakamoto W, Miyagishima S Y, Jarvis P.Chloroplast biogenesis: Control of plastid development, protein import, division and inheritance.Arabidopsis Book, 2008, 6: e110. |
| [19] | Webber A N, Malkin R.Photosystem I reaction-centre proteins contain leucine zipper motifs. A proposed role in dimer formation.FEBS Lett, 1990, 264: 1-4. |
| [20] | Rutherford A W, Faller P.Photosystem II: evolutionary perspectives.Philos Trans R SocLond B Biol Sci. 2003, 358: 245-253. |
| [21] | Santis-Maciossek G D, Kofer W, Bock A, Schoch S, Maier R M, Wanner G, Rüdiger W, Koop Hans-Ulrich, Herrmann R G. Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology biochemistry and ultrastructure. Plant J, 1999, 18: 477-489. |
| [22] | Peng L W, Yamamoto H, Shikanai T.Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex.Biochim Biophys Acta, 2011, 1807: 945-953. |
| [23] | Andersson I, Backlund A.Structure and function of Rubisco.Plant Physiol Biochem, 2008, 46: 275-291. |
| [24] | Livak K J, Schmittgen T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2 Delta Delta C (T)] method.Methods, 2001, 25: 402-408. |
| [25] | McCouch S R, Kochert G, Yu Z H, Wang Z Y, Khush G S, Coffman W R, Tanksley S D. Molecular mapping of rice chromosome.Theor Appl Genet, 1998, 76: 815-829. |
| [26] | Shi Y F, Chen J, Liu W Q, Huang Q N, Shen B, Leung H, Wu J L.Genetic analysis and gene mapping of a new rolled-leaf mutant in rice (Oryza sativa L.). Sci China C: Life Sci, 2009, 52: 885-890. |
| [27] | Kusumi K, Chono Y, Shimada H, Shimada H, Gotoh E, Tsuyama M, Iba K.Chloroplast biogenesis during the early stage of leaf development in rice.Plant Biotechnol, 2010, 27: 85-90. |
| [28] | Allen J F, Wilson B M, Puthiyaveetil S, Nield J.A structural phylogenetic map for chloroplast photosynthesis.Trends Plant Sci, 2011, 16: 645-655. |
| [29] | Pakrasi H B.Genetic analysis of the form, function of photosystem I and photosystem II.Annu Rev Genet, 1995, 29: 755-776. |
| [30] | 王兴春, 王敏, 季芝娟, 陈钊, 刘文真, 韩渊怀, 杨长登. 水稻糖苷水解酶基因OsBE1在叶绿体发育中的功能. 作物学报, 2014, 40(12): 2090-2097. |
| Wang X C, Wang M, Ji Z J, Chen Z, Liu W Z, Han Y H, Yang C D.Functional characterization of the glycoside hydrolase encoding gene OsBE1 during chloroplast development in Oryza sativa. Acta Agron Sin, 2014, 40(12): 2090-2097. (in Chinese with English abstract) | |
| [31] | Gothandam K M, Kim E S, Cho H, Chung Y Y.OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis.Plant Mol Biol, 2005, 58(3): 421-433. |
| [32] | Lin D, Jiang Q, Zheng K, Chen S, Zhou H, Gong X, Xu J, Teng S, Dong Y.Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol (Stuttg), 2015, 17(3): 599-607. |
| [33] | Zhang Z, Tan J, Shi Z, Xie Q, Xing Y, Liu C, Chen Q, Zhu H, Wang J, Zhang J, Zhang G.Albino Leaf1 that encodes the sole octotricopeptide repeat protein is responsible for chloroplast development. Plant Physiol, 2016, 171(2): 1182-1191. |
| [34] | Yoo S C, Cho S H, Sugimoto H, Li J, Kusumi K, Koh H J, Iba K, Paek N C.Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development.Plant Physiol, 2009, 150(1): 388-401. |
| [35] | Wang Y, Zhang J, Shi X, Peng Y, Li P, Lin D, Dong Y, Teng S.Temperature-sensitive albino geneTCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. J Exp Bot, 2016, 67(17): 5187-5202. |
| [36] | Su N, Hu M L, Wu D X, Wu F Q, Fei G L, Lan Y, Chen X L, Shu X L, Zhang X, Guo X P, Cheng Z J, Lei C L, Qi C K, Jiang L, Wang H, Wan J M.Disruption of a rice pentatricopeptide repeat protein causes a seedling- specific albino phenotype and its utilization to enhance seed purity in hybrid rice production.Plant Physiol, 2012, 159(1): 227-238. |
| [37] | Klinghammer M, Tenhaken R.Genome-wide analysis of the UDP-glucose dehydrogenase gene family in Arabidopsis, a key enzyme for matrix polysaccharides in cell walls. J Exp Bot, 2007, 58(13): 3609-3621. |
| [38] | 孙兰茜, 常平安, 曾鑫, 韩丽萍, 冉凤, 王超颖, 王玲, 黄飞飞. 甘油磷酸二酯酶家族蛋白的分子进化. 基因组学与应用生物学, 2015, 34(27): 172-178. |
| Sun L X, Chang P A, Zeng X, Han L P, Ran F, Wang C Y, Wang L, Huang F F.Molecular evolution of the glycerophospho- diesterase family proteins.Genomics Appl Boil, 2015, 34(27): 172-178. (in Chinese with English abstract) |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||